Abstract
In mammalian and budding yeast cells treated with genotoxic agents, different proteins implicated in detecting, signalling or repairing DNA lesions form nuclear foci. We studied foci formed by proteins involved in these processes in living fission yeast cells, which is amenable to genetic and molecular analysis. Using fluorescent tags, we analysed subnuclear localisations of the DNA damage checkpoint protein Rad9, of the homologous recombination protein Rad22 and of PCNA, which are implicated in many aspects of DNA metabolism. After inducing double strand breaks (DSBs) with ionising radiations, Rad22, Rad9 and PCNA form a low number of nuclear foci. Rad9 recruitment to foci depends on the presence of Rad1, Hus1 and Rad17, but is independent of downstream checkpoint effectors and of homologous recombination proteins. Likewise, Rad22 and PCNA form foci despite inactive homologous recombination repair and impaired DNA damage checkpoint. Rad22 and Rad9 foci co-localise completely, whereas PCNA co-localises with Rad22 and Rad9 only partially. Foci do not disassemble in cells unable to repair DNA by homologous recombination. Thus, in fission yeast, DSBs are detected by the DNA damage checkpoint and are repaired by homologous recombination at a few spatially confined subnuclear compartments where Rad22, Rad9 and PCNA concentrate independently.
INTRODUCTION
Genome stability is threatened by both exogenous and endogenous attacks. Cells have evolved mechanisms to repair DNA lesions and to coordinate DNA repair processes with cell cycle progression. Among the possible DNA lesions, double strand breaks (DSBs) are likely to be the most dangerous for genome stability. DSBs can arise after treatment with ionising radiations (IR), but also during normal cellular processes such as DNA replication. Two major mechanisms are involved in DSBs repair: homologous recombination repair (HRR) and non-homologous end joining (NHEJ), with HRR being the most accurate (reviewed in 1). Furthermore, DNA repair is regulated by DNA structure checkpoint pathways (2).
Repair of DSBs and checkpoint pathways have been extensively studied in both budding and fission yeast. In Saccharomyces cerevisiae, the RAD52 epistasis group of genes is required for HRR. This group includes RAD50, RAD51, RAD52, RAD54, RAD55, RAD57, RFA1, MRE11 and XRS2 (reviewed in 3). The analysis of double mutants has allowed the division of this group into two subgroups of ‘early’ and ‘late’ genes, according to the timing of respective involvement in the recombination process. Early genes include RAD50, MRE11 and XRS2 whose products form a complex functionally homologous to the Escherichia coli RecBCD complex (4). Processing of DNA ends at DSBs by the Rad50/Mre11/Xrs2 complex is required for later steps in the recombination process, which involve Rad51 and Rad52. In S.cerevisiae this complex is also required for NHEJ. Furthermore, integrity of the Rad50/Mre11/Xrs2 complex is essential for checkpoint activation (5).
Homologues of these genes have been found in different eukaryotes including Schizosaccharomyces pombe, suggesting conserved functions of the corresponding proteins. Fission yeast rad50+ and rad32+ genes are the counterparts of budding yeast ‘early genes’ RAD50 and MRE11, respectively. However, Rad50 and Rad32 are not required for NHEJ in fission yeast (6). Homologues of budding yeast RAD51, RAD52, RAD54 and RAD57 genes have also been identified in S.pombe and named rhp51+, rad22+, rhp54+ and rhp57+, respectively. Mutations of these fission yeast genes confer sensitivity to IR and to MMS, as is also the case for budding yeast mutants of the RAD52 gene group. However, fission yeast mutants, but not the corresponding budding yeast mutants, are also highly sensitive to UV irradiation and show clear morphological abnormalities suggesting a high degree of genomic instability even in the absence of genotoxic treatments (7).
Later steps in homologous recombination require proteins that are normally implicated in DNA synthesis. Among these proteins, PCNA, encoded by the fission yeast pcn1+ gene, appears to be the most versatile since it is involved in many aspects of DNA and chromatin metabolism (8).
Increasing evidence has pointed out a role for the DNA damage checkpoint pathway in promoting HRR in fission yeast (9,10). This pathway includes genes encoding the three subunits of the checkpoint sliding clamp (Rad9, Rad1 and Hus1), the sliding clamp loader (Rad17), the PI3-related kinase (Rad3) and its regulatory subunit (Rad26), and the downstream effectors (Mrc1 and Cds1 for the intra-S damage checkpoint; Crb2 and Chk1 for the late S and G2 damage checkpoint, reviewed in 2). However, how the different proteins involved in HRR are brought together and how they relate to the checkpoint pathway is not yet clear.
In eukaryotic cells, many proteins implicated in DNA repair cluster to form nuclear structures referred to as DNA repair factories or foci. The composition of these foci following different types of DNA lesions, the regulatory hierarchy of their assembly, and the molecular details of events occurring inside these structures remain poorly understood. Fission yeast Rhp51 is recruited in nuclear foci following IR, like its mammalian homologue Rad51 (9). In DNA repair foci, Rad51 co-localises with single-strand DNA binding protein, PCNA, Rad52, Rad54 and Rad55 (11,12). However, mutations in mammalian Rad52 do not block Rad51 foci formation (13). A critical role for recruiting proteins involved in homologous recombination at DNA repair foci is played by the phosphorylated form of histone H2AX. Phosphorylated H2AX also regulates the ability of 53BP1 to concentrate at IR-induced foci, which in turn facilitates phosphorylation of specific substrates by the checkpoint ATM kinase, including BRCA1 and p53 (14–16). In addition, it has been shown that Rad51, Rad52 and Rad54 foci are dynamic structures, of which Rad51 is a stably associated component (17). In vertebrates the study of the hierarchy of the assembly and disassembly of foci is complicated by the difficulty in obtaining full knock out of certain genes and difficulty in interpreting conflicting data obtained through the use of various cell lines, cancer cells and ES cells. In budding yeast, it has been shown that the protein Ddc1, which is homologous to fission yeast checkpoint protein Rad9, is recruited to DNA damage sites both in vitro and in vivo (18–20).
We decided to study DNA repair foci induced by IR in a simple model organism, the fission yeast S.pombe, whose cell cycle regulation has been extensively studied and whose complete genomic sequence was recently established (21). We studied foci in living cells formed by three different fluorescent fusion proteins: (i) Rad22, required for HRR; (ii) Rad9, required for the DNA damage checkpoint activation; (iii) PCNA, conserved in all eukaryotes and implicated in DNA replication and repair.
MATERIALS AND METHODS
Strains, plasmids and growth conditions
The strains used in this study are described in Table 1.
Table 1. Strains used in this study.
| Sp 78 | h– (972) [NYCY 1354] | a |
| Sp 1 | h+ leu1-32 [NYCY 1914] | a |
| Sp 25 | h+ leu1-32 ura4-D18 ade6-M216 | b |
| Sp 150 | h+ leu1-32 ura4::EGFP-pcn1+ | This study |
| Sp 208 | h+ leu1-32 ura4-D18 ade6-M216 rad9-YFP:ura4+ | This study |
| Sp 220 | h– rad22-YFP:KanR | This study |
| Sp 222 | h+ leu1-32 ura4::ECFP-pcn1+ rad22-YFP:KanR | This study |
| Sp 241 | h+ leu 1–32 rad9-YFP:ura4+ rad22-CFP:KanR | This study |
| Sp 254 | h? leu1-32 rad9-YFP:ura4+ ura4::ECFP-pcn1+ | This study |
| Sp 275 | h? leu1-32 ura4-D18 rad22-YFP:KanR Δcds1::ura4+ | This study |
| Sp 386 | h? leu1-32 ura4-D18 ade6-M210 rad9-YFP:ura4+ Δcds1::ura4+ | This study |
| Sp 338 | h? leu1-32 ura4-D18 rad22::YFP KanR Δchk1::ura4+ | This study |
| Sp 389 | h? leu1-32 ura4-D18 ade6-M210 rad9-YFP:ura4+ Δchk1::ura4+ | This study |
| Sp 296 | h? leu1-32 ura4-D18 rad22-YFP:KanR Δcrb2::ura4+ | This study |
| Sp 405 | h? leu1-32 ura4-D18 ade6 rad9-YFP:ura4+ Δcrb2::ura4+ | This study |
| Sp 280 | h? leu1-32 ura4-D18 rad22-YFP:KanR Δhus1::LEU2 | This study |
| Sp 397 | h? leu1-32 ura4-D18 ade6 rad9-YFP:ura4+ Δhus1::LEU2 | This study |
| Sp 516 | h? leu1-32 ura4-D18 ade6 rad22-YFP:KanR Δrad1::LEU2 | This study |
| Sp 537 | h? leu1-32 ura4-D18 ade6-M216 rad9-YFP:ura4+ Δrad1::LEU2 | This study |
| Sp 306 | h? leu1-32 ura4-D18 rad22-YFP:KanR Δrad17::ura4+ | This study |
| Sp 401 | h? leu1-32 ura4-D18 ade6 rad9-YFP:ura4+ Δrad17::ura4 | This study |
| Sp 535 | h? ura4-D18 rad9-YFP:ura4+ Δrad22::ura4+ | This study |
| Sp 413 | h? leu1-32 ura4-D18 ade6 rad9-YFP:ura4+Δrad26::ura4+ | This study |
| Sp 313 | h? leu1-32 ura4-D18 rad22-YFP:KanRΔrad26::ura4+ | This study |
| Sp 267 | h? leu1-32 ura4-D18 rad22-YFP:KanRΔrad3::ura4+ | This study |
| Sp 407 | h? leu1-32 ura4-D18 ade6 rad9-YFP:ura4+Δrad3::ura4+ | This study |
| Sp 326 | h? ura4-D18 rad22-YFP:KanR Δrad32::ura4+ | This study |
| Sp 411 | h? leu1-32 ura4-D18 rad9-YFP:ura4+Δrad32::ura4+ | This study |
| Sp 321 | h? leu1-32 ura4-D18 rad22-YFP:KanR Δrad50::KanR | This study |
| Sp 416 | h? leu1-32 ura4-D18 ade6-M210 rad9-YFP:ura4+Δrad50::KanR | This study |
| Sp 287 | h? leu1-32 ura4-D18 rad22-YFP:KanRΔrad9::ura4+ | This study |
| Sp 529 | h? ura4-D18 leu1-32 rad9-YFP:ura4+ Δrhp51::ura4+ | This study |
| Sp 622 | h? leu1-32 ura4-D18 ade6 rad22-YFP:KanR Δrhp51::KanR | This study |
aFrom National Collection of Yeast Cultures, Colney, Norwich, UK.
bFrom D. Beach, Cold Spring Harbour Laboratory, NY.
For Rad22-Yellow Fluorescent Protein (YFP) tagging, the YFP gene was cloned at the position of Cyan Fluorescent Protein (CFP) in plasmid pDH3 obtained from the Yeast Resource Center (YRC, University of Washington, USA) using the YFP ORF present in plasmid pDH5 (also from the YRC), excised with AscI and SmaI and cloned at the same sites in pDH3. Using these two plasmids, ∼700 bp from the 3′ end of the rad22+ gene was PCR-cloned in-frame with the genes encoding the fluorescent tags. PCR primers were 5′GCGCgtcgacAGCTCCTCGTAGAGCTGCAG3′ and 5′GCTcccgggCACGGGCACCACTTTTTTGGCTTTCTTATCC3′. The PCR product was cloned in-frame using AvaI and SalI cloning sites. The wild type S.pombe 972h– strain was transformed with these plasmids after linearisation by AflII. Transformants were selected for G418 resistance after 24 h of growth on complete medium.
For Rad9 YFP tagging, ∼800 bp from the 3′ end of the genomic rad9+ gene was PCR-cloned into the plasmid pSMUG (22). PCR primers were 5′GGgggcccTGGTCAGAGAACGGAAGAGC3′ and 5′GGctcgagCACGGGCACCATCTTCCTGAGAGAAAATGCC3′. The PCR product was cloned into pSMUG using ApaI and AvaI cloning sites. The GFP gene was then replaced with YFP present in plasmid pDH5 (YRC). The wild type SP808h+ strain was transformed with this MluI-linearised plasmid and recombinants were selected on minimal medium without uracil.
EGFP C1 (Clontech, Palo Alto, USA) was used to construct N-terminal EGFP-tagged PCNA, using BglII sites of PCNA and EGFP genes. A long and mobile linker composed of a stretch of alternating glycine and aspartic acid residues was inserted between EGFP and PCNA by cloning complementary oligos 5′GATCGGGTGAGGGTCAAGGTCAAGGTCAAGGTCCTGGTCGTGGTTACGCTTACCGTAGTA3′ and 5′GATCTACTACGGTAAGCGTAACCACGACCAGGACCTTGACCTTGACCTTGACCCTCACCC3′ at the BglII site located between EGFP and PCNA. For the pcn1+ gene promoter insertion, a PCR fragment of 2228 bp obtained with oligos 5′-GCGCATGCAGAACTGGATTATTTTTCGTTCTTC-3′ and 5′-CGCCATGGTCGTAATTATATTTTATACAAGCTA-3′ was inserted upstream of the EGFP–PCNA construct. Promoter and fusion protein sequences were cloned into the pSMUG ura4+ gene, which disrupted the ura4 gene. Transformation of the h+ leu1-32 S.pombe strain was carried out with the CelII–AvrII fragment of this plasmid. Transformants were selected for 5-FOA resistance after 24 h of growth at 30°C on rich medium followed by replica plating onto rich medium plus 0.1% 5-FOA.
For each construction, the PCR-obtained fragments were sequenced and correct insertion at the target genomic locus was determined by colony PCR.
Strains were grown at 30°C in YE-rich medium supplemented with uracil, leucine and adenine (23).
Elutriation and irradiation of S.pombe cells
About 2 × 1010 cells grown overnight in YE medium were used for elutriation in a Beckman J6-MC centrifuge using a single 45 ml elutriation chamber. After elutriation, small G2 cells were concentrated by filtration. For gamma irradiation, 108 cells were resuspended in 1 ml YE medium and irradiated with a 137Cs source, at a dose of 60 Gy/min. Cells were then transferred to 30 ml rich medium and incubated at 30°C. All time-lapse experiments were carried out at 30°C. At the indicated time points, aliquots were subjected to microscopic observations. For irradiation of asynchronous cultures, 108 exponentially growing cells were irradiated at 150 or 1000 Gy in 1 ml YE medium.
Microscopic observations
All microphotographs were taken using a Leica Microsystems DMRD Microscope with a 100× oil immersion objective and a Princeton CoolSnap fx cooled CCD camera. The light source for fluorescence excitation was an HBO 100 W Hg arc lamp. For observation of living cells, cultures were spread on a glass slide without fixation, covered with a coverslip and observed directly. For GFP expressing strains, typical exposure time was of ∼5 s. Image capture software was MetaView (Universal Imaging) whereas image processing software was Metamorph Offline (Universal Imaging). The latter was used to count cell types on pictures. About 200 cells were counted for each time point. When needed, final assembly of figures was performed using Adobe Illustrator and/or Photoshop.
RESULTS
SpRad22p forms foci following induction of DNA DSBs
To investigate whether Rad22 foci formation could be observed in fission yeast, we tagged the C-terminus of Rad22 with either YFP or CFP. Both YFP and CFP tags did not alter the function of Rad22. Indeed, we assayed for radioresistance and did not detect any differences between tagged and wild type strains (data not shown).
Rad22-YFP or -CFP proteins were visualised by epifluorescence microscopy. In asynchronous cultures, the majority of untreated cells displayed diffuse nuclear fluorescence (Fig. 1A), with lower fluorescence in the nucleolar area. About 15% of small mononucleate cells, which are in either late-S or early-G2 phase of cell cycle, displayed punctuate fluorescence (Fig. 1A, arrows). Punctuate patterns were never observed in M, G1 or S phase cells which are binucleate in S.pombe.
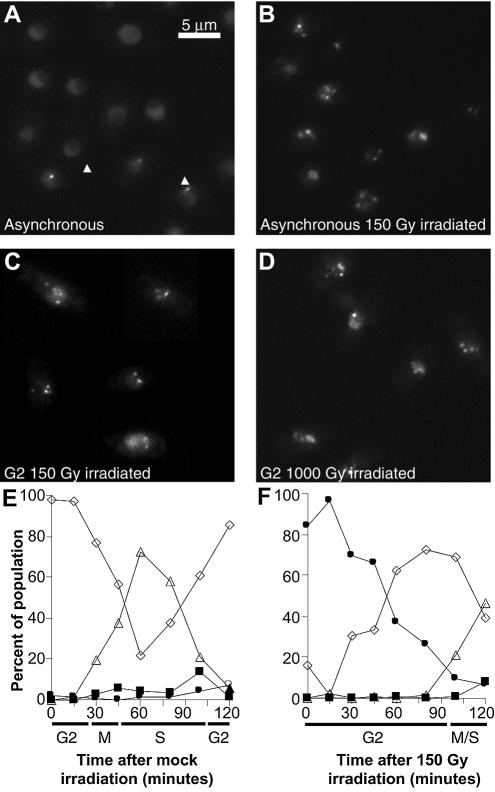
Figure 1.
Schizosaccharomyces pombe cells expressing Rad22-YFP. (A) Asynchronous unirradiated cells. Arrows indicate Rad22-YFP foci in small mononucleate cells. (B) Asynchronous cells 30 min after 150 Gy irradiation. (C) G2 synchronised cells 5 min after 150 Gy gamma irradiation. (D) G2 synchronised cells 5 min after 1000 Gy gamma irradiation. (E) Time course of cell type appearance following mock irradiation of G2 synchronised cells. White diamonds: mononucleate cells with homogenous nuclear Rad22-YFP fluorescence; white triangles: binucleate cells with homogenous nuclear Rad22-YFP fluorescence; black squares: binucleate cells showing Rad22-YFP foci; black circles: mononucleate cells showing Rad22-YFP foci. (F) Time course of cell type appearance following 150 Gy irradiation of G2 synchronised cells; symbols as in (E).
With gamma irradiation at 150 Gy, we generated DSBs in asynchronous cells expressing fluorescent Rad22 and investigated whether Rad22 foci were formed in the presence of DNA lesions. Cell survival was 99% at this IR dose (data not shown). Shortly after irradiation (5 min) we observed Rad22-YFP foci in nearly all the cells (Fig. 1B).
We then synchronised cells in G2 phase of the cell cycle and irradiated them at either 150 or 1000 Gy. At 1000 Gy, survival was ∼90%. The number of foci per cell was similar in cells treated at high or low IR doses (compare Fig. 1C and D).
After irradiating G2 synchronised cells with 150 Gy, we released them into the cell cycle and followed foci dynamics by monitoring the different cell types every 15 min: cells with one or two nuclei, showing or not showing Rad22 nuclear foci (Fig. 1E and F). The cell cycle progressed normally in mock-irradiated culture and cells entered mitosis without showing Rad22 foci (E). Very few unirradiated G2 cells harboured foci at the beginning of the time course (Fig. 1E) whereas ∼15% of the cells showed Rad22 foci in an asynchronous culture (Fig 1A). This difference is due to the time lapse between elutriation and observation, because Rad22 foci last only for a short time in asynchronous unirradiated cells. Indeed, a small population of unirradiated cells displaying Rad22 spots (∼15%) can be transiently observed after cytokinesis, at the end of S phase (Fig. 1E, 105 min). That 84% G2 cells showed Rad22 foci at time 0 in Figure 1F is due to the time lapse between cell retrieval after gamma irradiation and microphotography, which corresponded to 5 min under our experimental conditions. In other words, 5 min after irradiation 84% of G2 cells already showed Rad22 foci. The foci were detectable during the entire checkpoint delay and cells entered mitosis only when Rad22 foci had disappeared (Fig. 1F, 120 min).
Rad22 foci formation is independent of double strand break resection and checkpoint activation
Fission yeast proteins Rad50 and Rad32 are structurally related to S.cerevisiae Rad50 and Mre11, respectively. In budding yeast these proteins are required for ‘early’ steps of homologous recombination and for DNA damage checkpoint activation.
We analysed the presence of Rad22 foci in asynchronous cultures of either rad50 or rad32 null mutants before and after IR at 150 Gy (Fig. 2). Whereas only ∼15% of unirradiated wild type cells expressing Rad22-YFP showed nuclear foci, 62% of cells deleted for rad50 and 77% of cells deleted for rad32 showed Rad22 foci in unirradiated cultures. Following irradiation, both mutant strains showed Rad22 foci in almost all cells, indicating that neither Rad50 nor Rad32 are essential for Rad22 foci formation. Both deletion mutants are equally sensitive to IR, showing <5% survival after gamma irradiation at 150 Gy. After irradiation at 1000 Gy, both rad50 and rad32 mutants displayed Rad22 foci in all the cells, confirming that neither Rad50 nor Rad32 are essential for Rad22 foci formation (data not shown). Irradiated cultures showed a decreased number of septated cells and an increased number of elongated cells with a single hypertrophic nucleus (data not shown). These features indicate induction of cell cycle delay by the DNA integrity checkpoint in rad50 and rad32 mutant strains, which is in marked contrast with the situation in budding yeast (5).
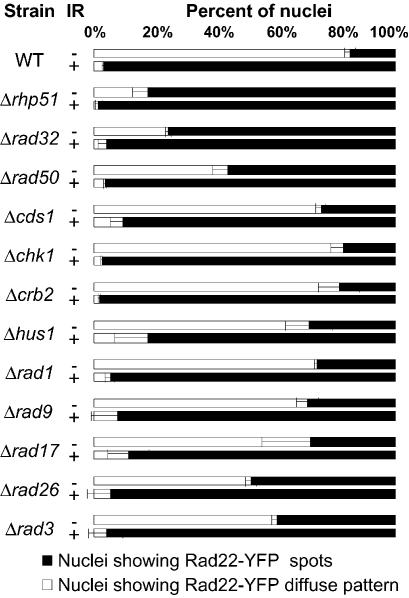
Figure 2.
Rad22-YFP foci appearance in unirradiated (–IR) or irradiated (+IR) asynchronously growing cells deleted of the indicated genes. Cells were irradiated at 150 Gy and imaged 30 min later.
We tested Rad22 foci formation in a strain lacking Rhp51, the S.pombe homologue of S.cerevisiae and mammalian Rad51, which are involved in late steps of homologous recombination. As we observed for deletions of early recombination genes, Rad22 foci were detectable in a high percentage (79%) of cells even in the absence of genotoxic treatment, and the proportion of cells with foci increased after irradiation (Fig. 2). Thus, like Rad32 and Rad50, Rhp51 protein is dispensable for Rad22 foci formation.
To test whether Rad22 foci formation was dependent upon a functional DNA structure checkpoint pathway, we tested for the presence of Rad22 foci in different checkpoint null mutants (cds1, chk1, crb2, hus, rad1, rad9, rad17, rad26, rad3; Fig. 2). Deletions of any checkpoint protein did not alter Rad22 foci formation following IR. However, all deleted strains displayed an increased number of cells harbouring Rad22 foci in the absence of IR, albeit to variable extents (Fig. 2). We conclude that the recruitment of Rad22 at DSBs repair foci can occur in checkpoint deficient cells.
SpRad9, a component of the checkpoint DNA sliding clamp, forms foci upon induction of DSBs
We tagged Rad9 protein, a component of the checkpoint sliding clamp, by fusing YFP to its C-terminus. The strain expressing Rad9-YFP showed normal sensitivity to several DNA damaging agents, including gamma- or UV-irradiation, MMS, CPT and HU treatments (data not shown). Rad9-YFP fluorescence appeared diffuse in the nucleus of almost all untreated cells (Fig. 3A). Following gamma-irradiation of asynchronous cells with 150 Gy, Rad9-YFP was quickly recruited to nuclear foci (5 min, Fig. 3B).
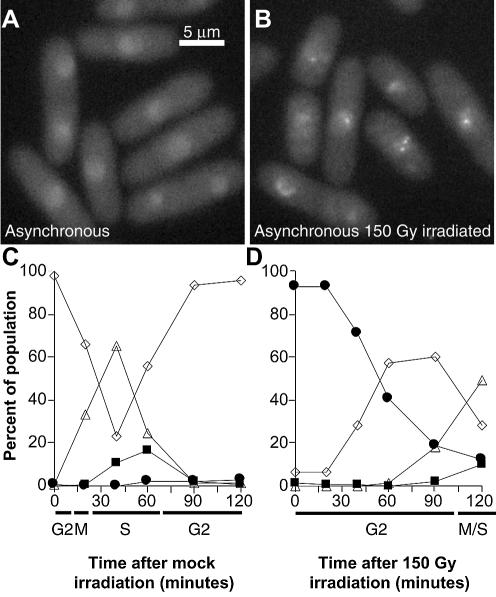
Figure 3.
Schizosaccharomyces pombe cells expressing Rad9-YFP. (A) Unirradiated asynchronous cells. (B) Asynchronous cells 30 min after 150 Gy irradiation. (C) Time course of cell type appearance following mock irradiation of G2 synchronised cells. White diamonds: mononucleate cells showing homogenous nuclear Rad9-YFP fluorescence; white triangles: binucleate cells with homogenous nuclear Rad9-YFP fluorescence; black squares: binucleate cells showing Rad9-YFP spots; black circles: mononucleate cells showing Rad9-YFP spots. (D) Time course of cell type appearance following 150 Gy irradiation of G2 synchronised cells; symbols as in (C).
We followed the dynamics of IR-induced Rad9 foci after synchronisation of cells in G2 (Fig. 3C and D). Again, different values at time zero in C and D of Figure 3 reflect the 5 min delay required to retrieve and photograph irradiated cells. Thus, 5 min after irradiation 92% of G2 cells showed Rad9-YFP foci. In the mock-irradiated sample, the cell cycle progressed normally (Fig. 3C). In the irradiated culture (Fig. 3D), Rad9 foci appeared rapidly, and were still present in 70% of cells 40 min after IR, after which they gradually disappeared before cells re-entered the cell cycle. Thus, under the same experimental conditions, Rad9 and Rad22 form foci at a similar rate following DSBs induction, since 90% of the cells displayed both Rad9 and Rad22 foci 5 min after IR treatment (Figs 1F and 3D).
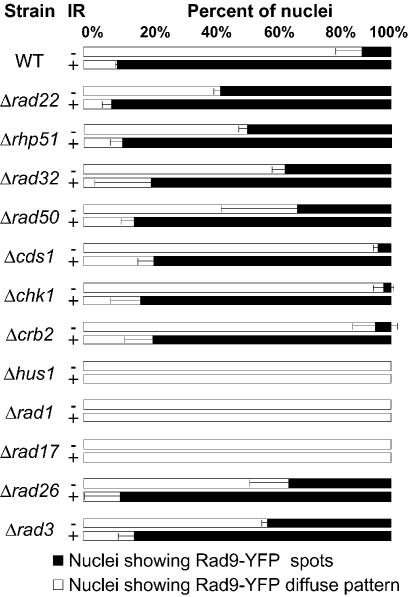
Next we studied Rad9 foci formation in strains deleted for different genes implicated in DSBs repair (Fig. 4). After IR, Rad9 foci formed rapidly in cells lacking Rad22, Rhp51, Rad50 or Rad32. Thus, absence of early or late DNA recombination genes did not affect Rad9 foci formation. Some DNA repair deficient cells displayed Rad9-YFP nuclear foci in the absence of IR (Fig. 4). However, their number was in general lower than that observed in Rad22-YFP foci analysis (compare Figs 2 and 4). Finally, Rad9 foci were not formed when Rad1, Hus1 or Rad17 proteins were absent, indicating that other members of the checkpoint sliding clamp and the sliding clamp loader are essential for Rad9 foci assembly (Fig. 4). However, Rad9-YFP still remained inside the nucleus, indicating that neither the other members of the checkpoint sliding clamp nor the checkpoint clamp loader itself are necessary for Rad9 nuclear localisation.
Figure 4.
Rad9-YFP foci appearance in unirradiated (–IR) or irradiated (+IR) asynchronously growing cells deleted of the indicated genes. Cells were irradiated at 150 Gy and imaged 30 min later.
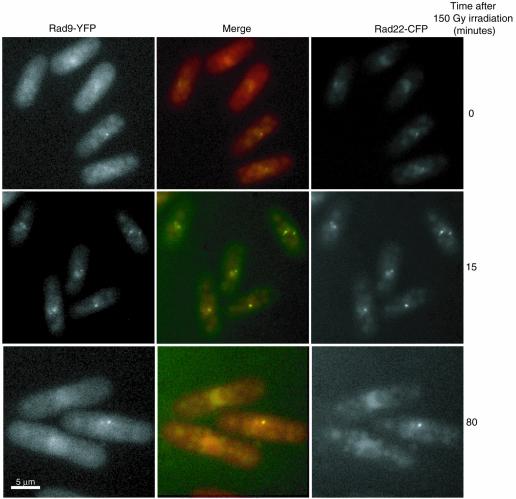
Rad9 and Rad22 foci overlap
To determine whether checkpoint complexes formed by Rad9-YFP are localised to the same DNA recombination foci revealed by Rad22-YFP, we performed co-localisation analysis in G2 cells expressing both Rad9-YFP and Rad22-CFP (Fig. 5). Following irradiation at 150 Gy, Rad22 and Rad9 fluorescence became punctuate and YFP and CFP patterns completely overlapped, indicating that Rad9 and Rad22 were present within the same foci (Fig. 5, time 0 corresponds to 5 min after irradiation). Also, concomitant disappearance of Rad9 and Rad22 foci was observed ∼80 min after irradiation (Fig. 5). Thus, the checkpoint sliding clamp and the homologous recombination complexes are located within the same subnuclear compartments after IR.
Figure 5.
In vivo co-localisation analysis of Rad9-YFP and Rad22-CFP foci induced with 150 Gy irradiation of G2 synchronised cells at different times after release into the cell cycle. Left panels: Rad9-YFP. Right panels: Rad22-CFP. Central panels: merge in yellow (red: Rad9-YFP; green: Rad22-CFP).
PCNA forms foci in the presence of DNA DSBs
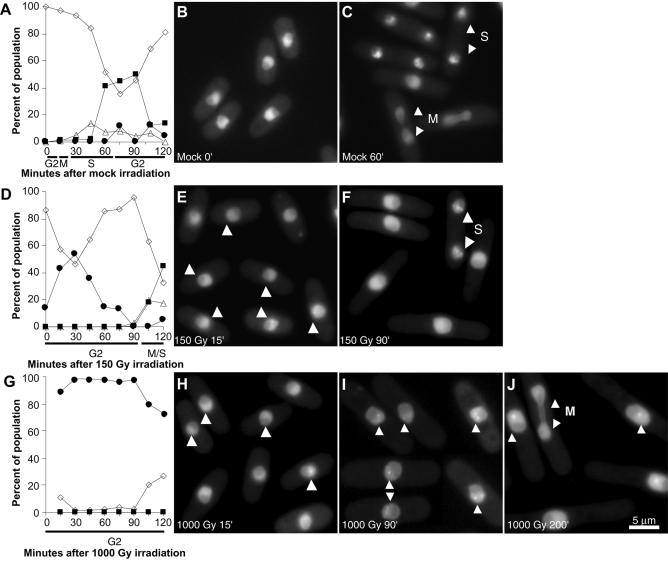
Rad9 has been shown to interact with PCNA after UV irradiation (24). PCNA could be a key member of the recombination/checkpoint complex because it is a conserved eukaryotic protein involved in many aspects of DNA metabolism, including recombination and checkpoint signalling. To determine whether PCNA is present at sites of DSBs repair in S.pombe, we fused PCNA to the C-terminus of EGFP or ECFP and expressed the fusion protein under transcriptional control of the wild type pcn1+ promoter. The EGFP–PCNA fusion protein complements the pcn1 gene deletion, but the strain is UV sensitive. Thus, we constructed a strain co-expressing both tagged and wild type PCNA to avoid side effects due to the fusion protein. Crosslink experiments of purified proteins or of total protein extracts with EGS [ethylene glycol-bis(succinic acid n-hydrosuccinimide ester)] showed that wild type and tagged PCNA interacted in vivo and in vitro, forming heterotrimers (unpublished results). In these conditions, the expression of tagged PCNA did not make the cells particularly sensitive to any tested genotoxic treatment (data not shown). During S phase, fluorescent PCNA forms foci which correspond to DNA replication factories, which will be described in detail elsewhere (Fig. 6A–C).
Figure 6.
In vivo dynamics of EGFP–PCNA foci in G2-synchronised unirradiated (A–C), 150 Gy irradiated (D–F) or 1000 Gy irradiated (G–J) S.pombe cells. (A) Time course of cell type appearance after mock irradiation. White diamonds: mononucleate cells with homogenous nuclear EGFP–PCNA fluorescence; white triangles: binucleate cells with homogenous nuclear EGFP–PCNA fluorescence; black squares: binucleate cells showing EGFP–PCNA foci; black circles: mononucleate cells showing EGFP–PCNA foci. (B) Microphotograph of mock-irradiated cells at time 0. (C) Microphotographs of mock-irradiated cells at 60 min. M indicates a mitotic cell; S indicates a binucleate cell in S phase showing DNA replication PCNA foci. (D) Time course of cell type appearance after 150 Gy irradiation. Symbols as in (A). (E) Microphotograph of cells 15 min after 150 Gy irradiation. Arrowheads indicate nuclei with DNA repair PCNA foci. (F) Microphotograph of cells 90 min after 150 Gy irradiation. S indicates a binucleate cell in S phase; arrowheads point to DNA replication PCNA foci. (G) Time course of cell type appearance after 1000 Gy irradiation. Symbols as in (A). (H) Microphotograph of cells 15 min after 1000 Gy irradiation. Arrowheads indicate nuclei with DNA repair PCNA foci. (I) Microphotograph of cells 90 min after 1000 Gy irradiation. Arrowheads indicate nuclei with DNA repair PCNA foci. (J) Microphotograph of cells 200 min after 1000 Gy irradiation. M indicates a mitotic cell; single arrowheads indicate nuclei with DNA repair PCNA foci.
When the strain expressing EGFP–PCNA was submitted to genotoxic treatment, PCNA foci became apparent in the vast majority of the cells. Thus, we decided to study these DNA repair foci in more detail. Synchronised G2 cells were irradiated with either 150 Gy (99% of cell survival) or 1000 Gy (90% of cell survival) and the different cell types were monitored every 15 min after release into the cell cycle (Fig. 6A, D and G). At the low dose (Fig. 6D, E and F), one to five bright fluorescent PCNA foci per cell appeared shortly after irradiation (Fig. 6E, arrows) and disappeared after 90 min (Fig. 6D and F). Once PCNA foci disappeared, cells entered mitosis followed by S phase (Fig. 6D and F). At the high dose (Fig. 6G–J), one to five bright fluorescent foci per cell could be observed in the great majority of the cells when the samples were taken out of the irradiator (irradiation with 1000 Gy lasted ∼15 min) (Fig. 6G and H). Ninety minutes after irradiation, foci were still present in many nuclei but were brighter and less abundant, most cells showing only one or two foci (Fig. 6I). After 105 min, the proportion of cells displaying foci began to decrease (Fig. 6G) and the cells contained only one or two large foci. Cells entering mitosis did not show PCNA foci (Fig. 6J, arrows).
PCNA foci formation is independent of checkpoint genes and homologous recombination
To investigate whether PCNA foci formation requires checkpoint activity, we analysed PCNA foci in G2-synchronised cells deleted for several checkpoint genes (chk1, crb2, rad3, rad26, rad17, rad1, cds1). In all these strains PCNA foci were detected before cells entered aberrant mitosis in the absence of a functional checkpoint (data not shown). We conclude that checkpoint activity is not required for PCNA foci formation.
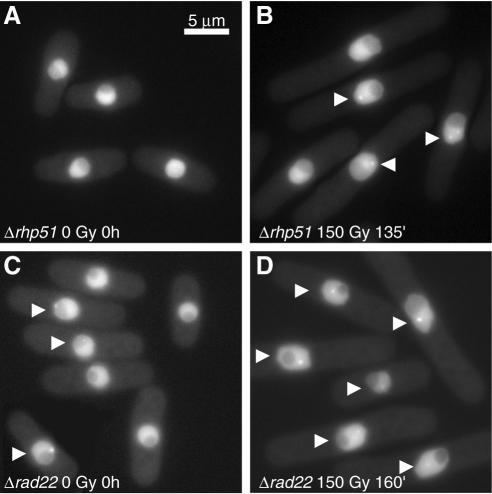
G2 phase cells expressing EGFP–PCNA and deleted for the rhp51 gene, which are unable to perform DNA repair by homologous recombination, were irradiated at 150 Gy (1% cell survival) and analysed for PCNA foci formation. The untreated sample showed a few mononucleate cells with PCNA foci (Fig. 7A). After IR, PCNA foci were formed and remained visible for >2 h during the sustained checkpoint delay due to the impossibility to repair DSBs through HRR (Fig. 7B). Four hours after IR some cells underwent aberrant mitosis. These observations indicate that PCNA foci induced by DSBs can form independently of active HRR but are not resolved when DNA recombination is prevented.
Figure 7.
In vivo detection of EGFP–PCNA foci in G2-synchronised cells of strains deleted of rhp51 (A and B) or rad22 (C and D) at the indicated times. Most cells show evident foci after irradiation in both strains (B and D, arrows). For the rad22 deletion, numerous cells show PCNA foci without irradiation in G2-synchronised cells (C, arrows).
We also tested EGFP–PCNA foci formation in irradiated G2 cells deleted for rad22 (Fig. 7C and D). Interestingly, ∼23% of mononucleate unirradiated cells showed PCNA foci (Fig. 7C). Shortly after irradiation at 150 Gy (<1% survival), most mononucleate cells showed a few bright fluorescent foci. During the 60 min period following IR the number of PCNA foci per cell decreased, as also observed in wild type and Δrhp51 cells. Irradiated cells gradually elongated and a large number still displayed PCNA foci after >2 h (Fig. 7D). Finally, after 4 h some cells underwent abnormal mitosis. In conclusion, PCNA foci dynamics is similar in both Δrhp51 and Δrad22 strains: foci are formed at low IR doses, are maintained during the G2 delay imposed by the DNA damage checkpoint and persist in cells unable to perform DNA recombination.
PCNA co-localises partially with Rad22 and Rad9 repair/checkpoint factories
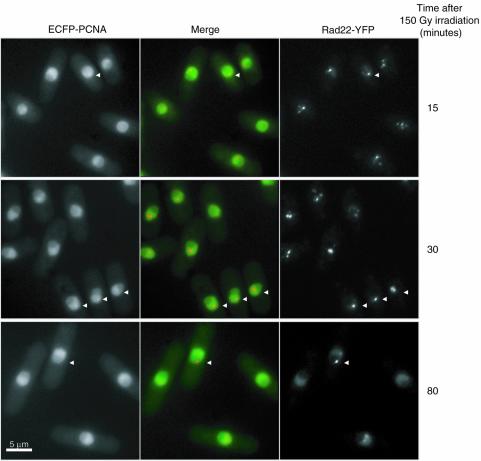
To determine if PCNA is present in checkpoint/recombination complexes revealed by Rad22 and Rad9 foci, we performed co-localisation experiments in irradiated cells expressing ECFP–PCNA and either Rad22-YFP or Rad9-YFP.
Figure 8 shows the results obtained for Rad22 and PCNA co-localisation. Cells showed partial but nonetheless clear co-localisation of the two proteins after irradiation. PCNA and Rad22 foci disappeared progressively, PCNA foci being the first ones to fade (Fig. 8, 80 min after irradiation). However, this could be due to the higher nuclear fluorescence background of EGFP–PCNA. We obtained similar results from co-localisation studies of Rad9-YFP and ECFP–PCNA (data not shown). Thus, we conclude that PCNA is also recruited at the intra-nuclear checkpoint/recombination structures induced by DSBs.
Figure 8.
In vivo co-localisation analysis of ECFP-PCNA and Rad22-YFP foci induced with 150 Gy irradiation of G2-synchronised cells at different times after release into the cell cycle. Most cells show co-localisation (arrows). Left panels: ECFP–PCNA. Right panels: Rad22-YFP. Central panels: merge in yellow (green: ECFP–PCNA; red: Rad22-YFP).
DISCUSSION
The results reported here show that fluorescent fusion proteins implicated in DNA damage checkpoint signalling (Rad9), in DNA repair by homologous recombination (Rad22) and in DNA synthesis (PCNA) co-localise in nuclear foci after induction of DSBs by gamma irradiation of living fission yeast cells. These foci are always limited in number, do not increase as a function of IR doses and are resolved when cells exit from DNA damage checkpoint delay. These observations imply that multiple DSBs are processed in the same recombination ‘factory’ which disappears when DNA repair is completed. It remains to be established how multiple DNA strand breaks are brought together within the same subnuclear location and what molecular mechanisms are responsible for foci assembly or disassembly.
Rad22, the major homologue of Rad52 protein family in S.pombe, is involved in homologous recombination and in other mechanisms of DSBs repair, such as single strand annealing (25). In S.pombe it has been shown that Rad22 interacts with the C-terminus of Rhp51, the homolog of Rad51 (26,27). Surprisingly, Rad22 recruitment to IR-induced foci is independent of functional DSBs repair, which cannot be performed when proteins Rad32, Rhp50 or Rhp51 are absent (Fig. 2). In addition, we also show that IR-induced Rad22 nuclear foci also contain the checkpoint signalling Rad9 protein. Despite complete co-localisation, Rad22 foci do not require the presence of Rad9 protein for their assembly, and vice versa. Also, Rad9 foci assembly does not depend on ongoing DSBs repair. However, it is worth noting that Rad9 cannot form IR-induced foci when Hus1, Rad1 or Rad17 are missing (Fig. 4). It is been shown in fixed cells that Hus1 and Rad9 nuclear localisation requires the presence of Rad17, the checkpoint clamp loader (9). This suggests that Rad9 foci may not be assembled in Δrad17 cells since the checkpoint sliding clamp cannot be formed when Rad9 and/or Hus1 are located outside the nucleus. However, in living S.pombe cells Rad9-YFP is nuclear despite the absence of Rad17, which excludes this hypothesis. Endogenous Rad9 or Rad22 foci are present in ∼15% of binucleate late-S phase cells in synchronised unirradiated wild type cultures (Figs 1E and 3C). Thus, Rad9 and Rad22 foci are probably assembled on abnormal DNA structures formed during DNA replication in the absence of induced DNA lesions. Strains deficient in DSBs repair also accumulate cells displaying Rad22 and Rad9 foci in the absence of induced DNA lesions (Figs 2 and 4). However, cells displaying endogenous Rad9 foci are less frequent than those containing Rad22 foci. This difference probably reflects the involvement of Rad22 in multiple DNA repair pathways, which could be activated when proteins critical for processing endogenous DNA lesions are absent in these strains (for a review, see 1). Thus, Rad9 and Rad22 foci in unirradiated DNA repair deficient cells indicate that abnormal DNA structures accumulate and elicit various cell reactions. We have shown that survival of Δrhp51 strain is dependent on the six checkpoint Rad genes, but not on the presence of the effector checkpoint kinases Chk1 or Cds1 (28). Thus, Rad9 foci in unirradiated Δrhp51 cells are not required for checkpoint kinase activation, which is dispensable for Δrhp51 cell survival, but probably for processing aberrant DNA structures accumulated in this strain.
PCNA trimers form a sliding clamp around DNA, a process that has been implicated in many DNA metabolism reactions, in particular replication and repair (8). Actually, live fission yeast cells expressing fluorescent PCNA show nuclear foci during S phase, corresponding to DNA replication foci (Fig. 6A and C). In contrast, G2 cells did not display PCNA foci in the absence of induced DNA lesions (Fig. 6A and B). This allowed us to study the assembly of IR-induced PCNA repair foci in G2, without confusing them with normal S phase DNA replication foci.
Since PCNA is implicated in late events in homologous recombination, because it is necessary for DNA synthesis occurring after strand invasion, we reasoned that PCNA foci induced by DSBs would be assembled only when the initial steps of recombination had occurred (8). Surprisingly, persistent PCNA foci are evident even at a low IR dose in the absence of either Rad22 or Rhp51 (Fig. 7B and D). Thus, active DNA repair involving the Rad52 group of genes is not needed for PCNA foci assembly. PCNA foci formation could precede or be concomitant with early recombination steps, functioning independently of actual DNA repair through homologous recombination. Since PCNA is able to interact with chromatin remodelling proteins, a possible role for early PCNA assembly at DSBs could be to promote chromatin modifications in the vicinity of the lesion (29). We also show that PCNA foci co-localise partially with Rad9/Rad22 foci, i.e. some PCNA foci do not include Rad9 or Rad22, probably because of the multiple functions of PCNA in DNA metabolism. Spontaneous PCNA foci are present in many non-irradiated G2 cells in which Rad22 was deleted, indicating that endogenous abnormal DNA structures are produced during DNA replication, remain unrepaired in the absence of Rad22, and stimulate PCNA foci formation. IR-induced PCNA foci, like Rad22 and Rad9 foci, disappear prior to entry into mitosis in wild type cells. They remain detectable in aberrant mitoses of checkpoint mutants unable to perform DNA repair. These observations indicate that foci disassembly depends on complete repair of DNA lesions and suggest that the presence of these foci is incompatible with normal mitosis.
In conclusion, we show for the first time in living yeast cells that, proteins implicated in DNA structure checkpoint, in homologous recombination DNA repair and in DNA synthesis, form nuclear foci quickly after DSBs induction. Foci persist until the end of cell cycle arrest induced by the DNA structure checkpoint. These observations in S.pombe, together with results obtained in budding yeast and mammalian cells, strengthen the idea that segregation of checkpoint, DNA repair and DNA synthesis factors to a few, specific nuclear territories is a general feature of cells with DNA lesions, and that their assembly and disassembly are strictly regulated.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Monique Smeets for helpful discussions and reagents and Dr Patrick Hughes for critical reading of the manuscript. This work was supported in part by ARC grant 5803 to G.B. and by Institut Curie PIC (Paramètres épigénétiques dans la réponse aux agents génotoxiques et le contrôle du cycle cellulaire). P.M. is the receiver of a scholarship from the French Ministry for Research and Technology.
REFERENCES
- 1.Jackson S.P. (2002) Sensing and repairing DNA double-strand breaks. Carcinogenesis 23, 687–696. [DOI] [PubMed] [Google Scholar]
- 2.Carr A.M. (2002) DNA structure dependent checkpoints as regulators of DNA repair. DNA Repair, 1, 983–994. [DOI] [PubMed] [Google Scholar]
- 3.Paques F. and Haber,J.E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev., 63, 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrini J.H. (1999) The mammalian Mre11-Rad50-nbs1 protein complex: integration of functions in the cellular DNA-damage response. Am. J. Hum. Genet., 64, 1264–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grenon M., Gilbert,C. and Lowndes,N.F. (2001) Checkpoint activation in response to double-strand breaks requires the Mre11/Rad50/Xrs2 complex. Nature Cell Biol., 3, 844–847. [DOI] [PubMed] [Google Scholar]
- 6.Manolis K.G., Nimmo,E.R., Hartsuiker,E., Carr,A.M., Jeggo,P.A. and Allshire,R.C. (2001) Novel functional requirements for non-homologous DNA end joining in Schizosaccharomyces pombe. EMBO J., 20, 210–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kolodner R.D., Putnam,C.D. and Myung,K. (2002) Maintenance of genome stability in Saccharomyces cerevisiae. Science, 297, 552–557. [DOI] [PubMed] [Google Scholar]
- 8.Warbrick E. (2000) The puzzle of PCNA’s many partners. Bioessays, 22, 997–1006. [DOI] [PubMed] [Google Scholar]
- 9.Caspari T., Murray,J.M. and Carr,A.M. (2002) Cdc2-cyclin B kinase activity links Crb2 and Rqh1-topoisomerase III. Genes Dev., 16, 1195–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Prudden J., Evans,J.S., Hussey,S.P., Deans,B., O’Neill,P., Thacker,J. and Humphrey,T. (2003) Pathway utilization in response to a site-specific DNA double-strand break in fission yeast. EMBO J., 22, 1419–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen L., Liu,T.H. and Walworth,N.C. (1999) Association of Chk1 with 14-3-3 proteins is stimulated by DNA damage. Genes Dev., 13, 675–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lisby M., Rothstein,R. and Mortensen,U.H. (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc. Natl Acad. Sci. USA, 98, 8276–8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastink A. and Lohman,P.H. (1999) Repair and consequences of double-strand breaks in DNA. Mutat. Res., 428, 141–156. [DOI] [PubMed] [Google Scholar]
- 14.Fernandez-Capetillo O., Chen,H.T., Celeste,A., Ward,I., Romanienko,P.J., Morales,J.C., Naka,K., Xia,Z., Camerini-Otero,R.D., Motoyama,N. et al. (2002) DNA damage-induced G2-M checkpoint activation by histone H2AX and 53BP1. Nature Cell Biol., 4, 993–997. [DOI] [PubMed] [Google Scholar]
- 15.DiTullio R.A. Jr, Mochan,T.A., Venere,M., Bartkova,J., Sehested,M., Bartek,J. and Halazonetis,T.D. (2002) 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nature Cell Biol., 4, 998–1002. [DOI] [PubMed] [Google Scholar]
- 16.Wang B., Matsuoka,S., Carpenter,P.B. and Elledge,S.J. (2002) 53BP1, a mediator of the DNA damage checkpoint. Science, 298, 1435–1438. [DOI] [PubMed] [Google Scholar]
- 17.Essers J., Houtsmuller,A.B., van Veelen,L., Paulusma,C., Nigg,A.L., Pastink,A., Vermeulen,W., Hoeijmakers,J.H. and Kanaar,R. (2002) Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J., 21, 2030–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kondo T., Wakayama,T., Naiki,T., Matsumoto,K. and Sugimoto,K. (2001) Recruitment of Mec1 and Ddc1 checkpoint proteins to double-strand breaks through distinct mechanisms. Science, 294, 867–870. [DOI] [PubMed] [Google Scholar]
- 19.Melo J.A., Cohen,J. and Toczyski,D.P. (2001) Two checkpoint complexes are independently recruited to sites of DNA damage in vivo. Genes Dev., 15, 2809–2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Majka J. and Burgers,P.M. (2003) Yeast Rad17/Mec3/Ddc1: a sliding clamp for the DNA damage checkpoint. Proc. Natl Acad. Sci. USA, 100, 2249–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wood V., Gwilliam,R., Rajandream,M.A., Lyne,M., Lyne,R., Stewart,A., Sgouros,J., Peat,N., Hayles,J., Baker,S. et al. (2002) The genome sequence of Schizosaccharomyces pombe. Nature, 415, 871–880. [DOI] [PubMed] [Google Scholar]
- 22.Kearsey S.E., Montgomery,S., Labib,K. and Lindner,K. (2000) Chromatin binding of the fission yeast replication factor mcm4 occurs during anaphase and requires ORC and cdc18. EMBO J., 19, 1681–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moreno S., Klar,A. and Nurse,P. (1991) Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol., 194, 795–823. [DOI] [PubMed] [Google Scholar]
- 24.Komatsu K., Wharton,W., Hang,H., Wu,C., Singh,S., Lieberman,H.B., Pledger,W.J. and Wang,H.G. (2000) PCNA interacts with hHus1/hRad9 in response to DNA damage and replication inhibition. Oncogene, 19, 5291–5297. [DOI] [PubMed] [Google Scholar]
- 25.van den Bosch M., Zonneveld,J.B., Vreeken,K., de Vries,F.A., Lohman,P.H. and Pastink,A. (2002) Differential expression and requirements for Schizosaccharomyces pombe RAD52 homologs in DNA repair and recombination. Nucleic Acids Res., 30, 1316–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsutsui Y., Khasanov,F.K., Shinagawa,H., Iwasaki,H. and Bashkirov,V.I. (2001) Multiple interactions among the components of the recombinational DNA repair system in Schizosaccharomyces pombe. Genetics, 159, 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim W.J., Lee,S., Park,M.S., Jang,Y.K., Kim,J.B. and Park,S.D. (2000) Rad22 protein, a rad52 homologue in Schizosaccharomyces pombe, binds to DNA double-strand breaks. J. Biol. Chem., 275, 35607–35611. [DOI] [PubMed] [Google Scholar]
- 28.Smeets M.F.M.A., Francesconi,S. and Baldacci,G. (2003) High dosage Rhp51 suppresion of the MMS sensitivity of DNA structure checkpoint mutants reveals a relationship between Crb2 and Rhp51. Genes Cells, 8, 573–586. [DOI] [PubMed] [Google Scholar]
- 29.Cromie G.A., Connelly,J.C. and Leach,D.R. (2001) Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell, 8, 1163–1174. [DOI] [PubMed] [Google Scholar]