Abstract
DNA single strand breaks (SSBs) are one of the most frequent DNA lesions in genomic DNA generated either by oxidative stress or during the base excision repair pathways. Here we established a new real-time assay to assess an imbalance of DNA SSB repair by indirectly measuring PARP-1 activation through the depletion of intracellular NAD(P)H. A water-soluble tetrazolium salt is used to monitor the amount of NAD(P)H in living cells through its reduction to a yellow colored water-soluble formazan dye. While this assay is not a direct method, it does not require DNA extraction or alkaline treatment, both of which could potentially cause an artifactual induction of SSBs. In addition, it takes only 4 h and requires less than a half million cells to perform this measurement. Using this assay, we demonstrated that the dose- and time-dependent depletion of NAD(P)H in XRCC1-deficient CHO cells exposed to methyl methanesulfonate. This decrease was almost completely blocked by a PARP inhibitor. Furthermore, methyl methanesulfonate reduced NAD(P)H in PARP-1+/+cells, whereas PARP-1–/– cells were more resistant to the decrease in NAD(P)H. These results indicate that the analysis of intracellular NAD(P)H level using water-soluble tetrazolium salt can assess an imbalance of SSB repair in living cells in real time.
INTRODUCTION
Cellular DNA is continuously exposed to insults from exposure to endogenous and exogenous electrophilic agents and oxidative stress. Base excision repair appears to be primarily responsible for the elimination of most of these deleterious DNA lesions (1). In addition to DNA lesions induced by these agents, spontaneous depurination/depyrimidination could introduce a significant amount of apurinic/apyrimidinic (AP) sites under physiological conditions (2). Using a combination of an aldehyde reactive probe and slot blot technique, we directly demonstrated the spontaneous depurination rate under physiological conditions to be 1.5 AP sites per 106 nucleotides per day, which corresponds to 9000 AP sites per cell per day (2). Modified bases and AP sites introduce DNA single strand breaks (SSBs) as intermediates of the base excision repair (BER) pathway (3). In this process, a DNA glycosylase cleaves the N-glycosylic bond between modified or even normal bases and deoxyriboses, leaving AP sites (3,4). The AP sites generated by the DNA glycosylase are subsequently incised by a class II AP endonuclease (5), resulting in a 3′-hydroxyl group and a 5′-deoxyribosephosphate (5′-dRp). After excision of 5′-dRp by DNA polymerase β (β-pol), repair is completed by the polymerase and ligase activities of β-pol and DNA ligase III, respectively. Furthermore, reactive oxygen species (ROS) also induce lesions by hydrogen abstraction from the deoxyribose, frequently producing oxidized AP sites as well as DNA SSBs (6,7). Therefore, DNA SSBs are one of the most frequent DNA lesions in mammalian cells even under physiological conditions. Single cell agarose gel electrophoresis, the so-called comet assay, is a well-established and highly sensitive assay to assess the amount of SSBs and their repair (8). However, this assay usually requires alkaline conditions to denature DNA for subsequent gel electrophoresis. It has been demonstrated that alkylating agents and oxidants introduce either alkaline-labile base lesions or AP sites leading to SSBs under basic conditions (9,10). Artifactual formation of SSBs might be introduced during DNA extraction. Therefore, it is difficult to accurately determine the number of SSBs and an imbalance in their repair using isolated cellular DNA. Accumulation of SSBs activates poly(ADP-ribose) polymerase-1 (PARP-1) that catalyzes the formation of polymers of poly(ADP-ribose), resulting in NAD+ depletion. In the present study, we established a new real-time assay to assess an imbalance of DNA SSB repair by indirectly measuring PARP-1 activation through the depletion of intracellular NAD(P)H following decrease in NAD+. Measurement was completed within 4 h requiring less than a half million living cells. A water-soluble tetrazolium salt is used to monitor the amount of NAD(P)H in living cells through its reduction to a yellow colored water-soluble formazan dye.
MATERIALS AND METHODS
Cell culture
XRCC1-proficient and -deficient CHO cells (11) were cultured as monolayers in alpha-minimal essential medium (Invitrogen) supplemented with 10% fetal bovine serum (Sigma), 100 µg/ml penicillin and 100 µg/ml streptomycin. Immortalized PARP-1+/+ and PARP-1–/– mouse embryonic fibroblasts were maintained in Dulbecco’s modified Eagle’s medium, 4.5 g/l glucose medium (Invitrogen) supplemented with 10% fetal bovine serum and 0.5% gentamicin (12). The cells utilized in this study were maintained in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
Determination of intracellular NAD(P)H using a water-soluble tetrazolium salt in living cells
A water-soluble tetrazolium salt was used to monitor the amount of NAD(P)H through its reduction to a yellow colored formazan dye. The total amount of NAD(P)H within viable cells in the medium was determined periodically by a spectrophotometer. Cells were seeded in 96-well plates (5 × 103 cells/well) and were cultured in 100 µl of medium containing fetal bovine serum and antibiotics as described above. After a 30-min incubation, cells were treated with methyl methanesulfonate (MMS) at indicated concentrations and 1/10 volume of CCK-8 solution (Dojindo Molecular Technology) which consisted of a water-soluble 2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium monosodium salt (WST-8, 5 mM) and 1-methoxy-5-methylphenazinium methylsulfate (1-methoxy PMS, 0.2 mM) as an electron mediator. Cells in each well were further cultured for up to 4 h. WST-8 produced a water-soluble yellow colored formazan dye through its bioreduction either in the presence of an electron carrier, 1-methoxy PMS and NAD(P)H or directly by NAD(P)H. Therefore, the reduction in tetrazolium salts mostly depends on the amount of intracellular NAD(P)H. The amount of formazan dye produced by the living cells in the medium was determined periodically (ca. every 30 min) by a spectrophotometer and compared to the values of control. Visible absorbance was recorded in a 96-well plate reader at 450 nm with 650 nm as a reference filter. A medium blank was prepared with only medium and CCK-8 reaction solution. The decrease in the intracellular NAD(P)H was assessed by comparing the absorbance of a well containing cells treated with MMS against that of a well with cells treated with PBS only. When necessary, specific PARP inhibitors, 3-aminobenzamide (3-AB, Sigma) (10 mM) and 3,4-dihydro-5-[4-(1-piperidinyl)butoxy]-1(2H)-isoquinolinone (DPQ, Sigma) (90 µM) were applied 1–2 h prior to the MMS treatment and kept in the medium during MMS exposure until the cells were analyzed.
Determination of intracellular NAD+
The cellular NAD+ level was determined by the enzyme cycling assay (13) with slight modification. Briefly, cells (1.5 × 106 cells/well) were treated with MMS at 0 and 2 mM for up to 4 h in 6-cm dishes. After washing with PBS, cells were trypsinized, harvested and transferred to microcentrifuge tubes followed by centrifugation. The supernatants were removed, and the pellets were resuspended in 100 mM potassium phosphate buffer containing 3% trichloroacetic acid. After 30 min on ice, the samples were centrifuged at 12 000 g. The acid-soluble fractions were neutralized with 800 mM KOH containing 200 mM Tris. Supernatants were mixed with a reaction medium containing 0.375 mM WST-8, 0.015 mM 1-methoxy PMS, 15 U/ml alcohol dehydrogenase (Sigma) and 120 mM ethanol in 100 mM potassium phosphate buffer. The reactions were performed in 96-well plates and the plates were incubated at 37°C in the dark for 10 min. The NAD+ content was determined using purified NAD+ (Sigma) as the standard. The decrease in the intracellular NAD+ was assessed by comparing the NAD+ content of a well containing cells treated with MMS against that of a well with cells treated with PBS only.
Determination of cell death
Cells were treated with MMS at 0, 0.75 and 1.5 mM for 1 and 4 h. After trypsinization, cells were exposed to the Trypan Blue dye-exclusion assay.
Comet assay
This assay was performed as described previously (14). Briefly, slides were first dipped into agarose and dried at 60°C. Low melting agarose (LMA) (0.5%) was prepared and held at 42°C. The cell suspension (1 × 104 cells/10 µl) was mixed with 190 µl of LMA. Ninety microliters of this suspension was pipetted onto each of the two slides and covered with a coverglass. Slides were then placed on ice for 5 min. The coverglass was removed, 90 µl of LMA was pipetted onto the slides, a coverglass was reapplied, and the slides were placed on ice again. After 5 min, the coverglass was removed and the slides were put in lysis buffer (pH 11) containing 2.5 M NaCl, 100 mM EDTA-2Na, 10 mM Tris, 1% Na sarcoinate, 1% Triton X-100 and 10% dimethyl sulfoxide for 20 min. Following lysis, slides were denatured in electrophoresis buffer (300 mM NaOH, 1 mM EDTA, pH 13). Electrophoresis was carried out in a large horizontal unit (Fisher Biotech) with a Bio-Rad 1000 power supply (Bio-Rad Labs) run for 20 min at 300 mV. Slides were removed and placed in a neutralizing buffer with 0.4 M Tris (pH 7.5) for 15 min, then placed in 95% ethanol for 5 min and allowed to air dry in a hood overnight.
Staining and analysis
Slides were stained using 35 µl of SYBR Green-1™ (10× in Tris-EDTA buffer, pH 8.0) (Molecular Probes, Eugene, OR, USA) and a coverglass was applied. Cells were viewed with a Leitz Orthoplan microscope with a 100 W Hg fluorescent light source and a Leitz I3 filter cube and a Dage CCD725 camera (Dage MTI, Michigan City, IA, USA) was used to capture images which were stored on a personal computer. Fifty cells from each of two slides were analyzed with the scorer being blinded to knowledge of the treatment group. The analysis was carried out using the Komet 5.0 (Kinetic Imaging, UK) comet analysis software.
Statistical analysis
Data were collected for tail length and percentage of DNA in the tail. The endpoints were used to calculate tail moment by the Komet 5.0 software using the formula: tail moment = tail length × percentage of DNA in the tail per 100 cells. Simple linear regression (one-tailed) was performed after checking for homogeneity of variance using Cochran’s C test and Bartlett’s test. If a statistically significant positive slope was obtained, a one-tailed Dunnett’s analysis was employed to compare each treatment mean to its concurrent control. The culture was the unit of experimentation and in all tests, the α was set at 0.05. All tests were performed using a Pentium based personal computer using Statgraphics™ Plus Version 5 statistical package.
RESULTS AND DISCUSSION
Intracellular NAD(P)H in DNA SSB repair deficient CHO cells
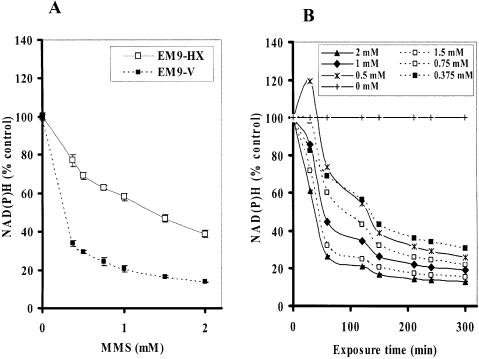
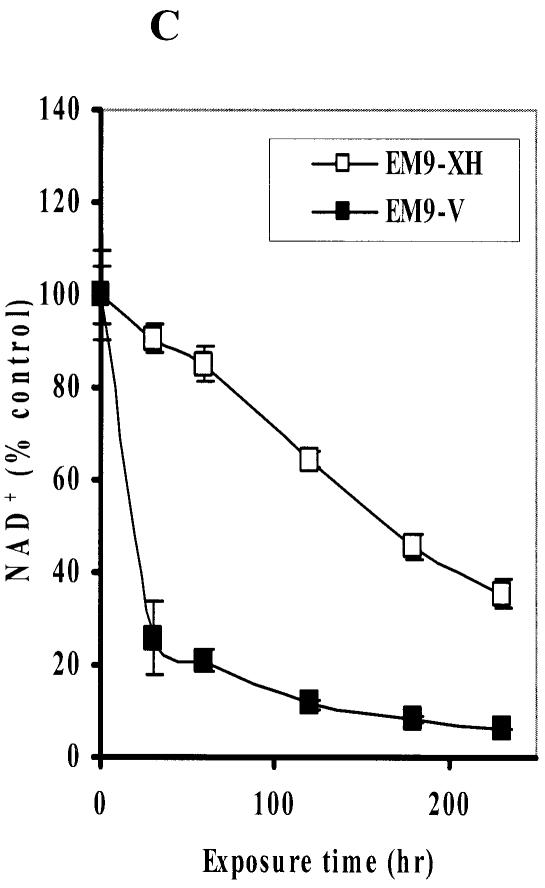
It has been widely accepted that DNA SSBs induce an activation of PARP-1 (15,16), which deplete intracellular NAD+ (17). While a dramatic decrease in the intracellular NAD+ further depletes ATP (18,19), depletion of NAD(P)H by PARP-1 activation has not been extensively studied. Therefore, we first tested whether a commercially available cell toxicity assay kit using water-soluble tetrazolium salts, which can monitor the amount of intracellular NAD(P)H level, could detect one of the early events induced by an imbalance of SSB repair. XRCC1 acts as a scaffold for interaction with other BER-associated proteins including human AP endonuclease (APE1), DNA Pol-β, DNA ligase III, polynucleotide kinase (PNK), and PARP-1 (20–24) and is required for efficient SSB repair and genetic stability in mammalian cells. Rodent cells lacking XRCC1 are hypersensitive to DNA damaging agents (21–23). Loss of XRCC1 also results in decreased genetic stability, including increased frequencies of spontaneous and/or induced chromosome translocations and deletions (24–29). We first utilized isogenic CHO cell lines either proficient or deficient in XRCC1 exposed to MMS. The cell lines used in this experiment were XRCC1-deficient CHO EM9 cells expressing an empty pcD2E vector (EM9-V) as a SSB repair-deficient cell and expressing wild-type human XRCC1 (pcD2EXH) (EM9-XH) as a SSB repair-proficient cell (21). These cells were exposed to MMS at different concentrations for 4 h. While the Trypan Blue exclusion assay demonstrated no major cell death during MMS treatment at any concentration (data not shown), the intracellular NAD(P)H in both cell lines was significantly reduced by treatment with MMS in a dose-dependent manner (Fig. 1A). These data indicate the decrement of NAD(P)H is not due to a reduction in the number of viable cells. These effects were more obvious in SSB repair-deficient EM9-V cells compared to those in EM9-XH cells. To further address earlier events in terms of the intracellular NAD(P)H, we monitored the reduction in NAD(P)H periodically from 30 min to 4 h during MMS exposure. Since CCK-8 reagent does not require cell lysis for the determination of NAD(P)H, we performed a real-time NAD(P)H assay using the same 96-well plate in a back and forth manner between the CO2 incubator and the plate reader. The reduction in NAD(P)H was detected at as early as 30 min during MMS exposure in EM9-V cells (Fig. 1B). Subsequently, we compared the degree of depletion of intracellular NAD+ in EM9-XH and EM9-V cells exposed to 2 mM MMS for up to 4 h. The depletion of intracellular NAD+ was determined by the enzyme cycling assay after cell lysis. EM9-XH cells were more resistant to the decrease in NAD+ compared to EM9-V cells. The decrease in NAD+ was detected slightly earlier and/or more extensively compared to that in intracellular NAD(P)H in EM9-V cells (Fig. 1B and C).
Figure 1.
Intracellular NAD(P)H and NAD+ levels in EM9 cells expressing empty vector (EM9-V) or human wild-type XRCC1 (EM9-XH). (A) EM9-XH and EM9-V cells were exposed to MMS for 4 h. Intracellular NAD(P)H levels in living cells were determined by adding CCK-8 solution. (B) EM9-V cells were exposed to MMS for up to 5 h. Intracellular NAD(P)H levels were monitored at 30, 60, 120, 150, 210, 240 and 300 min in real time. (C) EM9-XH and EM9-V cells were exposed to 2 mM MMS for 4 h. Intracellular NAD+ levels were determined by the enzyme cycling assay. Mean data and S.D. (bars) were from triplicate experiments using triplicate samples.
Effects of PARP inhibitor on the decrease in NAD(P)H in CHO cells
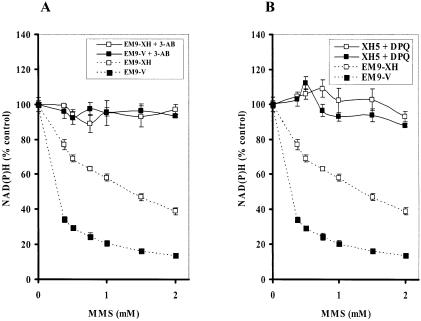
PARP-1 transfers hundreds of branched chains of ADP-ribose to a variety of nuclear proteins through its activation by DNA SSBs (22,23). Under massive DNA damage, activation of PARP-1 depletes its substrate, NAD+ (11). Since NADH is generated from NAD+ by the reaction of a dehydrogenase and its substrate, the decrease in the amount of NADH can follow the depletion of NAD+. To distinguish whether the reduction in NAD(P)H is due to a reduction in mitochondrial function or due to the depletion of NAD+ by PARP-1 activation, we co-exposed CHO cells to MMS and specific PARP inhibitors, i.e. 3-AB and DPQ. Either 10 mM 3-AB or 90 µM DPQ almost completely blocked the MMS-induced decrease in the amount of intracellular NAD(P)H in both cell lines (Fig. 2A and B). These results strongly suggest that the decrease in the intracellular NAD(P)H in CHO cells exposed to MMS for 4 h was primarily due to PARP-1 activation through formation of SSBs.
Figure 2.
Dose-dependency of the depletion of NAD(P)H in EM9-V and EM9-XH exposed to MMS for 4 h in the absence or presence of PARP inhibitor, 3-AB (10 mM) (A) or DPQ (90 µM) (B).
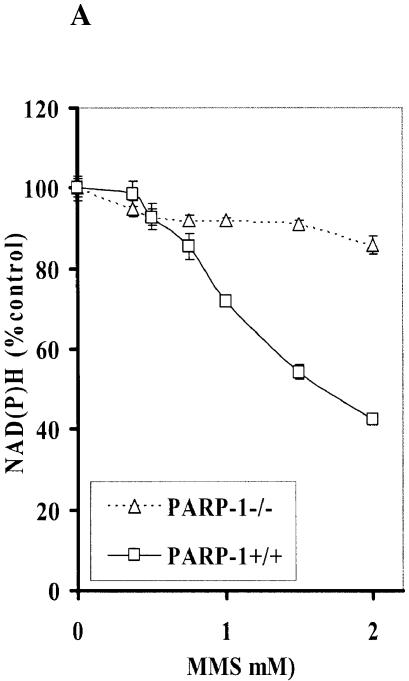
Intracellular NAD(P)H in PARP-1–/– cells exposed to MMS
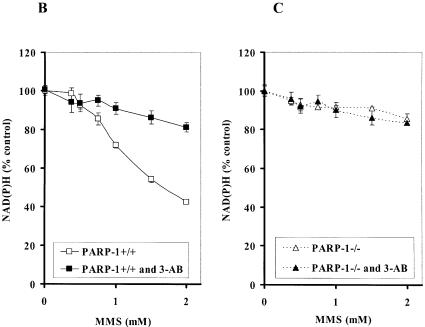
It has been reported that N-methyl-N′-nitro-N-nitrosoguanidine, an alkylating agent, caused a dramatic reduction in NAD+ levels in PARP-1+/+ fibroblasts, whereas NAD+ levels in PARP-1–/– fibroblasts were more resistant to the decrease in NAD+ (30). To vigorously confirm the decrement of NAD(P)H was due to PARP-1 activation, we exposed PARP-1–/– and PARP-1+/+ cells to MMS. While MMS reduced NAD(P)H in PARP-1+/+, PARP-1–/– cells were more resistant to the decrease in NAD(P)H (Fig. 3A). In addition, 3-AB mostly protected cells from the reduction in NAD(P)H in PARP-1+/+ cells exposed to MMS (Fig. 3B). We hypothesize that the slight decrease in NAD(P)H in PARP-1–/– cells may be due to a reduction in mitochondria function (Fig. 3C).
Figure 3.
Depletion of intracellular NAD(P)H in PARP-1+/+ and PARP-1–/– fibroblast cells. (A) NAD(P)H level in PARP-1+/+ and PARP-1–/– fibroblast cells exposed to MMS for 4 h. Depletion of NAD(P)H in PARP-1+/+ (B) and PARP-1–/– (C) fibroblast cells exposed to MMS for 4 h in the absence or presence of 3-AB (10 mM).
Comparison between sensitivity of SSB assay and comet assay
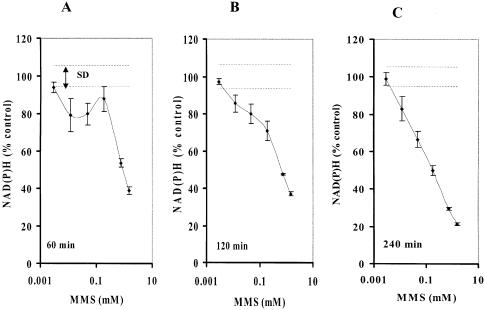
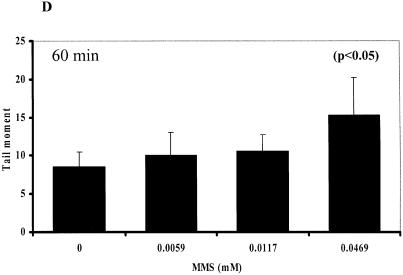
Although it is believed that the comet assay is the most sensitive method to determine SSBs in cells, there is still uncertainty that alkaline treatment (>pH 12.6) used to denature DNA may introduce artifactual formation of SSBs during the assay (8). We next compared the results from this newly developed real-time assay and the comet assay in XRCC1-deficient EM9-V cells exposed to MMS at concentrations as low as ∼5 µM for 4 h. The decrease in the amount of intracellular NAD(P)H was detected in cells exposed to MMS at concentrations as low as ∼10 µM at 1, 2 and 4 h, respectively (Fig. 4A–C). Using a regular comet assay (electrophoresis: pH 13), we also detected an increase in the amount of tail moment in EM9-V cells exposed to MMS at ∼50 µM for 1 h (Fig. 4D). These results indicate that this assay is reasonably sensitive to assess an imbalance of SSBs in mammalian cells.
Figure 4.
Comparison of the cellular effects induced by MMS detected by NAD(P)H assay and comet assay. Depletion of NAD(P)H in EM9-V cells exposed to MMS for 1 h (A), 2 h (B) and 4 h (C). (D) Tail moments were quantified by the comet assay in EM9-V cells immediately after MMS treatment at 0, 0.0059, 0.0117 or 0.0469 mM for 1 h. Results and S.D. are the mean from triplicate samples.
The present data indicate that in addition to NAD+ and ATP depletion, base excision repair, in an effort to counteract DNA alkylation, causes repair-mediated indirect SSB formation, further leading to NAD(P)H depletion through over-activation of PARP-1. By monitoring the intracellular NAD(P)H level, we established an indirect qualitative assay to assess an imbalance of SSB repair using less than a half million cells within 4 h. As far as we know, this is the first real-time assay to be able to monitor DNA damage present in living cells. The sensitivity of this assay appears to be equivalent to that of DNA damage induced by MMS determined by the comet assay. As described above, this assay can monitor PARP-1 activation through the depletion of intracellular NAD(P)H levels using living cells and does not require either DNA extraction or alkaline treatment, which could potentially introduce artifactual SSB formation during sample processing. However, it is noteworthy that this assay is an indirect method to monitor an accumulation of SSBs; therefore, it is necessary to confirm whether the decrement of intracellular NAD(P)H is due to NAD+ consumption by PARP-1. We believe that this assay may be applicable to elucidating the mechanism of action of DNA damaging agents and the screening for potential chemotherapeutic as well as chemopreventive agents. Furthermore, due to its convenience and high sensitivity, this assay can also be applicable to cell samples from large-scale population-based studies. While the BER pathway is one of the essential repair systems to eliminate DNA lesions induced by endogenous and exogenous agents, there is limited evidence indicating the association between the existence of a human population with a BER deficiency and genesis of cancer (31). This assay has the potential to be able to demonstrate that there is a wide variation in the capacity of the BER pathway in the human population through a large-scale study and could provide critical evidence regarding cancer-susceptible populations who repair oxidized and alkylated DNA lesions less efficiently.
REFERENCES
- 1.Wood R.D. (1996) DNA repair in eukaryotes. Annu. Rev. Biochem., 65, 135–167. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura J., Walker,V.E., Upton,P.B., Chiang,S.-Y., Kow,Y.W. and Swenberg,J.A. (1998) Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res., 58, 222–225. [PubMed] [Google Scholar]
- 3.Krokan H.E., Standal,R. and Slupphaug,G. (1997) DNA glycosylases in the base excision repair of DNA. Biochem. J., 325, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindahl T. (2000) Suppression of spontaneous mutagenesis in human cells by DNA base excision-repair. Mutat. Res., 462, 129–135. [DOI] [PubMed] [Google Scholar]
- 5.Demple B. and Harrison,L. (1994) Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem., 63, 915–948. [DOI] [PubMed] [Google Scholar]
- 6.Breen A.P. and Murphy,J.A. (1995) Reactions of oxyl radicals with DNA. Free Radic. Biol. Med., 18, 1033–1077. [DOI] [PubMed] [Google Scholar]
- 7.Von Sonntag C. (1987) The Chemical Basis of Radiation Biology. Taylor and Francis, London, pp. 238–249. [Google Scholar]
- 8.Tice R.R., Agurell,E., Anderson,D., Burlinson,B., Hartmann,A., Kobayashi,H., Miyamae,Y., Rojas,E., Ryu,J.C. and Sasaki,Y.F. (2000) Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environ. Mol. Mutagen., 35, 206–221. [DOI] [PubMed] [Google Scholar]
- 9.Burrows C.J. and Muller,J.G. (1998) Oxidative nucleobase modifications leading to strand scission. Chem. Rev., 98, 1109–1152. [DOI] [PubMed] [Google Scholar]
- 10.Miyamae Y., Iwasaki,K., Kinae,N., Tsuda,S., Murakami,M., Tanaka,M. and SasakiY.F. (1997) Detection of DNA lesions induced by chemical mutagens using the single-cell gel electrophoresis (comet) assay. 2. Relationship between DNA migration and alkaline condition. Mutat. Res., 393, 107–113. [DOI] [PubMed] [Google Scholar]
- 11.Taylor R.M., Thistlethwaite,A. and Caldecott,K.W. (2002) Central role for the XRCC1 BRCT I domain in mammalian DNA single-strand break repair. Mol. Cell. Biol., 22, 2556–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreiber V., Ame,J.C., Dolle,P., Schultz,I., Rinaldi,B., Fraulob,V., Menissier-de Murcia,J. and de Murcia,G. (2002) Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J. Biol. Chem., 277, 23028–23036. [DOI] [PubMed] [Google Scholar]
- 13.Tafani M., Karpinich,N.O., Hurster,K.A., Pastorino,J.G., Schneider,T., Russo,M.A. and Farber,J.L. (2002) Cytochrome c release upon Fas receptor activation depends on translocation of full-length bid and the induction of the mitochondrial permeability transition. J. Biol. Chem., 277, 10073–10082. [DOI] [PubMed] [Google Scholar]
- 14.Tennant A.H., Peng,B. and Kligerman,A.D. (2001) Genotoxicity studies of three triazine herbicides: in vivo studies using the alkaline single cell gel (SCG) assay. Mutat. Res., 493, 1–10. [DOI] [PubMed] [Google Scholar]
- 15.Lautier D., Lagueux,J., Thibodeau,J., Menard,L. and Poirier,G.G. (1993) Molecular and biochemical features of poly (ADP-ribose) metabolism. Mol. Cell. Biochem., 122, 171–193. [DOI] [PubMed] [Google Scholar]
- 16.de Murcia G., Schreiber,V., Molinete,M., Saulier,B., Poch,O., Masson,M., Niedergang,C. and Menissier de Murcia,J. (1994) Structure and function of poly(ADP-ribose) polymerase. Mol. Cell. Biochem., 138, 15–24. [DOI] [PubMed] [Google Scholar]
- 17.Berger N.A. (1985) Poly(ADP-ribose) in the cellular response to DNA damage. Radiat. Res., 101, 4–15. [PubMed] [Google Scholar]
- 18.Carson D.A., Seto,S., Wasson,D.B. and Carrera,C.J. (1986) DNA strand breaks, NAD metabolism, and programmed cell death. Exp. Cell Res. 164, 273–281. [DOI] [PubMed] [Google Scholar]
- 19.Oleinick N.L. and Evans,H.H. (1985) Poly(ADP-ribose) and the response of cells to ionizing radiation. Radiat. Res., 101, 29–46. [PubMed] [Google Scholar]
- 20.Caldecott K.W., Aoufouchi,S., Johnson,P. and Shall,S. (1996) XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res., 24, 4387–4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitehouse C.J., Taylor,R.M., Thistlethwaite,A., Zhang,H., Karimi-Busheri,F., Lasko,D.D., Weinfeld,M. and Caldecott,K.W. (2001) XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell, 104, 1–11. [DOI] [PubMed] [Google Scholar]
- 22.Kubota Y., Nash,R.A., Klungland,A., Schar,P., Barnes,D.E. and Lindahl,T. (1996) Reconstitution of DNA base excision-repair with purified human proteins: interaction between DNA polymerase beta and the XRCC1 protein. EMBO J., 15, 6662–6670. [PMC free article] [PubMed] [Google Scholar]
- 23.Caldecott K.W., Mckeown,C.K., Tucker,J.D., Ljungquist,S. and Thompson,L.H. (1994) An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol. Cell. Biol., 14, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Caldecott K.W., Tucker,J.D., Stanker,L.H. and Thompson,L.H. (1995) Characterization of the XRCC1-DNA ligase III complex in vitro and its absence from mutant hamster cells. Nucleic Acids Res., 23, 4836–4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carrano A.V., Minkler,J.L., Dillehay,L.E. and Thompson,L.H. (1986) Incorporated bromodeoxyuridine enhances the sister-chromatid exchange and chromosomal aberration frequencies in an EMS-sensitive Chinese hamster cell line. Mutat. Res., 162, 233–239. [DOI] [PubMed] [Google Scholar]
- 26.Dominguez I., Daza,P., Natarajan,A.T. and Cortes,F. (1998) A high yield of translocations parallels the high yield of sister chromatid exchanges in the CHO mutant EM9. Mutat. Res., 398, 67–73. [DOI] [PubMed] [Google Scholar]
- 27.Thompson L.H., Brookman,K.W., Dillehay,L.E., Carrano,A.V., Mazrimas,J.A., Mooney,C.L. and Minkler,J.L. (1982) A CHO-cell strain having hypersensitivity to mutagens, a defect in DNA strand-break repair, and an extraordinary baseline frequency of sister-chromatid exchange. Mutat. Res., 95, 427–440. [DOI] [PubMed] [Google Scholar]
- 28.Veld C.W.O.H., Jansen,J., Zdzienicka,M.Z., Vrieling,H. and vanZeeland,A.A. (1998) Methyl methanesulfonate-induced hprt mutation spectra in the Chinese hamster cell line CH09 and its xrccl-deficient derivative EM-C11. Mutat. Res., 398, 83–92. [DOI] [PubMed] [Google Scholar]
- 29.Zdzienicka M.Z., Vanderschans,G.P., Natarajan,A.T., Thompson,L.H., Neuteboom,I. and Simons,J.W.I.M. (1992) A Chinese-hamster ovary cell mutant (EM-C11) with sensitivity to simple alkylating agents and a very high level of sister chromatid exchanges. Mutagenesis, 7, 265–269. [DOI] [PubMed] [Google Scholar]
- 30.Yu S.W., Wang,H., Poitras,M.F., Coombs,C., Bowers,W.J., Federoff,H.J., Poirier,G.G., Dawson,T.M. and Dawson,V.L. (2002) Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science, 297, 259–263. [DOI] [PubMed] [Google Scholar]
- 31.Al-Tassan N., Chmiel,N.H., Maynard,J., Fleming,N., Livingston,A.L., Williams,G.T., Hodges,A.K., Davies,D.R., David,S.S., Sampson,J.R. and Cheadle,J.P. (2002) Inherited variants of MYH associated with somatic G:C->T:A mutations in colorectal tumors. Nature Genet., 30, 227–232. [DOI] [PubMed] [Google Scholar]