Abstract
APETALA2 (AP2) plays an important role in the control of Arabidopsis flower and seed development and encodes a putative transcription factor that is distinguished by a novel DNA binding motif referred to as the AP2 domain. In this study we show that the AP2 domain containing or RAP2 (related to AP2) family of proteins is encoded by a minimum of 12 genes in Arabidopsis. The RAP2 genes encode two classes of proteins, AP2-like and EREBP-like, that are defined by the number of AP2 domains in each polypeptide as well as by two sequence motifs referred to as the YRG and RAYD elements that are located within each AP2 domain. RAP2 genes are differentially expressed in flower, leaf, inflorescence stem, and root. Moreover, the expression of at least three RAP2 genes in vegetative tissues are controlled by AP2. Thus, unlike other floral homeotic genes, AP2 is active during both reproductive and vegetative development.
Keywords: homeotic gene, gene regulation, plant development
Genetic and molecular studies have defined an evolutionarily conserved network of genes that control flower development in Arabidopsis and in other plant species (1, 2). In Arabidopsis, the homeotic gene APETALA2 (AP2) has been shown to control three important processes during flower development: (i) the establishment of flower meristem identity (3, 4), (ii) the specification of flower organ identity and the regulation of floral organogenesis (5–9), and (iii) the temporal and spatial regulation of flower homeotic gene activity (10). Genetic studies have shown that AP2 is also required for normal ovule and seed development (9, 11, 12). Consistent with its genetic functions AP2 is expressed throughout flower development, in immature floral buds, in all four types of flower organ primordia, and in developing ovules and seeds (ref. 9; B. den Boer and K.D.J., unpublished data). Unlike most other flower homeotic genes, AP2 is also expressed at the mRNA level in both stem and leaf, suggesting that AP2 may also function during vegetative development (9). However, ap2 mutant plants show no dramatic defects during either stem or leaf development. One hypothesis to explain this apparent paradox is that the Arabidopsis genome may be genetically redundant for AP2 function during stem and leaf development.
Previous studies have suggested that AP2 functions as a nuclear transcription factor in plant cells (9). DNA sequence analysis showed that AP2 encodes a theoretical polypeptide of 432 amino acids that is distinct from known fungal and animal regulatory proteins (9). One important feature of the AP2 protein is a novel 68-amino acid repeated motif called the AP2 domain. The AP2 domain has been shown to be essential for AP2 functions and contains an 18-amino acid core region that is predicted to form an amphipathic α-helix (9). In tobacco, four AP2-like AP2 domain containing proteins have been identified (13). These proteins, referred to as the ethylene responsive element binding proteins (EREBPs), are thought to regulate gene expression at the transcriptional level. In vitro studies have shown that the EREBP AP2 domain is sufficient for sequence specific DNA binding activity (13). By analogy, these data suggest that the AP2 domains of AP2 also confer sequence specific DNA binding activity.
In Arabidopsis, recent genetic and molecular studies have identified two new genes that encode AP2 domain containing proteins, AINTEGUMENTA (ANT), a gene that regulates flower development and is essential for ovule formation (14, 15), and TINY, a gene that suppresses cell proliferation during both vegetative and floral organogenesis when overexpressed in transgenic plants (16). Although these genes have not been reported to be functionally redundant with AP2 during vegetative development, their existence suggests that such genes may be found in the Arabidopsis genome. In this study, we have identified and characterized 12 new AP2 domain containing or RAP2 (related to AP2) genes in Arabidopsis. Genetic and molecular analysis of RAP2 gene expression supports the hypothesis that AP2 does indeed function in tissues and organs other than flowers and seed.
MATERIALS AND METHODS
Plant Material.
Arabidopsis thaliana ecotype Landsberg erecta (L-er) and C24 were used as wild type. The ap2-10 mutant is in the C24 genetic background. Plants were grown at 22°C under a 16-hr light/8-hr dark photoperiod in a 1:1:1 mixture containing vermiculite/perlite/peat moss. Plants were watered with a one-fourth strength Peter’s solution (Grace-Sierra, Milpitas, CA). Root tissue was harvested from plants grown hydroponically in sterile flasks containing 1× Murashige and Skoog plant salts (GIBCO), 1 mg/liter thiamine, 0.5 mg/liter pyridoxine, 0.5 mg/liter nicotinic acid, 0.5 g/liter 2-(N-morpholino)ethanesulfonic acid (Mes), and 3% sucrose, with moderate shaking and 70 μmol⋅m−2⋅sec−1 of light.
Analysis of Cloned Arabidopsis cDNAs.
Arabidopsis expressed sequence tagged (EST) cDNA clones representing RAP2.1 and RAP2.9 were generated as described by Cooke et al. (17) and were kindly provided by Monique Raynal (Laboratoire de Physiologie et Biologie Moléculaire des Plantes, Centre National de la Recherche Scientifique, Université de Perpignan, Perpignan, France). EST cDNA clones representing RAP2.2 and RAP2.8 were generated as described by Höfte et al. (18) and were kindly provided by Yves Parmentier (Institut de Biologie Moléculaire des Plants, Centre National de la Recherche Scientifique, Strasbourg, France). EST cDNA clones representing all other RAP2 genes described in Fig. 1 were generated by Newman et al. (19) and provided by the Arabidopsis Biological Resource Center (Ohio State University). Plasmid DNAs were isolated and purified by anion exchange chromatography (Qiagen, Chatsworth, CA). DNA sequences were generated using fluorescence dye-based nucleotide terminators and analyzed as specified by the manufacturer (Applied Biosystems).
Figure 1.
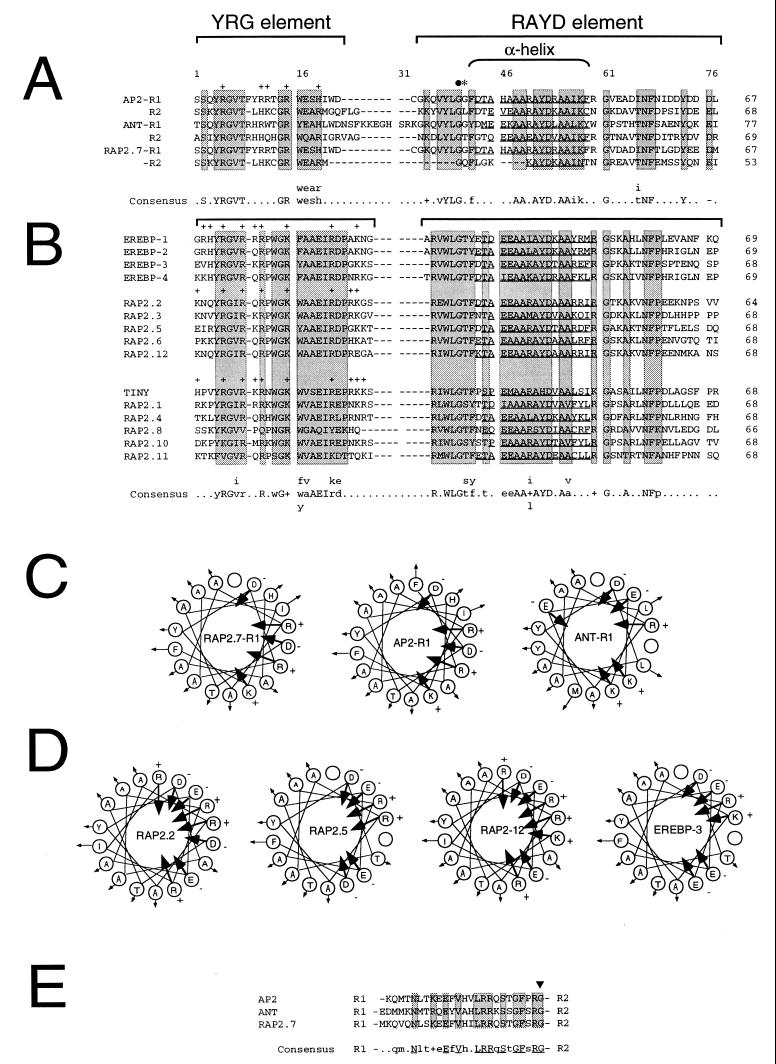
AP2 domain sequence and structure. Arabidopsis RAP2 amino acid sequences were determined and compared as described. Gene names are shown to the left of each sequence. The number of amino acid residues within each AP2 domain is shown to the right. Sequence gaps were introduced to maximize sequence alignments. The position of amino acid residues and sequence gaps within the AP2 domain alignments are numbered 1–77 for reference. The location of the conserved YRG and RAYD elements are indicated by brackets. Shaded boxes highlight regions of sequence similarity. Positively charged amino acids within the YRG element are indicated by + signs above the residues. The location of the 18-amino acid core region that is predicted to form an amphipathic α-helix in AP2 (9) is indicated by a bracket. Residues within the RAYD element of each AP2 domain that are predicted to form an amphipathic α-helix are underlined. (A) AP2-like AP2 domains. Amino acid sequence alignment between the AP2 domain repeats R1 and R2 contained within AP2 (9), ANT (14, 15), and RAP2.7 is shown. Brackets above the sequences designate the conserved YRG and RAYD blocks described above. The filled circle and asterisk indicate the positions of the ap2-1, and ap2-5 mutations, respectively (9). Amino acid residues that constitute a consensus AP2 domain motif for AP2, ANT, and RAP2.7 is shown below the alignment with invariant residues shown capitalized. The GenBank accession number for AP2 is U12546. (B) EREBP-like AP2 domains. Amino acid sequence alignment between the AP2 domains contained within the tobacco EREBPs and the Arabidopsis EREBP-like RAP2 proteins is shown. GenBank accession numbers for EREBP-1, EREBP-2, EREBP-3, and EREBP-4 are D38123, D38126, D38124, and D38125, respectively. (C) Schematic diagrams of the putative RAP2.7-R1, AP2-R1, and ANT-R1 amphipathic α-helices. Amino acid residues within the RAP2.7-R1, AP2-R1, and ANT-R1 motifs shown underlined in A that are predicted to form amphipathic α-helices are schematically displayed with residues rotating clockwise by 100° per residue to form helical structures. Arrows directed toward or away from the center of the helical wheel diagrams indicate the negative or positive degree of hydrophobicity as defined by Jones et al. (33). Positively and negatively charged amino acid residues are designated by + and − signs, respectively. (D) Schematic diagrams of the putative RAP2.2, RAP2.5, RAP2-12, and EREBP-3 amphipathic α-helices. Amino acid residues within the RAP2.2, RAP2.5, RAP2-12, and EREBP-3 motifs shown underlined in B that are predicted to form amphipathic α-helices are schematically displayed as described in C. (E) Sequence alignment between the 25–26 amino acid linker regions in AP2, ANT, and RAP2.7. R1 and R2 designate the positions of the R1 and R2 repeats within AP2, ANT, and RAP2.7 relative to the linker region sequences. Boxes designate invariant residues within the conserved linker regions. Amino acid residues that constitute a consensus linker region motif for AP2, ANT, and RAP2.7 are shown below the alignment with invariant residues shown capitalized. The arrowhead indicates the position of the ant-3 mutation described by Klucher et al. (15).
Nucleotide and Amino Acid Sequence Comparisons.
The tblastn program (20) and default parameter settings were used to search the Arabidopsis EST database (AAtDB 4-7) for genes that encode AP2 domain-containing proteins. Amino acid sequence alignments were generated using the clustal w multiple sequence alignment program (21). Secondary structure predictions were based on the principles and software programs described by Rost (22) and Rost and Sander (23, 24).
RAP2 Gene-Specific Probes.
RAP2 gene-specific fragments were generated by PCR using gene-specific primers and individual RAP2 plasmid DNAs as a template as specified by Perkin-Elmer (Roche Molecular Systems, Branchburg, NJ). The following primers were used to generate fragments representing each RAP2 gene: RAP2.1, 5′-AAGAGGACCATCTCTCAG-3′, 5′-AACACTCGCTAGCTTCTC-3′; RAP2.2, 5′-TGGTTCAGCAGCCAACAC-3′, 5′-CAATGCATAGAGCTTGAGG-3′; RAP2.3, 5′-TCATCGCCACGATCAACC-3′, 5′-AGCAGTCCAATGCGACGG-3′; RAP2.4, 5′-ACGGATTTCACATCGGAG-3′, 5′-CTAAGCTAGAATCGAATCC-3′; RAP2.7, 5′-CGATGGAGACGAAGACTC-3′, 5′-GTCGGAACCGGAGTTACC-3′; RAP2.8, 5′-TCACTCAAAGGCCGAGATC-3′, 5′-TAACAACATCACCGGCTCG-3′; RAP2.9, 5′-GTGAAGGCTTAGGAGGAG-3′, 5′-TGCCTCATATGAGTCAGAG-3′. PCR-synthesized DNA fragments were gel purified and radioactively labeled using random oligonucleotides (Amersham) for use as probes in gene mapping and RNA gel blot experiments.
Gene Mapping Experiments.
RAP2 genes were placed on the Arabidopsis genetic map by either restriction fragment length polymorphism segregation analysis using recombinant inbred lines as described by Reiter et al. (25) or by matrix-based analysis of pooled DNAs from the Arabidopsis yUP or CIC yeast artificial chromosome (YAC) genomic libraries (26, 27) using PCR (28, 29). Matrix pooled yUP and CIC DNAs were generously provided by Chris Somerville, Shauna Somerville, and Robin Buell (Carnegie Institute, Stanford University, Palo Alto, CA). Matrix based mapping results were confirmed by PCR using DNA from individual YAC clones (see Table 1).
Table 1.
Arabidopsis RAP2 genes
| Gene | RAP2 gene containing YAC clones* | Chromosome map position† |
|---|---|---|
| AINTEGUMENTA | ND | 4-73 |
| TINY | ND | 5-32 to 5-45 |
| RAP2.1 | yUP18H2, CIC11D10 | ND‡ |
| RAP2.2 | yUP6C1 | 38§ |
| RAP2.3 | yUP12G6, yUP24B8, yUP23E11, CIC4H5, CIC12C2 | 3-21 |
| RAP2.4 | CIC7D2, CIC10C4 | ND‡ |
| RAP2.7 | yUP10E1 | ND‡ |
| RAP2.8 | CIC10G7 | 1-94 to 1-103§ |
| RAP2.9 | CIC9E12 | 1-117§ |
| RAP2.10 | ND | 4-73¶ |
GenBank accession numbers for complete RAP2 EST sequences are given in parentheses following the designated gene names: AINTEGUMENTA (U40256/U41339); TINY, (X94598), RAP2.1 (AF003094), RAP2.2 (AF003095), RAP2.3 (AF003096), RAP2.4 (AF003097), RAP2.5 (AF003098), RAP2.6 (AF003099), RAP2.7 (AF003100), RAP2.8 (AF003101), RAP2.9 (AF003102), RAP2.10 (AF003103), RAP2.11 (AF003104), and RAP2.12 (AF003105). All RAP2 cDNA clones were originally reported with partial sequences and given GenBank accession numbers as shown in parentheses following each gene name: RAP2.1 (Z27045), RAP2.2 (Z26440), RAP2.3 (T04320 and T13104), RAP2.4 (T13774), RAP2.5 (T45365), RAP2.6 (T45770), RAP2.7 (T20443), RAP2.8 (Z33865), RAP2.9 (Z37270), RAP2.10 (T76017), RAP2.11 (T42962), and RAP2.12 (T42544). Due to the preliminary nature of the EST sequence data, the predicted amino acid sequences for EST Z27045, T04320, T13774, and T42544 contained several errors and were incorrectly reported (13, 15, 16, 31, 32). They are correctly given here. ND, not determined.
YAC clones were determined to contain the specified RAP2 gene by PCR-based DNA synthesis using gene-specific primers (28, 29).
Chromosome map positions are given with reference to the Arabidopsis unified genetic map (AAtDB 4-7).
YAC-based map position is ambiguous.
Preliminary map position is based on a single contact with the physical map.
R.V., N. Terryn, and M.V.M., unpublished results.
mRNA Isolation.
Polysomal poly(A) mRNAs from Arabidopsis flower, rosette leaf, inflorescence stem internode, and hydroponically-grown roots were isolated according to Cox and Goldberg (30).
RNA Gel Blot Studies.
RNA gel blot hybridizations were carried out as specified by the manufacturer (Amersham). mRNA sizes were estimated relative to known RNA standards (BRL). AP2 transcripts were detected using a labeled DNA fragment representing nucleotides 1–1371 of the AP2 cDNA plasmid clone pAP2c1 (9).
RESULTS
The AP2 Domain Defines a Large Family of Plant Proteins.
We carried out an extensive search for new genes that encode AP2 domain containing proteins in Arabidopsis. Using the AP2 domain as a sequence probe we identified 34 cDNA clones that encode putative RAP2 proteins in the Arabidopsis EST database (Materials and Methods). Several of these partial sequences have been reported previously (13–16, 31, 32). Based on nucleotide sequence comparison, we inferred that approximately half of the 34 RAP2 cDNA sequences were likely to represent redundant clones (data not shown). Therefore, we selected and generated a complete DNA sequence for 17 putative RAP2 cDNA clones that appeared to represent unique genes and which contained the largest cDNA inserts. We determined from the predicted amino acid sequences of these clones that the Arabidopsis RAP2 ESTs represent a minimum of 12 genes that we have designated RAP2.1–RAP2.12. As shown in Table 1, preliminary gene mapping experiments using restriction fragment length polymorphism analysis and PCR-based screening of the Arabidopsis yUP and CIC yeast artificial chromosome libraries (Materials and Methods) revealed that at least 7 members of the RAP2 gene family are distributed over 4 different chromosomes (Table 1). In addition, several family members are tightly linked in the genome. For example, RAP2.10 is only 10 kb away from AP2 (N. Terryn, R.V., and M.V.M., unpublished results), which is also closely linked to ANT on chromosome 4 (14, 15).
Sequence analysis also revealed that the proteins encoded by the RAP2 genes are highly divergent except for the presence of at least one AP2 domain. Fig. 1 shows a sequence comparison of 21 AP2 domains from 19 different polypeptides including RAP2.1–RAP2.12, AP2, ANT, TINY, and the tobacco EREBPs. From this comparison, we determined that there are 2 conserved sequence blocks within each AP2 domain. The first block, referred to as the YRG element, consists of 19–22 amino acids, is highly basic and contains the conserved YRG amino acid motif (Fig. 1 A and B). The second block, referred to as the RAYD element, is 42–43 amino acids in length and contains a highly conserved 18-amino acid core region that is predicted to form an amphipathic α-helix in the AP2 domains of AP2, ANT, TINY, and the EREBPs. In addition, there are several invariant amino acid residues within the YRG and RAYD elements that may also play a critical role in the structure or function of these proteins. For example, the glycine residue at position 40 within the RAYD element is invariant in all AP2 domain containing proteins (Fig. 1 A and B) and has been shown to be important for AP2 function (9).
RAP2 cDNA sequence comparison also shows that there are at least two branches to the RAP2 gene family tree. The AP2-like and EREBP-like branches are distinguished by the number of AP2 domains contained within each polypeptide and by sequences within the conserved YRG element. The AP2-like branch of the RAP2 gene family is comprised of three genes AP2, ANT, and RAP2.7, each of which encodes a protein containing two AP2 domains (Fig. 1A). In addition, these proteins possess a conserved WEAR/WESH amino acid sequence motif located in the YRG element of both AP2 domain repeats (Fig. 1A). By contrast, genes belonging to the EREBP-like branch of the RAP2 gene family encode proteins with only one AP2 domain and include RAP2.1–RAP2.6, RAP2.8–RAP2.12, and TINY (Fig. 1B). Proteins in this class possess a conserved 7-amino acid sequence motif referred to as the WAAEIRD box (Fig. 1B) in place of the WEAR/WESH motif located in the YRG element (Fig. 1A). Based on these comparisons, we have generated separate AP2 domain consensus sequences for both classes of RAP2 proteins (Fig. 1 A and B). Although the functions of these conserved sequences are not yet known, these results suggest that the AP2 domain and specific sequence elements within the AP2 domain are important for RAP2 protein functions.
The AP2-like class of RAP2 proteins is also characterized by the presence of a highly conserved 25–26 amino acid linker region that lies between the two AP2 domain repeats (15). This region is 40% identical and 48% similar between AP2, ANT and RAP2.7 (Fig. 1E) and is not found in proteins belonging to the EREBP-like branch of RAP2 proteins (data not shown). Molecular analysis of the ant-3 mutant allele showed that the invariant C-terminal glycine residue within this linker region is essential for ANT function in vivo (15), suggesting that the linker region may also play an important role in AP2 and RAP2.7 function.
Sequences Within the RAYD Element Are Predicted to Form Amphipathic α-Helices.
Previously we reported that the 18-amino acid core region within the RAYD element of the AP2 domain in AP2 is predicted to form an amphipathic α-helix that may be important for AP2 structure or function (9). We used secondary structure prediction analysis (Materials and Methods) to determine whether this structure has been conserved in RAP2 proteins. As shown in Fig. 1, the core region represents the most highly conserved sequence block in the RAYD element of AP2 and the RAP2 proteins. Secondary structure analysis predicts that all RAP2 proteins contain sequences within the RAYD element that are predicted to form amphipathic α-helices (Fig. 1 A and B). Fig. 1C shows that sequences in RAP2.7-R1 are predicted to form an amphipathic α-helix that is 100% identical to that predicted for AP2-R1 and 63% similar to that predicted for ANT-R1. Sequences within the AP2 domain of EREBP-like RAP2 proteins are predicted to form similar α-helical structures. Fig. 1D shows that the RAP2.2, RAP2.5, and RAP2.12 α-helices are 81, 100, and 81% similar to that predicted for EREBP-3, respectively. Together, these results strongly suggest that the predicted amphipathic α-helix in the RAYD element is a conserved structural motif that is important for AP2 domain function in all RAP2 proteins.
RAP2 Genes Are Expressed in Floral and Vegetative Tissues.
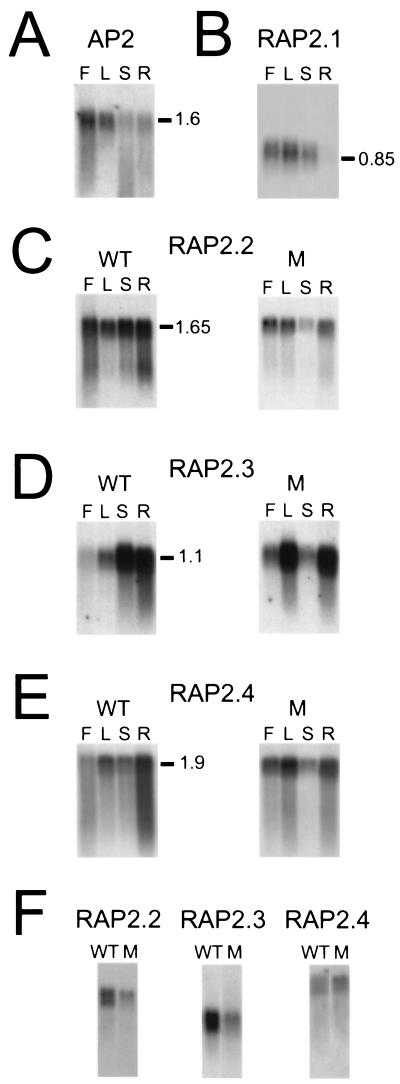
Previous studies have shown that AP2 and ANT are differentially expressed at the RNA level during plant development (9, 14, 15). As shown in Fig. 2A, AP2 is expressed at different levels in developing flowers, leaves, inflorescence stems, and roots. To determine where in plant development the EREBP-like class of RAP2 genes are expressed we reacted RAP2.1, RAP2.2, RAP2.3, and RAP2.4 gene-specific probes with a mRNA gel blot containing flower, leaf, inflorescence stem, and root polysomal poly(A) mRNA. Fig. 2 B–E shows that each RAP2 gene produces a uniquely sized mRNA transcript and displays a distinct pattern of gene expression in flowers, leaves, inflorescence stems, and roots. For example, the RAP2.1 gene is expressed at low levels in wild-type flower, leaf, and stem, and at very low levels in wild-type root (Fig. 2B). RAP2.2 gene expression appears to be constitutive in that RAP2.2 transcripts are detected at similar levels in wild-type flower, leaf, stem, and root (Fig. 2C). By contrast, the RAP2.3 gene is expressed at a low level in wild-type flowers, at a slightly higher level in leaves, and is relatively highly expressed in both stems and roots (Fig. 2D). Finally, the RAP2.4 gene is also expressed in wild-type flower, leaf, stem, and root and is most highly expressed in roots and leaves (Fig. 2E). We conclude from these data that individual members of the EREBP-like family of RAP2 genes are expressed at the mRNA level in both floral and vegetative tissues and show quantitatively different patterns of gene regulation.
Figure 2.
RAP2 gene expression in Arabidopsis. Wild-type (WT) and ap2-10 mutant (M) flower (F), leaf (L), stem (S), and root (R) polysomal poly(A) mRNAs were fractionated on denaturing agarose gels, transferred to nylon membranes, and reacted with labeled AP2, RAP2.1, RAP2.2, RAP2.3, and RAP2.4 gene-specific DNA probes as outlined in Materials and Methods. Flower mRNAs were isolated from inflorescences containing stages 1–10 floral buds (34). Leaf, stem, and root mRNAs were isolated from young expanding rosette leaves, inflorescence stem internode tissue, and hydroponically grown root tissue, respectively. mRNA sizes are shown to the right. Lanes F, L, S, and R contain 4 μg of mRNA. (A) AP2 gene expression in wild-type Arabidopsis. (B) RAP2.1 gene expression in wild-type L-er plants. (C) RAP2.2 gene expression in wild-type L-er and ap2-10 mutant plants. (D) RAP2.3 gene expression in wild-type L-er and ap2-10 mutant plants. (E) RAP2.4 gene expression in wild-type L-er and ap2-10 mutant plants. (F) RAP2 gene expression in wild-type C24 and ap2-10 mutant stems. Labeled RAP2.2, RAP2.3, and RAP2.4 gene-specific DNA fragments were hybridized to gel blots containing inflorescence stem mRNA from wild-type and mutant plants. Lanes WT and M contain 4 μg of mRNA.
RAP2 Gene Expression Patterns Are Affected by ap2.
We analyzed RAP2 gene expression in ap2-10 mutant plants by RNA gel blot analysis to determine whether AP2 is required for RAP2 gene expression. ap2-10 is a severe allele in the C24 background that produces no detectable AP2 protein (ref. 9; R. Khush and J.K.O., unpublished results). As shown in Fig. 2 C–E, the expression of three RAP2 genes are differentially affected by the loss of AP2 function. For example, Fig. 2C shows that RAP2.2 gene expression is not dramatically altered in mutant flowers, leaves, and roots compared to wild-type Landsberg erecta but is down-regulated in mutant stem. RAP2.3 gene expression appears unchanged in mutant roots but is up-regulated in mutant flowers and leaves and down-regulated in mutant stems (Fig. 2D). By contrast, RAP2.4 gene expression appears relatively unchanged in mutant stems and roots but is slightly up-regulated in mutant flowers and leaves (Fig. 2E). To control for possible secondary effects of ecotype on RAP2 gene expression we compared RAP2 gene expression levels in wild-type C24 and ap2-10 mutant stems. Fig. 2F shows that the differences in RAP2.2, RAP2.3, and RAP2.4 gene expression in C24 and ap2-10 stem are similar to those observed between wild-type Landsberg erecta and ap2-10 mutant stem (Fig. 2 B–E). Together these results suggest that AP2 directly or indirectly regulates the expression of at least three RAP2 genes. More importantly, these results suggest that AP2 is controlling gene expression during both reproductive and vegetative development.
DISCUSSION
RAP2 Genes Encode a New Family of Putative DNA Binding Proteins.
We have identified and characterized cDNA clones representing 12 new genes in Arabidopsis that encode AP2 domain containing proteins. One important conclusion from the characterization of these clones is that the AP2 domain has been evolutionarily conserved in both Arabidopsis and in tobacco (Fig. 1). Our analysis revealed that there are two subfamilies of AP2 domain containing proteins in Arabidopsis that we have designated as the AP2-like and the EREBP-like class of RAP2 proteins. In vitro studies have shown that both the EREBP and the AP2 proteins bind to DNA in a sequence specific manner (ref. 13; R. Khush and J.K.O., unpublished results) and that the AP2 domain is sufficient to confer EREBP DNA binding activity (13). From these results and the high degree of sequence similarity between the AP2 domain motifs in AP2, the EREBPs, and the RAP2 proteins, we propose that in general the RAP2 proteins may function as plant sequence specific DNA binding proteins. Although the exact amino acid residues within the AP2 domain required for DNA binding have not yet been identified, sequence comparisons have revealed two highly conserved motifs referred to as the YRG and RAYD elements (Fig. 1) within the AP2 domain.
The RAYD element is found in all known AP2 domains and contains a conserved core region that is predicted to form an amphipathic α-helix (Fig. 1). One hypothesis for the function of this α-helical structure is that it is involved in DNA binding, perhaps through the interaction of its hydrophobic face with the major groove of DNA (35). Alternatively, this structure may mediate protein–protein interactions important for RAP2 functions. These interactions may involve the ability to form homo- or heterodimers similar to that observed for the MADS (MCM1, AG and ARG80, DEF A, and SRF) box family of plant regulatory proteins (36, 37) and for the mammalian Fos-Jun family of transcription factors (38, 39).
Much less is known about the structure or function of the conserved YRG element. We postulate that this region may also function in DNA binding due to the highly basic nature of this region in all RAP2 proteins (Fig. 1). However, the YRG element also contains sequences that are specific for each class of RAP2 protein and may be functionally important for DNA binding. Specifically, the WAAIERD motif is highly conserved in tobacco EREBPs and in EREBP-like RAP2 proteins. By contrast, the WEAR/WESH motif replaces the WAAIERD box in AP2-like RAP2 proteins (Fig. 1). In vitro studies suggest that the EREBPs and AP2 recognize distinct DNA sequence elements (ref. 13; R. Khush and J.K.O., unpublished results). It is possible that the WAAIERD and WEAR/WESH motifs may be responsible for DNA binding sequence specificity. The presence of two AP2 domains in AP2 may also contribute to differences in sequence specificity. Although the molecular significance of having one or two AP2 domain motifs is not yet known, genetic and molecular studies have shown that mutations in either AP2 domain affect AP2 function, implying that both are required for wild-type AP2 activity (9).
In addition to Arabidopsis and tobacco, cDNAs that encode diverse AP2 domain-containing proteins have been found in maize, rice, castor bean, and several members of the Brassicaceae including canola (refs. 13–16 and 32; J.K.O., unpublished results). This strongly suggests that the AP2 domain is an important and evolutionarily conserved element necessary for the structure or function of these proteins.
RAP2 Gene Expression in Floral and Vegetative Tissues.
The AP2, RAP2.1, RAP2.2, RAP2.3, and RAP2.4 genes show overlapping patterns of gene expression at the mRNA level in flowers, leaves, inflorescence stems, and roots (Fig. 2). However, each gene appears to be differentially regulated in terms of its mRNA prevalence. The overlap in RAP2 gene activity could affect the genetic analysis of AP2 and RAP2 gene functions if these genes are also functionally redundant. For example, in flower development AP2 and ANT show partially overlapping patterns of gene expression at the organ and tissue levels (refs. 9, 14, and 15; W. Szeto, B. den Boer, and K.D.J., unpublished results). From single and double mutant analysis it has also been suggested that AP2 may be partially redundant in function with ANT (14). The phenomenon of genetic redundancy and its ability to mask the effects of gene mutation is more clearly demonstrated by the MADS domain containing floral regulatory genes APETALA1 (AP1) and CAULIFLOWER (CAL). Genetic studies have demonstrated that mutations in cal show no visible floral phenotype except when in double mutant combination with ap1 (4), indicating that AP1 is completely redundant in function for CAL. The hypothesis that the RAP2 genes may have genetically redundant functions is supported by the fact that the dominant gain-of-function mutation tiny is the only Arabidopsis RAP2 EREBP-like gene mutant isolated to date (16).
AP2 Activity Is Detectable in Vegetative Development.
Our analysis of RAP2 gene expression in wild-type and ap2-10 plants suggests that AP2 contributes to the regulation of RAP2 gene activity throughout Arabidopsis development. RAP2 gene expression is both positively and negatively affected by the absence of AP2 activity during development (Fig. 2). We believe that the observed differences in RAP2.2, RAP2.3, and RAP2.4 gene expression levels in wild-type and ap2-10 flowers and vegetative tissues are not due to differences in ecotype because similar changes in gene expression levels were observed for all three RAP2 genes in stems when ecotype was controlled (Fig. 2). The regulation of RAP2 gene expression by AP2 in stems clearly indicates that unlike other floral homeotic genes AP2 functions in both reproductive and vegetative development.
Acknowledgments
We thank Drs. Monique Raynal and Yves Parmentier for providing cDNA clones PAP204 and PAP582, and FAFD38 and FAFK31, respectively. We thank the Arabidopsis Biological Resource Center for providing RAP2 cDNA clones and Arabidopsis YAC clones. We express our appreciation to Pamela Wesley who helped initiate this project; Angus Murphy, Clara Bulla, Joseph Dutra, and Michelle Edgar for isolating the recombinant inbred DNAs; and Mr. Bob Kemp (San Lorenzo Valley High School) for isolating and characterizing the RAP2 plasmid DNAs. Our thanks to Dr. Pablo Scolnick for his assistance in restriction fragment length polymorphism mapping; Drs. Chris Somerville, Robin Buell, and Shauna Somerville for providing the pooled YAC DNA stocks; and Drs. Bob Fischer and John Harada for critical reading of this manuscript. This study was supported by National Institutes of Health Grant GM46309 to J.K.O. and National Institutes of Health–Minority Biomedical Research Program Grant RR08132. Support for studies done in Belgium was provided by the Vlaams Actieprogramma Biotechnologie, the Services of the Prime Minister, and a European Community Bridge Grant BIOT-CT92-0529 to M.V.M.
ABBREVIATIONS
- EST
expressed sequence tag
- YAC
yeast artificial chromosome
- MADS
MCM1, AG and ARG80, DEF A, and SRF
References
- 1.Weigel D. Annu Rev Genet. 1995;29:19–39. doi: 10.1146/annurev.ge.29.120195.000315. [DOI] [PubMed] [Google Scholar]
- 2.Yanofsky M F. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:167–188. [Google Scholar]
- 3.Irish V F, Sussex I M. Plant Cell. 1990;2:741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bowman J L, Alvarez J, Weigel D, Meyerowitz E M, Smyth D R. Development (Cambridge, UK) 1993;119:721–743. [Google Scholar]
- 5.Komaki M K, Okada K, Nishino E, Shimura Y. Development (Cambridge, UK) 1988;104:195–203. [Google Scholar]
- 6.Bowman J L, Smyth D R, Meyerowitz E M. Plant Cell. 1989;1:37–52. doi: 10.1105/tpc.1.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowman J L, Smyth D R, Meyerowitz E M. Development (Cambridge, UK) 1991;112:1–20. doi: 10.1242/dev.112.1.1. [DOI] [PubMed] [Google Scholar]
- 8.Kunst L, Klenz J E, Martinez-Zapater J, Haughn G W. Plant Cell. 1989;1:1195–1208. doi: 10.1105/tpc.1.12.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jofuku K D, den Boer B G W, Van Montagu M, Okamuro J K. Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Drews G N, Bowman J L, Meyerowitz E M. Cell. 1991;65:991–1002. doi: 10.1016/0092-8674(91)90551-9. [DOI] [PubMed] [Google Scholar]
- 11.Léon-Kloosterziel K M, Keijzer C J, Koornneef M. Plant Cell. 1994;6:385–392. doi: 10.1105/tpc.6.3.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Modrusan Z, Reiser L, Feldmann K A, Fischer R L, Haughn G W. Plant Cell. 1994;6:333–349. doi: 10.1105/tpc.6.3.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohme-Takagi M, Shinshi H. Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elliot R C, Betzner A S, Huttner E, Oakes M P, Tucker W Q J, Gerentes D, Perez P, Smyth D R. Plant Cell. 1996;8:155–168.18. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klucher K M, Chow H, Reiser L, Fischer R L. Plant Cell. 1996;8:137–153. doi: 10.1105/tpc.8.2.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson K, Long D, Swinburne J, Coupland G. Plant Cell. 1996;8:659–671. doi: 10.1105/tpc.8.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cooke R, Raynal M, Laudie M, Grellet F, Delseny M, et al. Plant J. 1996;9:101–124. [Google Scholar]
- 18.Höfte H, Desprez T, Amselem J, Chiapello H, Rouzé P, et al. Plant J. 1993;4:1051–1061. doi: 10.1046/j.1365-313x.1993.04061051.x. [DOI] [PubMed] [Google Scholar]
- 19.Newman T, de Bruijn F J, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, Retzel E, Somerville C. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rost B. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 23.Rost B, Sander C. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 24.Rost B, Sander C. Proteins. 1994;19:55–77. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 25.Reiter R S, Williams J G K, Feldmann K A, Rafalski J A, Tingey S V, Scolnik P A. Proc Natl Acad Sci USA. 1992;89:1477–1481. doi: 10.1073/pnas.89.4.1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ecker J R. Methods. 1990;1:186–194. [Google Scholar]
- 27.Creusot F, Fouilloux E, Dron M, Lafleuriel J, Picard G, Billault A, LePaslier D, Cohen D, Chaboute M-E, Durr A, Fleck J, Gigot C, Camilleri C, Bellini C, Caboche M, Bouchez D. Plant J. 1995;8:763–770. doi: 10.1046/j.1365-313x.1995.08050763.x. [DOI] [PubMed] [Google Scholar]
- 28.Green E D, Olson M V. Proc Natl Acad Sci USA. 1990;87:1213–1217. doi: 10.1073/pnas.87.3.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwiatkowski T J, Zoghbi H Y, Ledbetter S A, Ellison K A, Chinault A C. Nucleic Acids Res. 1990;18:7191–7192. doi: 10.1093/nar/18.23.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox K H, Goldberg R B. In: Plant Molecular Biology: A Practical Approach. Shaw C H, editor. Oxford: IRL; 1988. pp. 1–35. [Google Scholar]
- 31.Ecker J R. Science. 1995;268:667–675. doi: 10.1126/science.7732375. [DOI] [PubMed] [Google Scholar]
- 32.Weigel D. Plant Cell. 1995;7:388–389. doi: 10.1105/tpc.7.4.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones M K, Anantharamaiah G M, Segrest J P. J Lipid Res. 1992;33:287–296. [PubMed] [Google Scholar]
- 34.Smyth D R, Bowman J L, Meyerowitz E M. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zubay G, Doty P J. J Mol Biol. 1959;7:1–20. [Google Scholar]
- 36.Huang H, Tudor M, Su T, Zhang Y, Hu Y, Ma H. Plant Cell. 1996;8:81–94. doi: 10.1105/tpc.8.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Riechmann J L, Krizek B A, Meyerowitz E M. Proc Natl Acad Sci USA. 1996;93:4793–4798. doi: 10.1073/pnas.93.10.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hai T, Curran T. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Shea E K, Rutkowski R, Kim P S. Cell. 1992;68:699–708. doi: 10.1016/0092-8674(92)90145-3. [DOI] [PubMed] [Google Scholar]