SUMMARY
SUMOylation is a dynamic process, catalyzed by SUMO-specific E1, E2, and E3s and reversed by Sentrin/SUMO-specific proteases (SENPs). However, the physiologic consequence of SUMOylation and de-SUMOylation is not fully understood. Embryos with inactivated mouse SENP1 locus showed severe fetal anemia stemming from deficient erythropoietin (Epo) production and died in mid-gestation. SENP1 controls Epo production by regulating the stability of hypoxia-inducible factor 1α (HIF1α during hypoxia. Hypoxia induces SUMOylation of HIF1α which promotes its binding to an ubiquitin ligase, von Hippel-Lindau (VHL) protein, through a proline hydroxylation-independent mechanism, leading to ubiquitination and degradation. In the SENP1 −/− MEF cells, hypoxia-induced transcription of HIF1α-dependent genes, such as vascular endothelial growth factor (VEGF) and glucose transporter 1 (Glut-1) are markedly reduced. These results show that SENP1 plays a key role in the regulation of the hypoxic response through regulation of HIF1α stability and SUMOylation can serve as a direct signal for ubiquitin-dependent degradation.
INTRODUCTION
Conjugation of small ubiquitin-related modifier protein (SUMO) to a large number of substrates has been suggested to regulate numerous cellular processes from yeast to mammal (Hay, 2005; Yeh et al., 2000). Most of SUMO targets are in the nucleus; they include transcription factors, transcriptional co-regulators, and chromosome remodeling regulators (Gill, 2004). SUMOylation of these proteins can alter their cellular localization and biological activity.
SUMO conjugation is a dynamic process, in that it can be readily reversed by a family of Sentrin/SUMO-specific proteases SENPs (Hay, 2005; Yeh et al., 2000). Six SENPs have been identified in human, each with different cellular location and substrate specificity (Yeh et al., 2000). They can be divided into three subfamilies on the basis of their sequence homology, cellular location, and substrate specificity. The first subfamily consists of SENP1 and SENP2, which have broad substrate specificity. The second subfamily consists of SENP3 and SENP5, both of which are nucleolar proteins with preferences for SUMO-2/3 (Di Bacco et al., 2006; Gong and Yeh, 2006). The third subfamily consists of SENP6 and SENP7, which have an extra loop in their catalytic domain. Although SENPs are known to reverse SUMOylation in many different systems, their physiological role has not been precisely defined.
SENP1, a nuclear SUMO protease, has been shown to regulate androgen receptor transactivation by targeting histone deacetylase 1 and to induce c-Jun activity through de-SUMOylation of p300 (Cheng et al., 2005; Cheng et al., 2004). Yamaguchi et al., studied mice derived from an ES cell line with a retroviral vector that had been randomly inserted into the enhancer region on the SENP1 gene. This random insertion reduced expression of the SENP1 transcript, causing the mice to die between E12.5 and E14.5 (Yamaguchi et al., 2005). Although no specific histological abnormalities were found in the E13.5 embryos, there was a hint of an abnormality in the development of blood vessels in the placenta. Thus, it remains unclear how SENP1 contributes to normal development.
Therefore, we generated SENP1 knockout mice to delineate the contribution of SENP1 in development. Inactivation of the SENP1 gene causes severe fetal anemia in mid-gestation as a result of deficient Epo production. Epo is essential for growth and survival of erythroid progenitors during differentiation into red cells (Wu et al., 1995). We found that SENP1 controls Epo production by regulating the stability of hypoxia-inducible factor 1α (HIF1α). Hypoxia induces HIF1α SUMOylation, which promote HIF1α degradation through a VHL and proteasome-dependent mechanism. SENP1 de-conjugates SUMOylated HIF1α, and allows HIF1α to escape degradation during hypoxia. These results reveal an important physiological role of SENP1 in the hypoxic response through regulation of HIF1α stability, and that SUMOylation can also target a protein for ubiquitination and degradation.
RESULTS
Generation of SENP1 knockout mice
To generate SENP1 knockout mice, a gene-trapped vector was inserted into the mouse SENP1 open reading frame at codon 310 (Figure S1A). Specifically, the inserted β geo (β-galactosidase/neomycin-resistance fusion protein) was fused into the N-terminus of SENP1 at codon 310 to generate SENP1 (1–309)-β-geo fusion protein, which lacks the C-terminal catalytic domain of SENP1 (Figure S1A). This disruption was confirmed by the absence of transcripts that encode catalytic domain of SENP1 in SENP1−/− embryos (Figure S1B). The overall SUMOylation pattern in lysates of embryos revealed an increase in high-molecular-weight SUMO-1 and SUMO-2/3 conjugates in SENP1−/− embryos in comparison with wild-type or SENP1+/− embryos (Figure S1C). In addition, processing of the C-terminus of SUMO-1 but not SUMO-2/3 appeared to be decreased in SENP1−/− embryos (Figure S1C). These results indicated that disruption of the SENP1 locus reduced de-SUMOylation in SENP1−/− embryos.
Severe anemia in SENP1−/− embryos
Heterozygous mice carrying the inactivated SENP1 gene appeared normal and fertile. However, no live SENP1−/− mice were found among offsprings of SENP1+/− intercrosses, indicating that the SENP1−/− mutation leads to embryonic lethality. Examination of the SENP1−/− embryos at different stages of development revealed that the majority died between day 13 and 15 of gestation (Figure 1A). The SENP1−/−embryos appeared paler and smaller than their wild-type or SENP1+/− littermates (Figure 1B). The most dramatic morphological abnormality of these SENP1−/− embryos was severe fetal anemia. In particular, SENP1−/− embryos had more than 75% fewer erythrocytes than their wild-type or SENP1+/− littermates at E15.5 (Figure 1C).
Figure 1. Severe anemia in the SENP1−/− embryos.
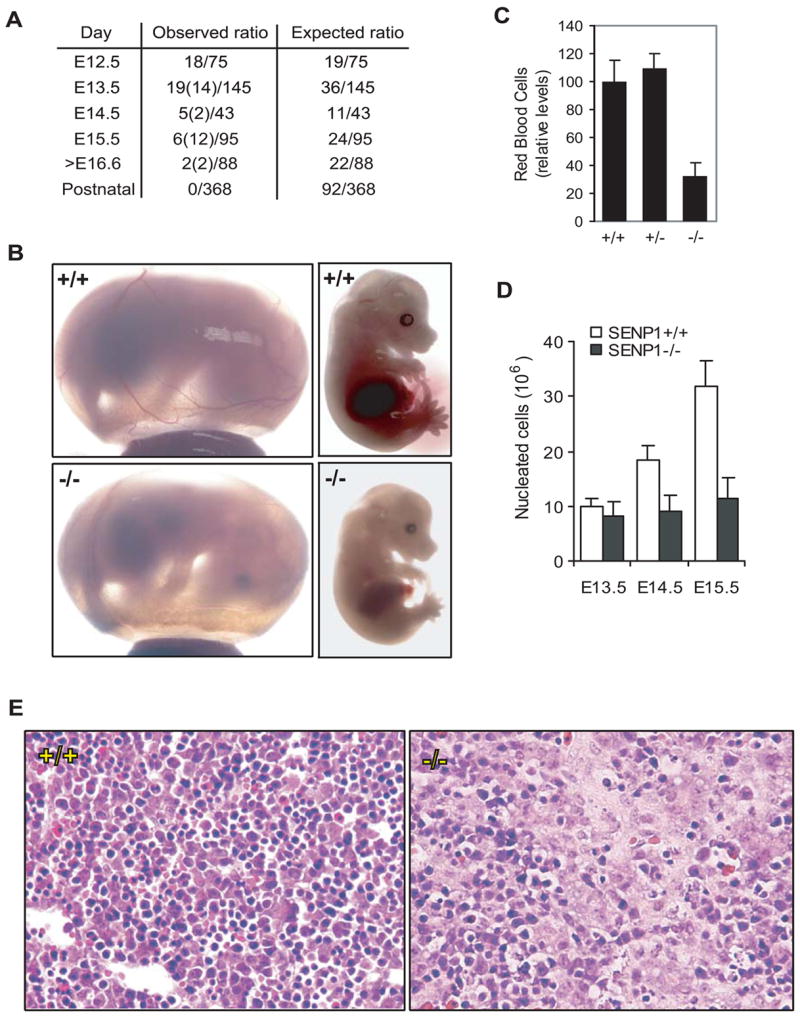
(A) The ratios of the observed live and dead (in parentheses) SENP1−/− embryos to the total number of embryos analyzed at different stages of embryonic development.
(B) Appearance of SENP1+/+ and SENP1−/− embryos at E15.5. The SENP1−/− embryo and yolk sac were paler and smaller than those of the wild-type embryo and appeared to contain fewer red blood cells in major blood vessels.
(C) Relative numbers of red blood cells in peripheral blood in E15.5 wild-type (+/+, n = 5), heterozygote (+/−, n = 9), and SENP1 (−/−, n = 4) embryos. Results shown are means ± SD. A significant decrease in the number of red blood cells was found in SENP1 (−/−) embryos (p < 0.007), when compared with those in wild-type or heterozygote embryos.
(D) Total number of nucleated cells per fetal liver in E13.5 (+/+: n = 4; −/−: n = 3), E14.5 (+/+: n = 4; −/−: n = 4), and E15.5 embryos (+/+: n = 5; −/−: n = 3). Results shown are means ± SD. Significant differences between wildtype and mutant embryos were found at E14.5 (p < 0.015) and E15.5 (p < 0.004).
(E) Hematoxylin and Eosin-stained sections of fetal liver from E12.5 SENP1+/+ and −/− embryos. The SENP1−/− fetal liver showed a marked decrease in the number of erythropoietic foci and increase in apoptotic cells.
Defective definitive erythropoiesis in the SENP1−/− embryos
During normal murine development, erythropoiesis is initiated in the yolk sac at around E8 (primitive erythropoiesis), followed by the aorta-gonadmesonephros (AGM) region. At midgestation, fetal liver becomes the major hematopoietic organ with hematopoietic activity starting around E11.0 to E12.5 until 1 week postnatal (definitive erythropoiesis) (Godin and Cumano, 2002). Because anemia was observed after E13.5 in the SENP1−/− embryos, we reasoned that these embryos likely had a defect in definitive erythropoiesis. Indeed, the number of nucleated cells in the fetal livers of the SENP1−/−embryos was markedly decreased compared to their wild-type littermates (Figure 1D). Histological examination of fetal liver sections also revealed a marked decrease in the number of erythropoietic foci in SENP1−/− embryos compared with their wild-type littermates (Figure 1E).
Increased apoptosis in erythroid precursors of SENP1−/− embryos
We next examined hematopoiesis in the fetal livers of both wild-type and SENP1−/− embryos at E13.5 by flow cytometry using markers characteristic of different types of hematopoietic cells and developmental stages, including CD34, CD44, c-kit, and Ter-119 (Kondo et al., 2003; Neubauer et al., 1998). There was no significant difference between SENP1−/− embryo and wildtype littermate, except that the population of Ter-119+, which denotes committed erythropoietic precursors, was markedly reduced in SENP1−/− fetal livers (Figure S2A). These data suggested that erythropoiesis was abnormal in SENP1−/−embryos as a result of a defect in erythroid differentiation.
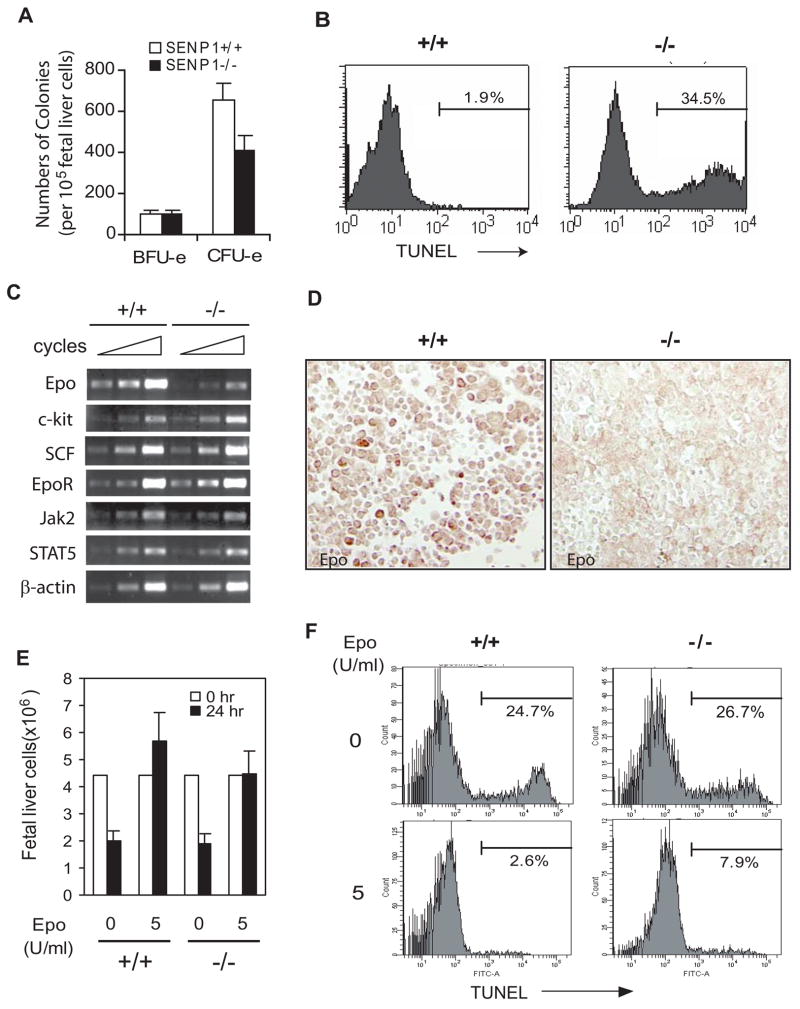
To determine at which stage of erythroid differentiation SENP1 plays a crucial role, we compared the ability of cells derived from wild-type and SENP1−/− E13.5 fetal livers to form erythroid colony-forming units (CFU-e) and more immature erythroid burst-forming units (BFU-e) (Wu et al., 1995). Although fetal livers from SENP1−/−embryo contained erythroid progenitors, the relative number of CFU-e arising from SENP1−/− fetal liver was over 40% fewer than that from wild-type fetal liver (Figure 2A). However, there was no significant difference in the numbers of the BFU-e between SENP1−/− and wild-type fetal livers (Figure 2A). Additionally, we observed no significant difference in the number of colony forming unit granulocyte-macrophage (CFU-GM) and pluripotent hematopoietic stem cells (CFU-GEMM) between SENP1−/− and wild-type fetal livers (Figure S2B). These results indicated that a developmental defect in CFU-e was a prominent feature in the SENP1−/− embryo.
Figure 2. SENP1−/− erythroid progenitors undergo apoptosis due to Epo deficiency.
(A) Analysis of CFU-e- and BFU-e-forming ability of fetal liver cells from E13.5 SENP1+/+ and SENP1−/− embryos. Results shown are means ± SD determined from three embryos. The number of CFU-e from liver cells of SENP1−/− embryos was significantly less than that from SENP1+/+ embryos (p<0.05). (B) Liver cells isolated from E13.5 SENP1+/+ and SENP1−/− embryos were stained with Ter-119 antibody and analyzed by the TUNEL procedure. Histograms show TUNEL staining profile of the Ter-119-positive population identified by flow cytometry. The percentages in the histograms are the percentages of TUNEL-positive cells.
(C) RT–PCR analysis of a variety of genes that are involved in erythroid differentiation in fetal livers of SENP1+/+ and SENP1−/− embryos at E11.5. Samples were amplified for 36, 39, and 42 cycles for Epo; 25, 28, and 32 cycles for EpoR, c-kit, SCF, Jak2, and STAT5. β-Actin mRNA levels (amplified for 18, 21, and 24 cycles) were measured as control.
(D) Fetal liver sections from E12.5 SENP1+/+ and SENP1−/− embryos were stained with anti-Epo antibody (brown).
(E and F) Fetal liver cells isolated from E13.5 SENP1+/+ and SENP1−/− embryos were cultured for 24 hours in the absence or presence of Epo (5 U/ml). Cell numbers were counted under microscope (E). Results shown in (E) are means ± SD determined from three embryos. TUNEL-positive cells were quantitated by flow cytometry. Percentages in histograms are the percentage of TUNEL-positive cells (F).
The reduced ratio of CFU-e to BFU-e progenitors and the decrease in the number of Ter-119+ erythroid cell suggested a reduction in the net growth of SENP1−/− erythroid progenitors resulting from either decreased cell proliferation or increased apoptosis. No differences were observed in the proliferation of fetal liver cells between SENP1−/−embryos and their wild-type littermates, as shown by Ki-67 staining (upper panel, Figure S2C). However, TUNEL staining showed that the number of TUNEL-positive cells was much greater in the liver sections from SENP1−/− embryos than their wild-type littermates (lower panel, Figure S2C). To validate further, we stained fetal liver cells with the anti-Ter-119 antibody and then analyzed cells by flow cytometry. This showed that the percentage of Ter-119-positive cells was decreased in SENP−/− embryos, from 81% in E13.5 wild-type fetal livers to 37% in their SENP1−/− littermates (data not shown). Among the Ter-119-positive cells, about 34.5% SENP1−/− cells were stained by TUNEL; in contrast, few TUNEL-positive cells (1.9%) were observed from the wild-type liver (Figure 2B). These results suggested that the erythroid cells in fetal liver were more apoptotic in SENP1−/− embryos than their wild-type counterparts, which account for the decrease in CFU-e progenitors, smaller fetal liver size, and anemia in the SENP1−/− embryos.
Epo production is reduced in SENP1−/− fetal liver
The hematopoietic defect in SENP1−/− embryos described above was similar to that observed in Epo−/− embryos or in embryos with mutation of the genes critical to the Epo signaling pathway (Neubauer et al., 1998; Parganas et al., 1998; Socolovsky et al., 1999; Wu et al., 1995). Since fetal liver erythroid progenitors depend on Epo for growth and survival during terminal differentiation into red blood cells (Wu et al., 1995), we reasoned that the increased apoptosis of erythroid cells in SENP1−/− fetal liver could be due to either a defect in Epo production or blockage in the Epo signaling pathway. To assess these possibilities, we first examined the expression of genes involved in the Epo signaling pathway (specifically, EpoR, Jak2, and STAT5) and other growth factors associated with erythroid progenitors in fetal livers. A dramatic reduction in the level of Epo mRNA expression was shown in SENP1−/− fetal livers (Figure 2C). In contrast, the expression of EpoR, Jak2, STAT5, and growth factors c-kit and SCF was not significantly different between SENP1−/− and wild-type fetal livers (Figure 2C). The reduced Epo expression in E12.5 fetal liver cells of SENP1−/− embryos was further confirmed by immunohistochemistry (Figure 2D). These results demonstrated that Epo production was significantly reduced in SENP1−/− fetal livers.
Epo prevents apoptosis of SENP1−/− fetal liver cells
To determine whether apoptosis of cells in the fetal liver of SENP1 −/− embryo was caused by a deficiency in Epo production, we examined the ability of Epo to prevent apoptosis of the erythroid progenitors from SENP1−/− fetal liver. As shown by rates of apoptosis in vitro, E13.5 wild-type fetal liver cells, which were grown in a medium lacking Epo, showed a reduction in cell number and increase in apoptosis similar to those observed in fetal liver cells of their SENP1−/− littermates. Addition of Epo to the culture medium prevented reduction in cell number and apoptosis in SENP1−/− fetal liver cells similar to their wild-type littermates (Figure 2E and 2F). These results demonstrated that the increased apoptosis of erythroid progenitors was due to deficiency in Epo production in SENP1−/− fetal liver.
SENP1 regulates Epo transcription through HIF1α
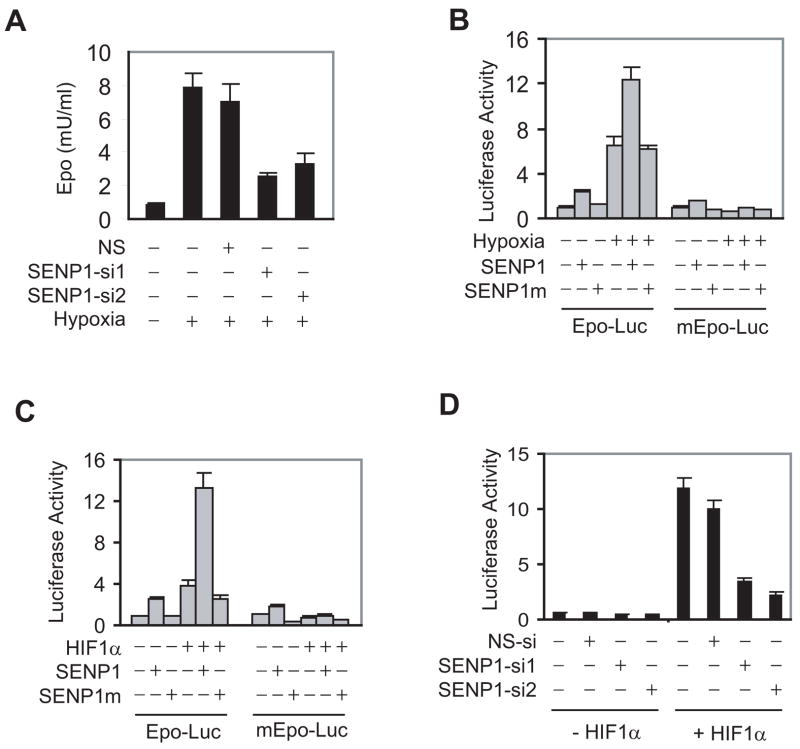
The Epo gene is expressed primarily in fetal liver and adult kidney and is regulated in response to oxygen availability (Ebert and Bunn, 1999). Since SENP1 is expressed in mouse fetal liver at midgestation stage (Figure S3), we reasoned that SENP1 might directly regulate Epo expression in fetal liver. A hepatoma cell line Hep 3B was used to determine whether SENP1 regulated Epo production. Two SENP1-specific siRNAs were able to efficiently knock down SENP1 expression (Figure S4) and significantly reduced Epo expression in Hep 3B cells in response to treatment with hypoxia (Figure 3A), indicating that SENP1 played an important role in Epo production under hypoxia condition.
Figure 3. SENP1 regulates Epo production through HIF1α.
(A) ELISA analysis of Epo production in Hep 3B cells. Data are presented as means ± SD of the results of three independent experiments for Figure 3A, B, C, and D. (NS = Non-specific siRNA).
(B) SENP1 enhanced Epo transcription induced by hypoxia. The indicated reporter gene, HIF1α, and SENP1 expression plasmids were co-transfected into Hep 3B cells. The cells were treated with hypoxia (1% O2) for 12 hr before luciferase assay. The Epo reporter gene with mutation of HIF1α binding sites on the Epo enhancer is indicated by mEpo-Luc. SENP1m is the catalytically inactive SENP1.
(C) SENP1 enhanced HIF1α-dependent Epo transcription. The indicated reporter gene, HIF1α, and SENP1 expression plasmids were co-transfected into Hep 3B cells.
(D) SENP1 was essential for HIF1α-dependent Epo transcription. Epo-Luc and the indicated siRNA expression plasmids were co-transfected into Hep 3B cells without or with HIF1α plasmids.
Since SENP1 regulates the transcription of numerous genes by targeting transcription factors or co-regulators (Cheng et al., 2005; Cheng et al., 2004), we next examined whether Epo transcription was also regulated by SENP1. In this study, the luciferase reporter gene driven by the Epo promoter plus enhancer (Epo-Luc) was transfected into Hep 3B cells with SENP1 or SENP1 catalytic inactive mutant (SENP1m), in which a conserved amino acid Cysteine603 in catalytic domain of SENP1 was substituted by Alanine. In the presence of hypoxia, SENP1, but not the SENP1 mutant, significantly induced Epo reporter gene transcription (Figure 3B), suggesting that SENP1 regulated Epo transcription and the regulation required the de-SUMOylation activity of SENP1. Interestingly, without hypoxia treatment, SENP1 had only marginal effect on Epo transcription.
Since HIF1α is a major transcription factor for hypoxia-regulated Epo expression (Giaccia et al., 2004), we asked whether HIF1α was required for SENP1-regulated Epo transcription. Co-expression of SENP1 markedly enhanced HIF1α-dependent Epo transcription (Figure 3C). However, mutation of HIF1α binding sites on the Epo enhancer (mEpo-Luc) completely abolished SENP1 activity (Figure 3C). Furthermore, silencing of endogenous SENP1 expression in Hep 3B cells using SENP1 siRNA (SENP1-si1 and SENP1-si2) also reduced Epo transcription in response to HIF1α by 80%(Figure 3D). These results strongly suggested that Epo expression was regulated by SENP1 through its regulation of HIF1α activity.
SENP1 is essential for ensuring HIF1α stability during hypoxia
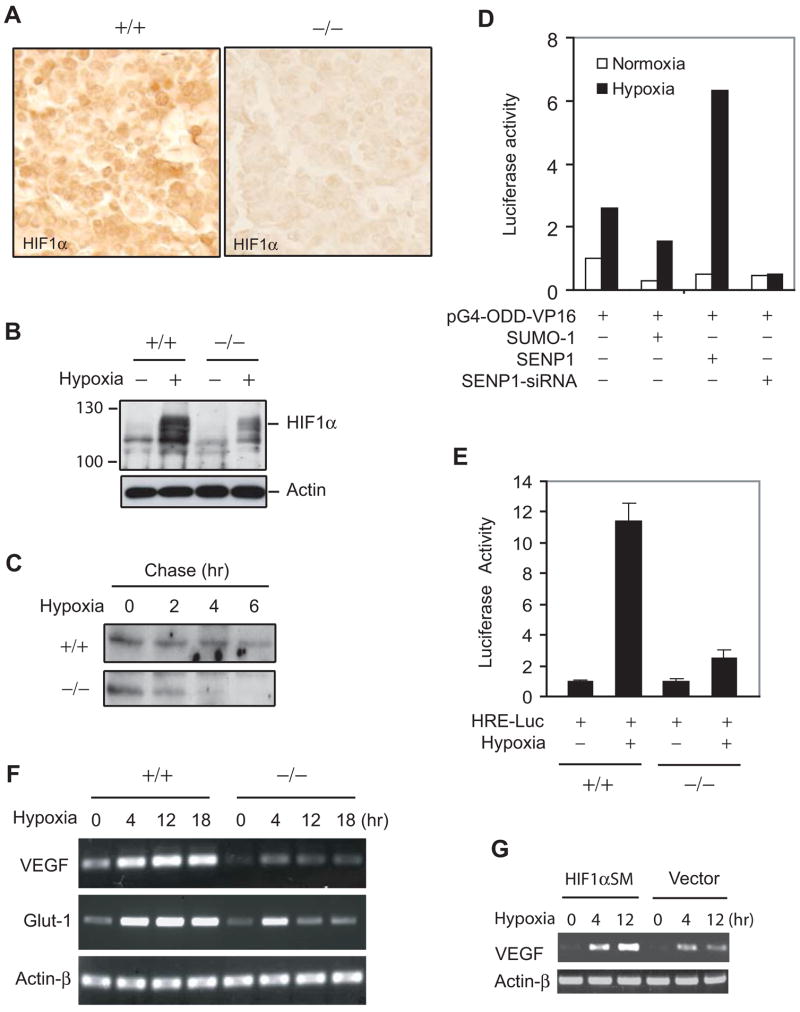
It is well-known that regulation of HIF1α occurs mostly at the protein level. In SENP1−/− mice, HIF1α protein level was significantly decreased in section of SENP1−/−fetal liver compared with SENP1+/+ littermate (Figure 4A). However, HIF1α mRNA level in fetal livers of SENP1−/− or wild-type embryo was similar, suggesting that the decrease in HIF1α protein level resulted from protein degradation but not a reduction of HIF1α transcription in SENP1−/− embryo (Figure S5).
Figure 4. Defect in hypoxia-induced stabilization of HIF1α in SENP1−/− embryos.
(A) HIF1α expression was decreased in SENP1−/− fetal liver. Fetal liver sections from E12.5 SENP1+/+ and SENP1−/− embryos were stained with anti-HIF1α antibody (brown).
(B) Hypoxia-induced HIF1α protein expression was decreased in SENP1−/− MEF cells. SENP1+/+ and −/− MEF cells were treated with hypoxia (1% O2) for 3 hrs before harvest. The whole cell lysate was analyzed by western blotting with anti-HIF1α and Actin antibodies.
(C) The half-life of HIF1α protein under hypoxia was decrease in SENP1−/− MEF cells.
(D) SENP1 was essential for hypoxia-induced HIF1α ODD (344–698) activity. 293 cells were transfected with pGal4-VP16 or pGal4-ODD (344–698)-VP16 plus other plasmids as indicated. Cells were incubated for 12 hrs in normoxia or hypoxia (1% O2) before harvest. The data are presented as the corrected (by internal control) pGal4-ODD-VP16 luciferase activity that normalized to the counts of pGal4-VP16.
(E) Hypoxia-induced HIF1α activity was significantly reduced in SENP1−/− MEF cells. Data are presented as means ± SD of three independent experiments
(F) Hypoxia-induced expression of VEGF and Glut-1 was reduced in SENP1−/− MEF cells. Expression of VEGF, Glut-1, and Actin-β was determined in SENP1+/+ or −/− MEF cells by RT-PCR.
(G) HIF1α restored hypoxia-induced expression of VEGF in SENP1−/− MEF cells. SENP1−/− MEF cells were transfected with HIF1α SM (SUMOylation mutant), or vector control plasmids and treated with hypoxia (1% O2) for different time as indicated. Expression of VEGF and Actin-β was determined by RT-PCR.
HIF1α is degraded by a proteasome-dependent mechanism under normoxia (Huang et al., 1998; Maxwell et al., 1999; Wang et al., 1995). Hypoxia was believed to stabilize HIF1α protein and therefore increased its activity (Giaccia et al., 2004; Huang et al., 1998; Maxwell et al., 1999). We hypothesized that regulation of HIF1α protein to hypoxia was defective in SENP1−/− cells. To test this possibility, we determined changes in HIF1α protein level in SENP1−/− and wild type MEF cells when exposed to hypoxia. As shown in Figure 4B, hypoxia markedly increased protein level of HIF1α in the wild type MEF cells, but much less in the SENP1 −/− MEF cells. Furthermore, the half-life of HIF1α protein was markedly reduced in the SENP1−/− MEF cells under hypoxia condition in a pulse-chase experiment (Figure 4C). Normoxia-induced HIF1α degradation is dependent on its internal oxygen-depend-degradation (ODD) domain (Huang et al., 1998). We then determined whether the ODD domain was a direct target of SENP1 regulation of HIF1α stability under hypoxia condition. We used HIF1α ODD domain (344–698) fused Gal4 (pG4-ODD-VP16), which was previously demonstrated to have normoxic- and VHL-dependent degradation and activity (Huang et al., 1998; Maxwell et al., 1999). Indeed, co-expression of SENP1 markedly enhanced hypoxia-induced ODD activity. However, hypoxia-induced activity of ODD domain was completely abolished by SENP1-siRNA (Figure 4D). These results strongly indicated that SENP1 was essential for HIF1α stabilization under hypoxia condition.
Consistent with the decrease in HIF1α expression during hypoxia, HIF1α activity was also significantly reduced in SENP1−/− MEF cells as measured by the response of HRE-luciferase reporter gene to hypoxia in SENP1−/− and wild type MEF cells (Figure 4E). As HIF1α is a major transcription factor in the regulation of a large number of hypoxia-inducible genes, we also showed that hypoxia-induced transcription of VEGF and Glut-1 genes were dramatically decreased in SENP1−/− MEF cells (Figure 4F). This defect was similar to the response of HIF1α −/− ES upon hypoxia exposure (Iyer et al., 1998; Ryan et al., 1998). We further confirmed that this defect in SENP1−/− MEF cells was mediated by HIF1α, as transfected HIF1α SM (SUMOylation mutant) could restore hypoxia-induced expression of VEGF gene in the transfected cells (Figure 4G). These results indicated that SENP1 played a critical role in regulating the stability and activity of HIF1α protein during hypoxia.
SUMOylated HIF1α is degraded in a proteasome-dependent manner
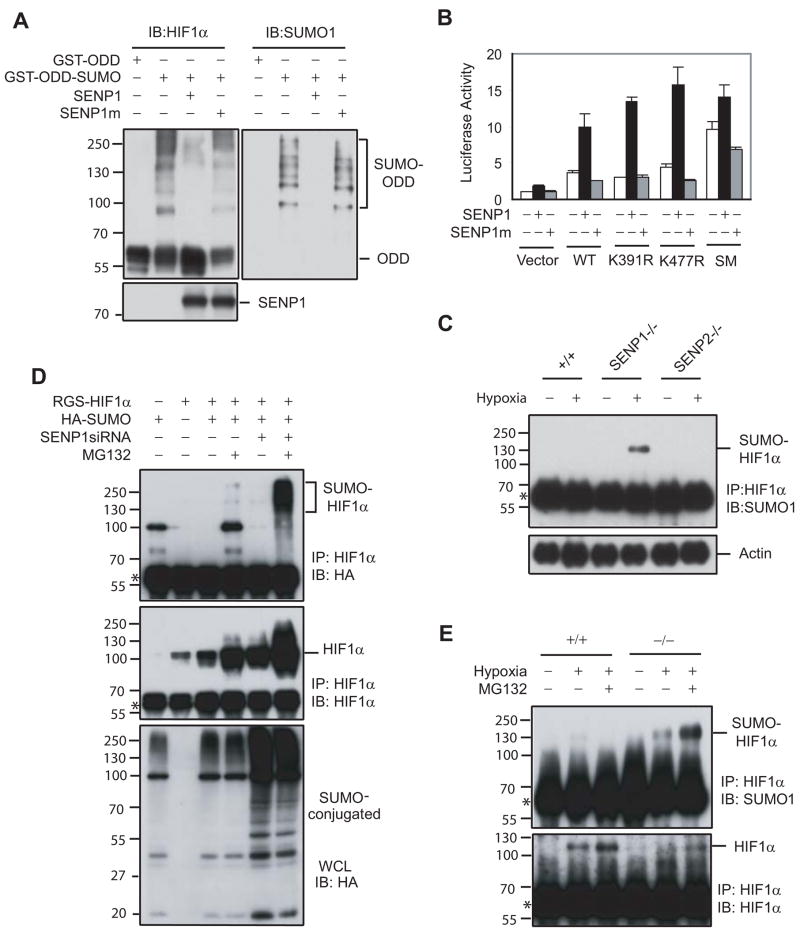
Since SENP1 could de-SUMOylate HIF1α SENP1 might be directly responsible for the decrease in HIF1α activity and stability in the SENP1 −/− embryo (Figure 5A and S6). The effect of de-SUMOylation on HIF1α activity was directly confirmed by using HIF1α SUMOylated site mutants: K391R, K477R, and K391/477R (SM) (Bae et al., 2004). As shown in Figure 5B, mutation of the two SUMOylation sites of HIF1α (SM) significantly increased the transcriptional activity of HIF1α and also reduced the ability of SENP1 to enhance HIF1α-dependent Epo transcription. Furthermore, hypoxia-induced accumulation of SUMOylated HIF1α only occurred in SENP1−/− MEF cells, but not in wildtype cells (Figure 5C). Interestingly, this accumulation did not appear in SENP2−/−MEF cells (Figure S7), suggesting that SENP1 specifically targeted HIF1α for de-SUMOylation (Figure 5C).
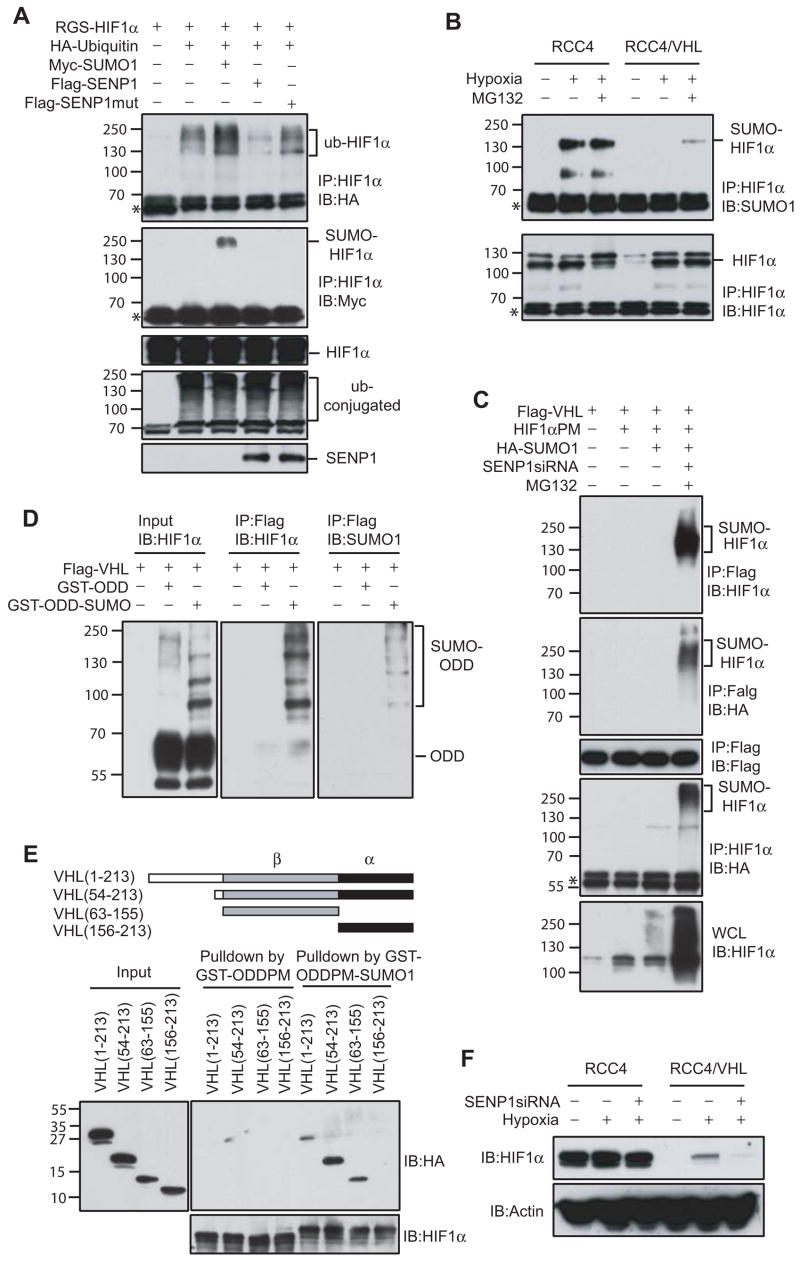
Figure 5. SUMOylated HIF1α accumulated in SENP1−/− MEF cells and underwent proteasomal-dependent degradation in the hypoxic condition.
(A) SENP1 de-conjugated SUMOylated HIF1α in vitro. SUMOylated GST-ODD (344–698) PM recombinant protein was produced by in vitro SUMOylation. Flag-SENP1 or SENP1m was generated by in vitro translation. SUMOylated GST-ODDPM and SENP1 (or SENP1m) were incubated for 1 hr at 37 °C. The reaction mixtures were detected by western blot with anti-HIF1α (left panel), anti-SUMO-1 (right panel), and anti-Flag antibodies (bottom panel).
(B) Mutation of SUMOylation sites increased HIF1α-dependent Epo transcription and reduced HIF1α response to SENP1. Epo-Luc and the indicated plasmids were co-transfected into Hep-3B cells. Data are presented as means ± SD of three independent experiments.
(C) SUMOylated HIF1α accumulated in SENP1−/− MEF cells, but not in wildtype or SENP2−/−MEF cells after exposure to hypoxia. Wildtype (+/+), SENP1−/−, or SENP2−/− MEF cells were treated with or without hypoxia (1% O2) for 4 hrs as indicated. HIF1α was immunoprecipitated with anti-HIF1α antibody from these cell lysates. The precipitates were immunoblotted (IB) with anti-SUMO-1 antibody (top panel). Asterisk indicates IgG band.
(D) SUMOylated HIF1α level was controlled by SENP1 and proteasome-dependent degradation. COS-7 cells were transfected with indicated plasmids and then treated without or with MG132 (10 μM) for 4 hours before harvesting and immunoprecipitated with anti-HIF1α (IP). Bound proteins were detected by anti-HA and anti-HIF1α immunoblotting (IB). Whole-cell lysates (WCL) were immunoblotted (IB) with anti-HA. Asterisk indicates IgG band.
(E) SUMOylated HIF1α accumulated in SENP1−/− MEF cells after exposure to hypoxia and undergoes proteasome-dependent degradation. SENP1+/+ or −/− MEF cells were treated by hypoxia (1% O2) and MG132 (10 μM) for 4 hrs as indicated. HIF1α was immunoprecipitated with anti-HIF1α antibody from cell lysates. The precipitates were immunoblotted (IB) with anti-SUMO-1 and HIF1α antibodies. Asterisk indicates IgG band.
The simultaneous accumulation of SUMOylated HIF1α and the decrease in HIF1α stability in SENP1−/− cells during hypoxia prompted us to study whether SUMOylation might lead to HIF1α degradation. Indeed, SUMOylated bands of HIF1α resulted from co-expression of HIF1α and HA-SUMO-1 were difficult to detect in the absence of a proteasome inhibitor MG132, but was readily detectable when both SENP1 siRNA and MG132 were used (Figure 5D). These results were further confirmed by examining endogenous HIF1α in SENP1 −/− and wild type MEF cells with exposure to both hypoxia and MG132. As expected, hypoxia-induced SUMOylated HIF1α was readily detected in SENP1−/− MEF, but not in wild-type MEF cells. Furthermore, MG132 treatment markedly increased hypoxia-induced SUMOylated bands of HIF1α in SENP1−/− MEF cells, suggesting that the level of SUMOylated HIF1α was controlled by a proteasome-dependent mechanism (Figure 5E, upper panel). Meanwhile, HIF1α protein level was significantly increased in SENP1 wild type MEF cells, but not in the SENP1 −/−MEF cells with exposure to hypoxia (Figure 5E, lower panel). Taken together, these results suggested that SUMOylated HIF1α was degraded in a proteasome dependent manner during hypoxia in the absence of SENP1.
Degradation of SUMOylated HIF1α is dependent on VHL
The above results suggested that SUMO and SENP1 might regulate ubiquitination of HIF1α. As shown in Figure 6A, ubiquitination of HIF1α was significantly increased when SUMO-1 was over-expressed (lane 3 vs 2 in Figure 6A). In contrast, co-transfection of SENP1, but not SENP1 mutant, significantly decreased ubiquitination of HIF1α (lane 4 and 5 vs. 2 in Figure 6A). We asked whether VHL, a well-known E3 ligase for HIF1α ubiquitination (Ivan et al., 2001; Jaakkola et al., 2001; Maxwell et al., 1999; Ohh et al., 2000), could also serve as an E3 ligase for SUMOylated HIF1α. To address this possibility, we first examined hypoxia-induced SUMOylation of HIF1α in RCC4 (renal cancer cells with inactivated VHL) and RCC4/VHL (VHL-restored RCC4 cells) (Maxwell et al., 1999). SUMOylated HIF1α was easily detected in VHL-null RCC4 cells even without MG132 treatment (lane 2 vs. 3 in top panel, Figure 6B). In contrast, this band was only weakly detectable in RCC4/VHL cells exposed to hypoxia and MG132 treatment. Consistent with other reports, hypoxia-induced increase in HIF1α protein was only observed in VHL-restored RCC4 cells (bottom panel, Figure 6B). These results suggested that SUMOylated HIF1α was degraded through a VHL-dependent mechanism.
Figure 6. VHL is required for degradation of SUMOylated HIF1α.
(A) HIF1α ubiquitination was regulated by SUMO-1 and SENP1. COS-7 cells were co-transfected with indicated plasmids and treated with MG132 (10 μM) for 9 hours before harvesting. HIF1α was immunoprecipitated with anti-HIF1α (IP) and bound proteins were detected by anti-HA (top panel), anti-Myc (second panel), and anti-HIF1α immunoblotting (third panel) (IB). Whole-cell lysates (WCL) were immunoblotted (IB) with anti-HA (fourth panel) or anti-Flag (bottom panel) antibodies. Asterisk indicates IgG band.
(B) RCC4 or RCC4/VHL cells were treated by hypoxia (1% O2) and/or MG132 (10 μM) for 4 hrs as indicated. HIF1α was immunoprecipitated with anti-HIF1α antibody from cell lysates. The precipitates were immunoblotted (IB) with anti-SUMO-1 or HIF1α antibodies. Asterisk indicates IgG band.
(C) VHL bound to SUMOylated HIF1α proline mutant (HIF1α PM) in vivo. COS-7 cells were co-transfected with indicated plasmids and treated without or with MG132 (10 μM) for 4 hrs before harvesting. VHL was immunoprecipitated with anti-Flag (IP) from the nuclear fraction of the transfected cells and bound proteins were detected by anti-HIF1α (top panel), anti-HA (second panel), or anti-Flag immunoblotting (third panel) (IB). HIF1α was also immunoprecipitated with anti-HIF1α (IP) from the nuclear fraction of transfected cells and bound proteins were detected by anti-HA antibody (IB) (fourth panel). Whole-cell lysates (WCL) were immunoblotted (IB) with anti-HIF1α (bottom panel). Asterisk indicates IgG band.
(D) VHL specifically bound to SUMOylated HIF1α ODD with proline mutation (GST-ODD (344–698) PM) in vitro. GST-ODD (344–698) PM recombinant protein and SUMOylated GST-ODD (344–698) PM produced by in vitro SUMOylation was incubated with Flag-VHL produced by in vitro translation. After washing and eluting with Flag peptide, the precipitates were immunoblotted with ant-HIF1α (middle panel) or anti-SUMO-1(right panel) antibodies.
(E) Mapping of VHL domain that bound to SUMO-1-fused ODD. GST-ODD (344–698) PM and SUMO-1-fused GST-ODD (344–698) PM recombinant proteins were incubated with HA-VHL and its mutant produced by in vitro translation for two hr. The precipitates with glutathione-agarose beads were detected by immunoblotting with ant-HA (top and right panel) or anti-HIF1α antibodies (bottom panel).
(F) HIF1α degradation induced by SENP1 silencing was VHL-dependent. RCC4 or RCC4/VHL cells and SENP1-siRNA stable transfected RCC4 or RCC4/VHL cells were treated by hypoxia (1% O2) for 4 hrs as indicated. Cell lysates were detected by immunoblotted with anti-HIF1α or anti-actin antibodies.
SUMO provides an alternative signal for HIF1α to bind to VHL
We further speculated that SUMOylation served as an alternative signal for HIF1α to bind to VHL during hypoxic condition. The interaction of SUMOylated HIF1α and VHL in vivo was examined first by using a HIF1α proline residues 402/564 mutant (PM), which would decrease background binding resulted from hydroxylation of HIF1α. As shown in Figure 6C, SUMOylation of HIF1α PM was greatly enhanced by expression of SENP1siRNA plus MG132 treatment (lane 4, bottom panel, Figure 6C). SUMOylated HIF1α from the nuclear fraction was efficiently co-precipitated by VHL (land 4 in top and second panels, Figure 6C). To directly confirm the binding of VHL and SUMOylated HIF1α we performed an in vitro binding assay. As shown in Figure 6D, VHL pull-downed SUMOylated GST-ODD (344–698) PM, detected by anti-HIF1α antibody (Figure 6D, middle panel). These bands were confirmed as SUMOylated HIF1α by anti-SUMO-1 antibody (Figure 6D, right panel). Importantly, un-modified GST-ODD (344–698) PM, even though presented in a much larger quantity than SUMOylated GST-ODD (344–698) PM, could not be co-precipitated by VHL. We further mapped the SUMO binding domain on VHL protein by using recombinant SUMO-fused GST-ODD (344–698) PM to pull down in vitro translated VHL deletion mutants. As shown in Figure 6E, SUMO-fused GST-ODDPM protein could pull down VHL deletion mutants that contain the β-domain. However, SUMO-fused GST-ODDPM protein could not pull down the VHL deletion mutant that only contained the α-domain. This was dependent on SUMO-1 because GST-ODDPM could not pull down any VHL deletion mutants. Interestingly, β-domain is well documented as a substrate binding region when VHL acts as a component of E3 ligase complex (Ohh et al., 2000; Stebbins et al., 1999). Taken together, these results clearly suggested that SUMOylation of HIF1α could serve as an alternate signal for binding to VHL in the absence of proline hydroxylation.
To directly demonstrate the role of SENP1 in VHL-dependent degradation of SUMOylated HIF1α, we generated SENP1 siRNA stably transfected RCC4 and RCC4/VHL cell lines. The expression of endogenous SENP1 was significantly decreased in these two cell lines (Figure S8). Silencing of SENP1 markedly decreased HIF1α expression in RCC4/VHL, but not in RCC4 cells (Figure 6F), confirming that HIF1α turnover promoted by inactivation of SENP1 was VHL-dependent.
DISCUSSION
Biological consequence of SUMOylation and de-SUMOylation
SUMOylation and de-SUMOylation have been shown to regulate a large number of biological processes, including transcription, cell signaling, cell cycle progression, and cancer pathogenesis(Cheng et al., 2006; Yeh et al., 2000). However, many of these studies were shown in cell culture systems and by over-expression of enzymes involved in SUMOylation and de-SUMOylation. There were a limited number of studies that using knock-out mouse demonstrating the importance of this emerging pathway of biological regulation. For example, knock-out of the SUMO conjugation enzyme Ubc9, in mice results in embryonic lethality at the early post-implantation stage (Nacerddine et al., 2005). Ubc9-deficient cells showed severe defects in nuclear organization including chromosome condensation and segregation. This study demonstrates that global disruption of the SUMOylation pathway is incompatible with life in the mammals. Only a single study on the role of de-SUMOylation in mouse has been reported. Yamaguchi et al demonstrated that mouse with an incomplete SENP1 knock-out died between E12.5 and E14.5 (Yamaguchi et al., 2005).
In the present study, we show that SENP1 plays an essential role in the de-conjugation of SUMOylated HIF1α. This de-conjugation has important biological consequence because the SENP1 −/− embryos are severely anemic, which is most likely the cause of fetal death. This is surprising because in the over-expression system, both SENP1 and SENP2 have identical substrate specificity. Here, SUMOylated HIF1α is only regulated by SENP1, but not SENP2 in vivo. Thus, SENPs clearly perform non-redundant biological function in both MEF cells and in animals.
Regulation of HIF1α stability during hypoxia
HIF1α is mainly regulated at the level of protein stability (Huang et al., 1998; Jiang et al., 1996). During normoxia, HIF1α is hydroxylated at two critical proline residues by a family of oxygen-sensitive enzymes prolyl 4-hydroxylases (PHD) (Bruick and McKnight, 2001; Epstein et al., 2001; Yu et al., 2001). Proline hydoxylated HIF1α then binds to VHL, a component of the ubiquitin E3 ligase complex consists of Cul-2, VHL, elongin B, and elongin C (Bruick and McKnight, 2001; Ivan et al., 2001; Jaakkola et al., 2001; Maxwell et al., 1999). Subsequently, HIF1α is ubiquitinated and degraded by the proteasome. It was assumed that during hypoxia, proline hydroxylation occurs inefficiently, thus allowing HIF1α to escape binding to VHL and proteasomal degradation. Most of studies about HIF1α stability primarily focus on the mechanism involving the regulation of PHD enzymatic activities (Epstein et al., 2001; Gerald et al., 2004; Ivan et al., 2001; Jaakkola et al., 2001; Nakayama et al., 2004). However, a few reports have shown that there are other factors that can regulate HIF1α stability in VHL-independent manner. For example, Liu et al. recently showed that RACK1 could replace HSP90’s binding of HIF1α and target the unmodified HIF1α to the elongin C complex for ubiquitination and degradation (Liu et al., 2007). There are also modifications other than hydroxylation that regulate HIF1α stability and activity (Brahimi-Horn et al., 2005).
Our SENP1 −/− animal model reveals a more complex picture for the regulation of HIF1α stability during hypoxia. As shown in Figure 7, hypoxia induces nuclear translocation and SUMOylation of HIF1α, which binds to VHL in a hydroxyl proline-independent manner, leading to ubiquitination and proteasomal degradation. It should be noted that SUMOylation is not required for hypoxia-induced nuclear translocation of HIF1α (Kang and Yeh, unpublished results). However, HIF1α is quickly SUMOylated once it enters the nucleus. Thus, SENP1, which is predominately a nuclear protein, is well-positioned to regulate the activity and stability of HIF1α in the nucleus by removing SUMO (Gong et al., 2000). Un-modified HIF1α would escape VHL/proteasome-dependent degradation to participate in the regulation of hypoxia-responsive genes. When SENP1 is absent, as in the case of our SENP1 −/− embryo and MEF cells, SUMOylated HIF1α will be subjected to degradation through a VHL/proteasome-dependent mechanism. Thus, SENP1 plays a critical role to control HIF1α stability during hypoxia.
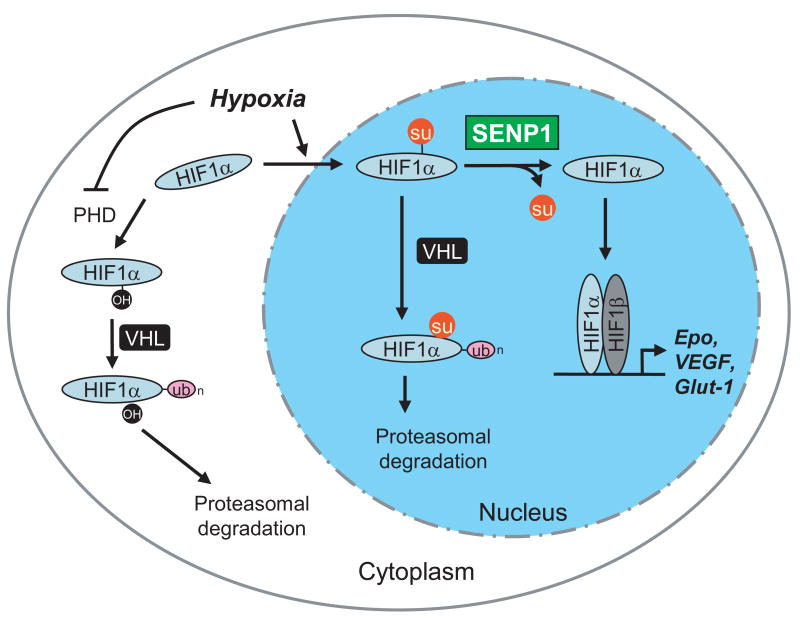
Figure 7. SENP1 regulates of HIF1α stability in hypoxia.
Hypoxia blocks the activity of PHD, preventing hydroxylation of HIF1α and its subsequent degradation in a VHL- and ubiquitin-dependent manner. On the other hand, hypoxia induces nuclear translocation and SUMOylation of HIF1α, which provides an alternative signal for VHL- and ubiquitin-dependent degradation. SENP1 stabilizes HIF1α by removing the alternative VHL-binding signal.
Bae et al. showed that over-expression of SUMO-1 increased HIF1α protein level and its activity not only under hypoxia but also in normoxia condition (Bae et al., 2004). This report, unlike the present study, was conducted entirely with over-expression system without the benefit of the SENP1 knock out mice. In the presence of SENP1, SUMOylated HIF1α will be de-conjugated and indeed stabilized.
We currently do not know how hypoxia induces SUMOylation of HIF1α. This may be due to an increase in specific SUMO E3 ligase activity (Kang and Yeh, unpublished results). Moreover, the level of SUMOylated HIF1α is also regulated by VHL-dependent degradation and SENP1-mediated stabilization. So, the absolute level of SUMOylated HIF1α is regulated at multiple levels. Groulx et al. have proposed a model of VHL shuttling between the nucleus and cytosol and HIF1α degradation mainly in the cytosol (Groulx and Lee, 2002). However, we do not know whether SUMOylated and ubiquitinated HIF1α is also degraded in the cytosol. Further studies are required to resolve this issue.
SUMOylation can also target a protein for proteasomal degradation
Our results provide direct evidence that SUMOylation can induce degradation of a target protein in a proteasome-dependent manner. It is well-accepted that SUMOylation can stabilize target protein as SUMO can conjugate to the same lysine sites on target protein as ubiquitination (Hay, 2005). Although ubiquitin-conjugated sites on the HIF1α protein have not been defined precisely, the reported ubiquitin-conjugated region of HIF1α indicates that at least lysine 391 and 477, two SUMOylation sites on the HIF1α protein, are not the major sites for ubiquitination (Ivan et al., 2001; Masson et al., 2001; Ohh et al., 2000). We also found that level of HIF1α SUMOylation correlated with the level of ubiquitination, suggesting that SUMO conjugation of HIF1α is not on the same sites as ubiquitination. More importantly, we identified an E3 ubiquitin ligase VHL for SUMOylated HIF1α degradation. It was previously shown that PML could be SUMOylated and ubiquitinated when exposed to arsenic trioxide (Lallemand-Breitenbach et al., 2001). However, it is not know whether SUMOylated PML is required for binding to an ubiquitin-ligase to allow for ubiquitination and degradation. Currently, we also do not know whether SUMO-mediated binding of substrates to ubiquitin E3 can be generalized to more SUMOylated substrates.
In conclusion, our results support a model in which SUMOylated HIF1α isunstable but can be stabilized when SUMO is removed by SENP1 (Figure 7). When SENP1 is deleted, SUMOylated HIF1α is degraded, in a VHL- and ubiquitin/proteasome-dependent manner, resulting in decreased Epo production and severe fetal anemia. To our knowledge, this study provides the first evidence for the physiological role of de-SUMOylation, as demonstrated in an animal model; our study also revealed for the first time that SUMOylation could also target a protein for ubiquitination and proteasomal degradation.
EXPERIMENTIAL PROCEDURES
Generation of SENP1−/− mice
The SENP1+/− ES cell line XG001 was obtained from BayGenomics. XG001 cells were generated by using a gene trap protocol with the trapping construct pGT1Lxf containing the intron from the engrailed-2 gene upstream of the gene encoding the β-galactosidase/neomycin-resistance fusion protein (see http://baygenomics.ucsf.edu). The vector was inserted into intron 8 of the SENP1 locus. A male chimeric mouse was generated from the ES cell line. C57 BL6 mice were obtained from The Jackson Laboratory.
Plasmids and antibodies
The Epo enhancer (Epo-Luc) and the HIF1α binding site mutant (mEpo-Luc) were provided by Dr. H.M. Sucov. HRE-Luc was provided by Dr. J. Yi. pGal4-VP16 and pGal4-ODD(344–698)-VP16 were provided by Dr. P.J. Ratcliffe. Flag-VHL was provided by Dr. T. Kamitani. VHL(1–213), VHL(54–213), VHL(63–155), and VHL(156–213) were provided by Dr. M. Ohh. HA-SUMO-1, Flag-SENP1, and Flag-SENP1 catalytic mutant were previously described (1, 2). RGS-HIF1α, RGS-HIF1αK391R, RGS-HIF1αK477R, RGS-HIF1αSM (K391R, K477R), RGS-HIF1α PM (P402A, P564A), pET-ODD (344–698)PM(p402A, P564A), and pET-ODD (344–698)PM-SUMO1 were generated using standard cloning procedures and PCR-based mutagenesis. We used antibodies against Flag (M2, Sigma), HA (HA-7, Sigma), Myc (Santa Cruz), SUMO-1 (Zymed), SUMO-2/3 (gifted from Dr. Mike Matunis), mouse HIF1α (Novus), and human HIF1α (BD).
Histopathologic and immunohistochemistry analysis
Embryos were fixed in 10% buffered formalin (Sigma) and embedded in paraffin. Five-micrometer-thick sections were stained with hematoxylin and eosin. For proliferation studies, the sections were stained with Ki67-specific antibodies (Dako). Apoptotic cells were detected in sections using the TUNEL stain (TACS TdT DAB kit, R&D).
Colony-formation assays
Cells were prepared from the livers of E13.5 embryos in α-MEM (GIBCO–BRL) and counted in the presence of 3% acetic acid, which lysed erythrocytes. Cell suspensions were mixed with MethoCult M3334 to detect BFU-e or MethoCult M3434 to detect CFU-e (StemCell Technologies). Cells were plated in 35-mm dishes and cultured at 37°C in an atmosphere containing 5% CO2. For the CFU-e assay, benzidine-positive CFU-e colonies were scored on day 3. For the BFU-E assay, benzidine-positive BFU-e colonies were scored on day 8.
Flow cytometry
Single-cell suspensions were obtained from E13.5 wild-type and mutant fetal livers. Cell suspensions were first incubated on ice with rat anti-mouse CD16/CD32 (PharMingen) to block non-specific binding to Fc receptors. Subsequently, cells were incubated with rat anti-mouse PE-conjugated anti-c-kit and anti-CD44 and FITC–conjugated anti-CD34 and anti-Ter-119 (all from PharMingen). Appropriate isotype control antibodies were used. Cell surface expression of different markers was analyzed in a Becton Dickinson FACScan using CellQuest software. For the TUNEL assay, cells were stained using a TUNEL kit from Roche and then analyzed by flow cytometry.
RNA interference
Two 21-nucleotide SENP1 siRNAs (si-1: AACTACATCTTCGTGTACCTC; si-2: CTAAACCATCTGAATTGGCTC) were synthesized (Dharmacon). The same sequence of the si-1-inverted orientation was used as a non-specific siRNA control. The SENP1 and non-specific siRNA oligos were inserted into a pSuppressorNeo vector (IMGENEX Corporation) according to manufacturer’s instructions. Hep 3B cells were transfected with the siRNA plasmid using lipofectamine 2000 (Invitrogen). RCC4 and RCC4/VHL (purchased from ECACC) were infected by retrovirus based SENP1si-1 virus particles produced from pSuppressorNeo-SENP1si-1 and selected by G418. Silencing efficiency of the siRNA was confirmed by performing real-time PCR (for Hep 3B) or RT-PCR (for RCC4 and RCC4/VHL) to examine SENP1 expression. TaqMan Master Mix Reagents (Applied Biosystems) were utilized for quantitative real-time PCR (QRT-PCR) reaction. The TaqMan ABI PRISM 7000 Sequence Detector System (PE Applied Biosystems) was used for the analysis. Primers for SENP1 (forward: 5′-TTGGCCA GAGTGCAAATGG-3′; reverse: 5′-TCGGCTGTTTCTTGA TTTTTGTAA-3′) and the housekeeping 18S rRNA (ABI) were utilized.
In vitro SUMOylation assay
In vitro SUMOylation kit was purchased from LAE Biotech International (Rockville, MD). The reaction was carried out at 37°C, for 1 hr with the mixture including E1 (150 ng), E2 (5 μg), SUMO-1 (5 μg), ATP (2 mM), and GST-ODD (344–698)PM (300 ng).
Supplementary Material
Acknowledgments
We thank J. Ni, J. Ren, and L. Zhao for expert technical assistance. Supported in part by NIH R01 CA 80089 (E.T.H.Y), Department of Defense-CDMRP PC040121 (E.T.H.Y.), and CA-16672 (MDACC).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bae SH, Jeong JW, Park JA, Kim SH, Bae MK, Choi SJ, Kim KW. Sumoylation increases HIF-1alpha stability and its transcriptional activity. Biochem Biophys Res Commun. 2004;324:394–400. doi: 10.1016/j.bbrc.2004.09.068. [DOI] [PubMed] [Google Scholar]
- Brahimi-Horn C, Mazure N, Pouyssegur J. Signalling via the hypoxia-inducible factor-1alpha requires multiple posttranslational modifications. Cell Signal. 2005;17:1–9. doi: 10.1016/j.cellsig.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Cheng J, Bawa T, Lee P, Gong L, Yeh ET. Role of desumoylation in the development of prostate cancer. Neoplasia. 2006;8:667–676. doi: 10.1593/neo.06445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Perkins ND, Yeh ET. Differential Regulation of c-Jun-dependent Transcription by SUMO-specific Proteases. J Biol Chem. 2005;280:14492–14498. doi: 10.1074/jbc.M412185200. [DOI] [PubMed] [Google Scholar]
- Cheng J, Wang D, Wang Z, Yeh ET. SENP1 enhances androgen receptor-dependent transcription through desumoylation of histone deacetylase 1. Mol Cell Biol. 2004;24:6021–6028. doi: 10.1128/MCB.24.13.6021-6028.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bacco A, Ouyang J, Lee HY, Catic A, Ploegh H, Gill G. The SUMO-specific protease SENP5 is required for cell division. Mol Cell Biol. 2006;26:4489–4498. doi: 10.1128/MCB.02301-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert BL, Bunn HF. Regulation of the erythropoietin gene. Blood. 1999;94:1864–1877. [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Gerald D, Berra E, Frapart YM, Chan DA, Giaccia AJ, Mansuy D, Pouyssegur J, Yaniv M, Mechta-Grigoriou F. JunD reduces tumor angiogenesis by protecting cells from oxidative stress. Cell. 2004;118:781–794. doi: 10.1016/j.cell.2004.08.025. [DOI] [PubMed] [Google Scholar]
- Giaccia AJ, Simon MC, Johnson R. The biology of hypoxia: the role of oxygen sensing in development, normal function, and disease. Genes Dev. 2004;18:2183–2194. doi: 10.1101/gad.1243304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 2004;18:2046–2059. doi: 10.1101/gad.1214604. [DOI] [PubMed] [Google Scholar]
- Godin I, Cumano A. The hare and the tortoise: an embryonic haematopoietic race. Nat Rev Immunol. 2002;2:593–604. doi: 10.1038/nri857. [DOI] [PubMed] [Google Scholar]
- Gong L, Millas S, Maul GG, Yeh ET. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J Biol Chem. 2000;275:3355–3359. doi: 10.1074/jbc.275.5.3355. [DOI] [PubMed] [Google Scholar]
- Gong L, Yeh ET. Characterization of a family of nucleolar SUMO-specific proteases with preference for SUMO-2 or SUMO-3. J Biol Chem. 2006;281:15869–15877. doi: 10.1074/jbc.M511658200. [DOI] [PubMed] [Google Scholar]
- Groulx I, Lee S. Oxygen-dependent ubiquitination and degradation of hypoxia-inducible factor requires nuclear-cytoplasmic trfficking of von Hippel-Lindau tumor suppressor protein. Mol Cell Biol. 2002;22:519–5336. doi: 10.1128/MCB.22.15.5319-5336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1–12. doi: 10.1016/j.molcel.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci U S A. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaakkola P, Mole DR, Tian YM, Wilson MI, Gielbert J, Gaskell SJ, Kriegsheim A, Hebestreit HF, Mukherji M, Schofield CJ, et al. Targeting of HIF-alpha to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science. 2001;292:468–472. doi: 10.1126/science.1059796. [DOI] [PubMed] [Google Scholar]
- Jiang BH, Semenza GL, Bauer C, Marti HH. Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am J Physiol. 1996;271:C1172–1180. doi: 10.1152/ajpcell.1996.271.4.C1172. [DOI] [PubMed] [Google Scholar]
- Kondo M, Wagers AJ, Manz MG, Prohaska SS, Scherer DC, Beilhack GF, Shizuru JA, Weissman IL. Biology of hematopoietic stem cells and progenitors: implications for clinical application. Annu Rev Immunol. 2003;21:759–806. doi: 10.1146/annurev.immunol.21.120601.141007. [DOI] [PubMed] [Google Scholar]
- Lallemand-Breitenbach V, Zhu J, Puvion F, Koken M, Honore N, Doubeikovsky A, Duprez E, Pandolfi PP, Puvion E, Freemont P, de The H. Role of promyelocytic leukemia (PML) sumolation in nuclear body formation, 11S proteasome recruitment, and As2O3-induced PML or PML/retinoic acid receptor alpha degradation. J Exp Med. 2001;193:1361–1371. doi: 10.1084/jem.193.12.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YV, Baek JH, Zhang H, Diez R, Cole RN, Semenza GL. RACK1 competes with HSP90 for binding to HIF-1alpha and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1alpha. Mol Cell. 2007;25:207–217. doi: 10.1016/j.molcel.2007.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson N, Willam C, Maxwell PH, Pugh CW, Ratcliffe PJ. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. Embo J. 2001;20:5197–5206. doi: 10.1093/emboj/20.18.5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Nacerddine K, Lehembre F, Bhaumik M, Artus J, Cohen-Tannoudji M, Babinet C, Pandolfi PP, Dejean A. The SUMO pathway is essential for nuclear integrity and chromosome segregation in mice. Dev Cell. 2005;9:769–779. doi: 10.1016/j.devcel.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromage H, Tempst P, Frappell PB, et al. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117:941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93:397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, Pavletich N, Chau V, Kaelin WG. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol. 2000;2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, et al. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93:385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. Embo J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Socolovsky M, Fallon AE, Wang S, Brugnara C, Lodish HF. Fetal anemia and apoptosis of red cell progenitors in Stat5a−/− 5b−/− mice: a direct role for Stat5 in Bcl-X(L) induction. Cell. 1999;98:181–191. doi: 10.1016/s0092-8674(00)81013-2. [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Kaelin WG, Jr, Pavletich NP. Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science. 1999;284:455–461. doi: 10.1126/science.284.5413.455. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Liu X, Jaenisch R, Lodish HF. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 1995;83:59–67. doi: 10.1016/0092-8674(95)90234-1. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T, Sharma P, Athanasiou M, Kumar A, Yamada S, Kuehn MR. Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol Cell Biol. 2005;25:5171–5182. doi: 10.1128/MCB.25.12.5171-5182.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh ET, Gong L, Kamitani T. Ubiquitin-like proteins: new wines in new bottles. Gene. 2000;248:1–14. doi: 10.1016/s0378-1119(00)00139-6. [DOI] [PubMed] [Google Scholar]
- Yu F, White SB, Zhao Q, Lee FS. HIF-1alpha binding to VHL is regulated by stimulus-sensitive proline hydroxylation. Proc Natl Acad Sci U S A. 2001;98:9630–9635. doi: 10.1073/pnas.181341498. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.