Abstract
Primary cilia play critical roles in many aspects of biology. Specialized versions of primary cilia are involved in many aspects of sensation. The single photoreceptor sensory cilium (PSC) or outer segment elaborated by each rod and cone photoreceptor cell of the retina is a classic example. Mutations in genes that encode cilia components are common causes of disease, including retinal degenerations. The protein components of mammalian primary and sensory cilia have not been defined previously. Here we report a detailed proteomics analysis of the mouse PSC complex. The PSC complex comprises the outer segment and its cytoskeleton, including the axoneme, basal body, and ciliary rootlet, which extends into the inner segment of photoreceptor cells. The PSC complex proteome contains 1968 proteins represented by three or more unique peptides, including ∼1500 proteins not detected in cilia from lower organisms. This includes 105 hypothetical proteins and 60 proteins encoded by genes that map within the critical intervals for 23 inherited cilia-related disorders, increasing their priority as candidate genes. The PSC complex proteome also contains many cilia proteins not identified previously in photoreceptors, including 13 proteins produced by genes that harbor mutations that cause cilia disease and seven intraflagellar transport proteins. Analyses of PSC complexes from rootletin knock-out mice, which lack ciliary rootlets, confirmed that 1185 of the identified PSC complex proteins are derived from the outer segment. The mass spectrometry data, bench-marked by 15 well characterized outer segment proteins, were used to quantify the copy number of each protein in a mouse rod outer segment. These results reveal mammalian cilia to be several times more complex than the cilia of unicellular organisms and open novel avenues for studies of how cilia are built and maintained and how these processes are disrupted in human disease.
Primary cilia are present on most cells in the human body. These structures are typically sensory organelles and are involved in many critical aspects of cell biology (1, 2). For example, sensation of flow by primary cilia is required for maintenance of renal nephron structure and body axis determination. Recent evidence has also revealed that primary cilia play important roles in various aspects of development, such as planar cell polarity and Hedgehog signaling (3, 4). All cilia are composed of a microtubule-based axoneme surrounded by a distinct domain of the plasma membrane. The axonemes are derived from and anchored to the cell via basal bodies (5).
Cilia are involved in many aspects of sensation, including vision, smell, and hearing (2, 3). The sensory cilium elaborated by each rod and cone photoreceptor cell of the retina is a classic example. These photoreceptor sensory cilia or outer segments are among the largest of mammalian cilia (2, 6). Like other cilia, the outer segments contains an axoneme, which begins at the basal bodies and passes through a transition zone (the so-called “connecting cilium”) and into the outer segment (7) (see Fig. 1). The basal bodies also nucleate the ciliary rootlet, which extends into the inner segment. The rootlet serves to anchor the cilium to the cell and functions as a channel for proteins destined for the outer segment (6, 8). The photoreceptor sensory cilium (PSC)1 complex comprises the outer segment and its cytoskeleton, including the rootlet, basal body, and axoneme (see Fig. 1). The outer segment membrane domain of the PSC complex is highly specialized with discs (lamellar membranes) stacked in tight order at 30 per μm along the axoneme. The proteins required for phototransduction are located in or associated with these discs.
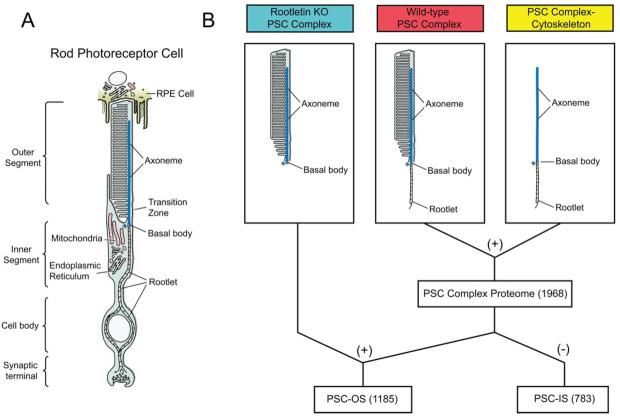
Fig. 1. Illustration of rod photoreceptor cell and fractions isolated for analyses.
A, schematic of a rod photoreceptor cell. Note that the rootlet extends from the basal bodies down into the cell body, and the axoneme extends from the basal bodies up into the outer segment. A transition zone (the so-called connecting cilium) connects the outer and inner segments. B, PSC complex preparations analyzed by LC-MS/MS. The combination (+) of the PSC complex and PSC complex-cytoskeleton data is defined as the PSC complex proteome. The overlap (+) between PSC complex proteome and rootletin KO PSC complexes identifies the proteins located in the PSC-OS. The difference (−) between the PSC complex and the PSC-OS identifies proteins associated with the PSC-IS. Numbers in parentheses are number of proteins detected; see text and Fig. 3. Colors of labels correspond to those used in Figs. 2 and 3.
Mutations in genes that encode cilia components cause many different types of disease. To date, mutations that cause inherited retinal degenerations, which are common causes of blindness, have been identified in genes encoding 38 PSC complex proteins (RetNet: www.sph.uth.tmc.edu/Retnet/). Furthermore mutations in genes encoding proteins expressed both in photoreceptors and other cilia result in systemic diseases, such as Usher syndrome, Bardet-Biedl syndrome (BBS), Senior-Loken syndrome, and Joubert syndrome that involve retinal degeneration along with other disorders consequent to cilia dysfunction such as deafness and polycystic kidney disease (2, 3).
The proteomes of mammalian primary and sensory cilia have not been defined to date. Such datasets have the potential to accelerate investigations of the cell biology of sensory and primary cilia and facilitate understanding of cilia disruption and dysfunction in disease. A proteomics analysis of axonemes isolated from the cilia of cultured human respiratory epithelium was reported several years ago, but it identified a relatively small number of proteins (86 by ≥2 peptides) (9). Proteomics analysis of cilia, flagella, and basal bodies from three unicellular organisms and several in silico analyses designed to identify cilia genes based on conservation among ciliated organisms have been performed (10-15). In aggregate, these analyses have identified ∼1200 non-redundant putative cilia-related proteins (16, 17). It remains unclear, however, how many of these proteins have homologs that are components of mammalian cilia and associated structures.
To identify the proteins that make up an important mammalian cilium, we undertook a detailed proteomics analysis of the mouse PSC complex. Here we report this PSC complex to consist of ∼2000 proteins. To differentiate proteins associated with the inner and outer segment portions of the PSC complex, we also analyzed PSCs from rootletin knock-out (KO) mice, which lack ciliary rootlets (6). We used the MS/MS spectral count data, normalized for detectable peptides and benchmarked by 15 well characterized outer segment proteins, to estimate the copy number of each protein in a mouse photoreceptor outer segment.
EXPERIMENTAL PROCEDURES
Animals
This research was approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Wild-type C57BL/6J mice were obtained from The Jackson Laboratory or bred from commercially obtained founders. Rootletin KO mice were described previously (6).
PSC Complex and PSC Complex-Cytoskeleton Isolation
PSC complexes were isolated from the retinas of 4-week-old wild-type or rootletin KO mice using modifications of established techniques (18, 19). Briefly fresh mouse retinas were gently vortexed for 1 min in 500 μl of buffer A (10 mm Pipes, pH 7.0, 5 mm MgCl2, and 1× proteinase inhibitor mixture) with 50% (w/v) sucrose. PSC complexes were separated from the remainder of the retina by centrifugation for 20 min at 13,000 × g. The PSC complexes were collected from the top of the 50% sucrose buffer, diluted 1:1 in buffer A without sucrose, and subjected to a second round of centrifugation on a 50% sucrose cushion. Purified PSC complexes were collected at the interface and sedimented by decreasing the sucrose concentration. A portion (20%) of the purified wild-type PSC complex was placed in SDS-PAGE sample buffer for the PSC complex proteomics analysis (see below).
The PSC complex-cytoskeletons were then isolated by extracting the remaining 80% of the purified wild-type PSC complexes with detergent (buffer B: 10 mm Pipes, pH 7.0, 5 mm MgCl2, 1% Triton X-100, 1 mm DTT, and 1× proteinase inhibitor mixture) for 1 h on ice to remove the plasma membrane and outer segment discs (19, 20). The PSC complex-cytoskeleton fraction was separated by discontinuous sucrose gradient centrifugation (40, 50, and 60% sucrose) in buffer B for1hat 13,000 × g. The enriched cytoskeleton fraction was collected at the 50/60% sucrose interface and pelleted by decreasing the sucrose concentration in buffer B. The PSC complex and cytoskeleton preparations were viewed by differential interference contrast microscopy on a Zeiss LSM 510 Meta confocal microscope and immunostained as described below.
Proteomics Analyses
For the proteomics analyses, purified PSC complexes and cytoskeletons were extracted with SDS sample buffer, and the insoluble material was removed by centrifugation. The wild-type PSC complex (45 μg), PSC complex-cytoskeleton (40 μg), and rootletin KO PSC complex (45 μg) proteins were then separated by electrophoresis for 6 cm on a 4–12% bis-Tris minigel (NuPage), and the gel was stained with colloidal Coomassie Blue.
Each 60-mm lane was subsequently cut into uniform 1-mm slices with the MEF-1.5 Gel Cutter (The Gel Co., San Francisco, CA). Two adjacent gel slices were combined per digestion tube, and the samples were digested in-gel with trypsin as described previously (21). Tryptic digests (10 μl of a total of 30 μl) were separated by reverse phase HPLC on a nanocapillary column, 75-μm-inner diameter × 20-cm PicoFrit (New Objective, Woburn, MA), packed with MAGIC C18 resin, 5-μm particle size (Michrom BioResources, Auburn, CA), using a NanoLC pump (Eksigent Technologies, Livermore, CA). Solvent A was 0.58% acetic acid in Milli-Q water, and solvent B was 0.58% acetic acid in acetonitrile. Peptides were eluted into an LTQ-FT mass spectrometer (ThermoElectron, San Jose, CA) at 200 nl/min using an acetonitrile gradient. Each reverse phase LC run consisted of a 10-min sample load at 1% B and a 75-min total gradient consisting of 1–28% B over 50 min, 28–50% B over 14 min, 50–80% B over 5 min, and 80% B for 5 min before returning to 1% B in 1 min. To minimize sample carryover to the next LC-MS/MS run, a 28-min blank cycle was run between each sample. Sample injection was ∼7 min, resulting in a total sample to sample cycle time of about 120 min. The mass spectrometers were set to repetitively scan m/z from 400 to 1600 followed by data-dependent MS/MS scans on the six most intense ions with dynamic exclusion enabled.
MS/MS Data Analyses
Peptides from each LC-MS/MS run were identified from the MS/MS spectra using the Bioworks Browser 3.3 program (ThermoElectron). DTA files were generated from MS/MS spectra using an intensity threshold of 3000 and a minimum ion count of 30. The DTA files generated were searched against the National Center for Biotechnology Information (NCBI) non-redundant (July 2006) database with a reversed copy of the database placed in front of the forward copy of the database. To reduce database search time, the databases were indexed with the following parameters: monoisotopic mass range of 750–3500, length of 6–100, half-tryptic specificity with up to two internal missed cleavage sites, static modification of cysteine using iodoacetamide modification (+57.02150 Da), and dynamic modification of Met to methionine sulfoxide (+15.9949 Da). The DTA files were searched with a 2.5-Da peptide mass tolerance and a 1.0-Da fragment ion mass tolerance using the Sequest algorithm within Bioworks 3.3. Other search parameters were identical to those used for database indexing. Peptide identification false positive rates were estimated after applying appropriate data filters as the number of redundant peptide hits to the reverse database divided by the number of redundant peptide hits to the forward database times 100. After estimating false positive rates, all hits to the reverse database were removed prior to further analysis.
DTASelect and in-house custom programs were used to combine and filter the outputs from the Sequest searches for all 30 LC-MS/MS runs from each sample into a single summary (22). Peptide hits from database searches were filtered using the following: mass accuracy, <10 ppm; ΔCn ≥ 0.08; and full tryptic boundaries. When large datasets of LC-MS/MS data from LTQ-FT mass spectrometers are searched using a wide mass window and half-tryptic constraints followed by filtering using full tryptic boundaries and a tight mass tolerance, much lower false positive rates are obtained than when stringent cross-correlation scores or probability scores are used.2
The identified peptides were grouped into the smallest set of non-redundant proteins possible. Different forms (charge states and modifications) of the same peptide were compressed into a single hit. Java and Perl scripts were developed to perform additional processing steps such as retrieving gene names, comparing datasets, and removing peptide redundancy. The corresponding mouse gene was identified for each protein. The protein and gene identifications were reviewed manually to ascertain their accuracy and to ensure that consistent database entries were reported in the different samples. This included reviewing the identifications of related proteins that shared common peptides. When proteins could not be reliably distinguished by unique peptides, they were reported as a protein group and identified in the Note column of the PSC complex proteome spreadsheet (Supplemental Table S1) by an M (for multiple). Supplemental Tables S2-S4 contain the DTASelect summaries with peptide sequences for the PSC complex, PSC complex-cytoskeleton, and rootletin KO PSC complex LC-MS/MS analyses, respectively.
Prior to additional analyses, obvious contaminants such as keratins, histones, and serum proteins were removed from the proteome. As in other cilia, mitochondria and ribosomes, which are not part of photoreceptor outer segments, can segregate with PSC complexes due to their high abundance in inner segments (9, 13, 23). We therefore removed mitochondrial and ribosomal proteins from the PSC proteome. Mitochondrial proteins were identified using a curated list of known mitochondrial proteins derived from the MitoP2 database (24). Keratins, histones, and ribosomal and serum proteins were identified based on protein names and annotations in NCBI protein and gene records.
Immunofluorescence Microscopy
Eyes from wild-type adult mice were processed in two different ways for immunostaining experiments. For preparation of sections from unfixed retina, eyes were enucleated, snap frozen, embedded in OCT, and cryosectioned at 10 μm. Sections were postfixed with 1% paraformaldehyde in PBS for 10 min before performing immunostaining. For preparation of sections from fixed retina, eyes were enucleated after cardiac perfusion with 4% paraformaldehyde in PBS (pH 7.4), fixed in 4% paraformaldehyde for 3 h, embedded in OCT freezing medium, and cryosectioned at 10 μm. Both types of frozen sections were then immunostained as described previously (25). For staining of the isolated PSC complex and cytoskeleton preparations, 10 μl of each suspension was spotted on glass slides and immunostained as described previously (19). Cy3-, Alexa 488-, and Alexa 633-conjugated secondary antibodies were obtained from Jackson ImmunoResearch Laboratories or Molecular Probes. Stained sections were viewed with a Zeiss LSM 510 Meta confocal microscope, and the images were processed with Zeiss Meta 510 software (Carl Zeiss MicroImaging).
Antibodies
The primary antibodies used for these studies are described in Supplemental Table S5. We requested antibodies to 200 proteins whose location in the PSC complex had not been reported previously. These 200 antibodies were identified by literature searches and were directed against proteins that were distributed throughout the range of relative protein abundance in the PSC complex proteome. We received or purchased 51 of these antibodies, including 33 directed against the outer segment portion of the PSC complex (PSC-OS) proteins and 18 against the inner segment portion of the PSC complex (PSC-IS) proteins (Supplemental Table S5).
Quantitative Analysis
To estimate the relative abundance of proteins in the PSC-OS, we used a measure of total spectra observed per detected protein (26-28). Spectral counts derived from the MS/MS data were normalized by dividing the total number of spectra detected by the predicted number of observable peptides per protein. The predicted number of observable peptides per protein was determined by identifying predicted complete tryptic peptides with detectable masses (750–3500 Da). For the 208 proteins in the PSC-OS group where one or more peptides were shared between related proteins, the number of spectra from the shared peptides assigned to each protein was determined based on the ratios of spectra derived from the unique peptides that identified each protein.
The concentrations of 15 known photoreceptor proteins were obtained from the literature (Supplemental Table S7). These are expressed relative to rhodopsin of which 7 × 107 molecules are present on average in the mouse rod sensory cilium (29). Where appropriate, protein levels were corrected for the portion of total protein present in the outer segment (Supplemental Table S7, Notes).
Because the amounts of transducin, arrestin, and recoverin in the outer segment vary with light exposure, the locations of transducin and arrestin under the conditions of illumination used to harvest retinas were determined using immunofluorescence assays (30). Frozen sections of retinas were prepared from mice that were 1) completely dark-adapted, 2) exposed to the same light regimen as mice from which retinas were harvested for PSC complex isolation, or 3) bleached with 15,000 lux of light for 30 min with dilated pupils prior to sacrifice. These sections were embedded adjacent to each other and probed with antibodies to arrestin and α-transducin. The percentage of the total photoreceptor fluorescence signal in the outer segment was calculated using Zeiss LSM 510 software. As shown in Supplemental Fig. S1, only minimal translocation of arrestin into the outer segment occurred in the mice exposed to the dissecting microscope; ∼5% of the photoreceptor arrestin fluorescence signal was located in the outer segment. The distribution of transducin under the lighting conditions used was the same as that observed in dark-adapted animals (data not shown). Based on these data, it was assumed that recoverin also retained its dark-adapted distribution.
Functional Analysis
We evaluated the functional categories and domains of proteins in the PSC complex proteome based on the Database for Annotation, Visualization, and Integrated Discovery (DAVID) 2006 (31). We used the Expression Analysis Systematic Explorer (EASE) program to determine which functional categories are statistically overrepresented in PSC complex proteome with respect to total protein set annotated in mouse (32).
Comparisons with Published Datasets and Identification of Candidate Genes
To allow integration of data from heterogeneous databases such as GenBankTM, UniGene, and Ensembl, protein identities were extracted from published articles and mapped to Entrez gene identities (July 2006) using Java and Perl scripts. To compare proteomes derived from different organisms, mouse or human homologs for all proteins were found using Homologene (July 2006). To identify candidate genes, the human homologs of the mouse genes that encode the proteins in the PSC complex proteome were compared electronically to the Online Mendelian Inheritance in Man (OMIM) and RetNet (www.sph.uth.tmc.edu/Retnet/) databases. Potential candidate genes were evaluated manually to determine whether they fell within published intervals for disease loci.
RESULTS
Identification of the PSC Complex Proteome
To identify the proteome of the mouse PSC, we performed LC-MS/MS analyses on three distinct protein preparations: wild-type PSC complexes, PSC complex-cytoskeletons, and rootletin KO PSC complexes (Fig. 1). Photoreceptor cells are classically described as having an outer segment, where phototransduction takes place, and an inner segment, where the biosynthetic functions of the cell are concentrated (Fig. 1). Although the inner and outer segment compartments are distinguishable by morphology, by function, and no doubt by their protein complements, they cannot be completely separated using standard biochemical techniques because the outer segment is an elaborated cilium, and its ciliary rootlet extends far into the inner segment (2, 33). PSC complexes isolated from wild-type mice thus contain outer segments plus the portions of their cytoskeletons that extend into the inner segments of photoreceptor cells, including the basal body and ciliary rootlet (Fig. 1).
Ciliary rootlets are a polymer of rootletin, and in its absence rootlets do not form (6). PSC complexes isolated from rootletin KO mice therefore lack ciliary rootlets and separate from the inner segments of photoreceptor cells easily and without the major inner segment component of the PSC complex-cytoskeleton (Fig. 1). Despite the lack of rootlets, PSCs in rootletin KO mice have apparently normal structure and function (6). The overlap between wild-type PSC complex proteome and rootletin KO PSC complexes thus identifies the proteins located in the PSC-OS (Fig. 1).
We also prepared PSC complex-cytoskeletons to improve our ability to detect lower abundance proteins in these structures because phototransduction proteins in the PSC membrane discs are very abundant. The combination of the PSC complex and PSC complex-cytoskeleton data is defined as the PSC complex proteome (Fig. 1). The difference between the PSC complex and the PSC-OS identifies proteins associated with the PSC-IS (Fig. 1).
We isolated PSC complexes and cytoskeletons from wild-type mouse retinas and PSC complexes from rootletin KO mouse retinas using established techniques (6, 18, 19). Microscopic and immunofluorescence analyses showed that these three preparations were highly purified (Fig. 2). At lower magnification, it is evident that the wild-type and rootletin KO PSC complex preparations consist almost entirely of PSC complexes with minimal contamination by the other structures noted (Fig. 2, B and J). At higher magnification, it can be seen that the wild-type PSC complexes consist of outer segments with thin extensions at their bases (Fig. 2, C and D). The extensions are the portions of the cytoplasmic cytoskeleton that were attached to the basal bodies as indicated by staining with antibodies to rootletin. Although a small amount of abnormal rootletin protein is produced from the targeted Crocc allele, PSC complexes isolated from rootletin KO mice do not contain ciliary rootlets (Fig. 2, J and K). The PSC complex-cytoskeletons were composed of axonemes, basal bodies, and rootlets and were also highly purified (Fig. 2, F-H).
Fig. 2. Purified PSC complexes and fractions.
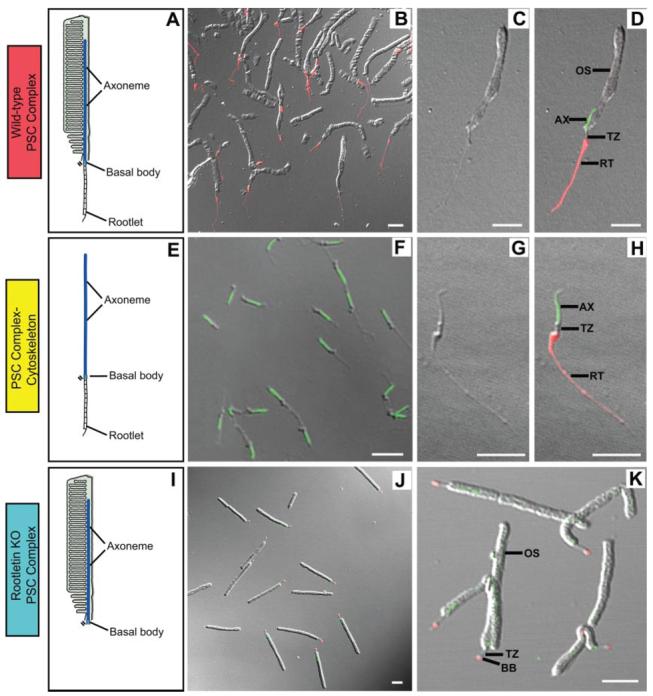
A, schematic of a wild-type PSC complex showing that it is comprised of the outer segment and its cytoskeleton, including the rootlet, basal body, and axoneme. B–D, wild-type PSC complex preparation viewed with differential interference contrast and fluorescence microscopy. At higher magnification (C and D) it can be seen that the PSC complexes consist of outer segments with thin extensions at their bases. The extensions are the portions of the cytoplasmic cytoskeleton that were attached to the basal bodies as indicated by staining with antibodies to rootletin (red; B and D). The axoneme in the outer segment is demonstrated by staining with antibodies to Rp1 (green; D). At lower magnification (B), it is evident that the preparation consists of highly enriched PSC complexes with minimal contamination by the other structures noted. E–H, drawing of PSC complex-cytoskeleton (E) and isolated PSC complex-cytoskeleton preparation viewed with differential interference contrast (G) and fluorescence microscopy (F and H) using antibodies to rootletin (red) and Rp1 (green). The cytoskeletons consist of rootlets, basal bodies, and axonemes and are highly purified as illustrated in the images.I–K, drawing of a rootletin KO PSC complex (I) and rootletin KO PSC complex preparation (J and K) viewed with differential interference contrast and fluorescence microscopy and stained with antibodies to rootletin (red) and Rp1 (green). In these PSC complexes, small dots of rootletin staining are noted at the basal bodies, but no formed rootlets are present. Colors of labels correspond to those used in Figs. 1 and 3. BB, basal body; RT, ciliary rootlet; TZ, transition zone; AX, axoneme. Bars = 5 μm.
Electrophoretic analysis showed that the wild-type PSC complex preparation contained a dense group of bands at ∼35– 40 kDa consisting mainly of rhodopsin (Fig. 3A). Consistent with successful removal of membranes, these bands were greatly diminished in the PSC complex-cytoskeleton preparation. The rhodopsin band remained prominent in the rootletin KO PSC complexes, but many of the less abundant protein bands were missing, consistent with lack of the inner segment portion of the PSC complex (Fig. 3A). The proteins in the wild-type and rootletin KO PSC complexes and the PSC complex-cytoskeleton preparations were identified by LC-MS/MS analyses (Table I and Supplemental Tables S2-S4). Stringent data filters applied during the analyses resulted in about 0.2% false positive peptide identifications as estimated from a concatenated reverse database.
Fig. 3. Proteomics analyses of wild-type and rootletin KO PSC complexes and PSC complex-cytoskeleton proteins.
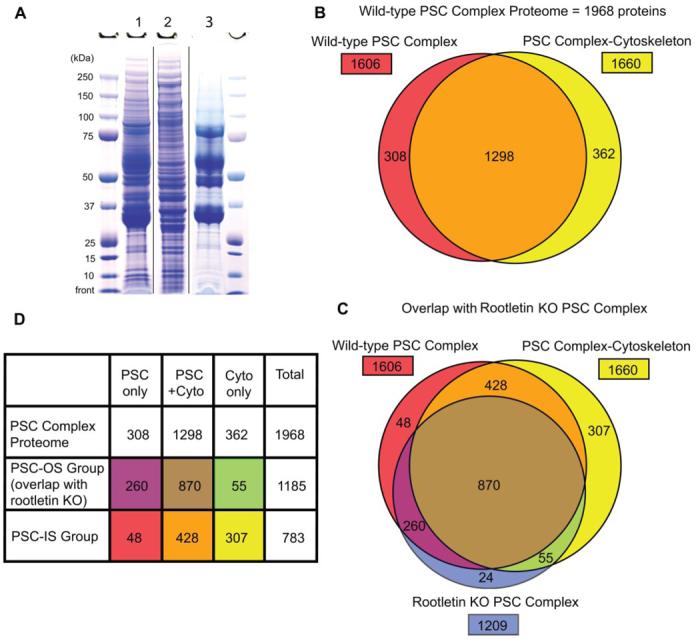
A, wild-type PSC complex (lane 1), PSC complex-cytoskeleton (lane 2), and rootletin KO PSC complex (lane 3) proteins were prepared and electrophoresed as described. The sizes of the molecular mass markers are indicated. B, Venn diagram of wild-type PSC complex proteome showing contributions from PSC complex (red) and PSC complex-cytoskeleton preparations (yellow) and overlap (orange). C, Venn diagram showing comparison of wild-type PSC complex proteome with proteins from the rootletin KO PSC complex analysis (blue). D, summary of proteomics analyses with PSC-OS and PSC-IS groups identified. Colors match the Venn diagram in C. Cyto, cytoskeleton.
Table I.
PSC complex proteomics analysis summary
| PSC complex |
PSC complex- cytoskeleton |
Rootletin KO PSC complex |
|
|---|---|---|---|
| Spectra | 232,578 | 181,752 | 169,817 |
| Unique peptides | 18,436 | 24,612 | 13,599 |
| False positive pate (%) | 0.2 | 0.2 | 0.21 |
| Proteins, ≥3 peptides | 1,606 | 1,660 | 1,209 |
The PSC complex proteome identified by three or more peptides, generated by combining the results of the wild-type PSC complex and PSC complex-cytoskeleton analyses, contains a total of 1968 proteins (Fig. 3 and Supplemental Table S1A). Combination of the data from the PSC complex and PSC complex-cytoskeleton preparations enriched the proteome. Because the PSC complex-cytoskeleton is a subset of the PSC complex, proteins identified only in the PSC complex-cytoskeleton preparation reflect the improved capacity to detect lower abundance proteins in this further purified sample. For example, all of the 12 molecularly characterized intraflagellar transport (IFT) proteins detected in these analyses were enriched in the PSC complex-cytoskeleton preparation, including five that were not detected in the intact PSC complex (Supplemental Table S1). IFT is a highly conserved process by which groups of proteins are moved in and out of cilia by active transport along the axoneme (34).
An additional 1322 proteins were identified by one or two peptides. These proteins were not included in the analyses described below because the proportion of proteins that are contaminants is expected to increase at very low abundance levels. However, it is worth noting that some proteins detected by one or two peptides are PSC complex components, such as whirlin, which has been detected in the transition zone of photoreceptors (35).
Assessment of the Quality of the PSC Complex Proteome
Several measures show that the PSC complex proteome is relatively comprehensive and has minimal contamination by non-PSC complex proteins. Proteins produced by genes that harbor mutations known to cause inherited retinal degenerations provide a basis for estimating the percentage of false negatives in the proteome as the locations of many of them in the retina have been well characterized. They also represent multiple types of photoreceptor proteins. Thus, of the retinal disease genes whose protein products are known to be located in the PSC complex, 36 of 38 were present in our proteome (Table II), providing an estimate of about 5% for false negatives. To estimate the false positive rate in the PSC complex proteome, we used two measures. First, we examined whether proteins known to be located in proximity to but not in the PSC complex were detected in the proteome. None of the five nuclear proteins and only one of the 12 retinal pigment epithelium (RPE) proteins produced by retinal degeneration disease genes were detected (RetNet: www.sph.uth.tmc.edu/Retnet/). Second, immunofluorescence analysis using antibodies to 51 proteins detected in our analyses whose location in photoreceptor cells has not been reported previously confirmed the location of 45 of these proteins in the PSC complex (Supplemental Table S5). The 51 antibodies used for these analyses were directed against proteins distributed throughout the range of relative protein abundance in the PSC complex proteome and thus were a good subset for estimation of the false positive rate. Additional investigation of the six proteins that were not detected is warranted as they may be present at very low abundance in PSC complexes instead of being contaminants from other structures. Based on these data, we estimate the maximum false positive rate to be ∼10% (0/5 + 1/12 + 6/51 = 7/68).
Table II.
Known PSC complex disease proteins
| Protein/gene | Diseasesa | Protein locationb | Detected | Ref. |
|---|---|---|---|---|
| OS | ||||
| ABCA4 | CD, MD, RP | OS | Yes | 66 |
| AIPL1 | CD, LCA | ONL, IS, OS | Yes | 67 |
| CNGA1 | RP | OS | Yes | 68 |
| CNGA3 | O | Cone OS | Yes | 69 |
| CNGB1 | RP | OS | Yes | 68 |
| CNGB3 | O | Cone OS | No | 70 |
| GNAT1 | CSNB | OS | Yes | 71 |
| GNAT2 | O | Cone OS | Yes | 72 |
| GRK1 | CSNB | OS | Yes | 73 |
| GUCA1A | CD | OS | Yes | 74 |
| GUCA1B | MD, RP | IS, OS | Yes | 75 |
| GUCY2D | CD, LCA | OS | Yes | 76 |
| OPN1MW | O | Cone OS | Yes | 77 |
| OPN1SW | O | Cone OS | Yes | 77 |
| PCDH15 | USH | OS | Yes | 53 |
| PDE6A | RP | OS | Yes | 78 |
| PDE6B | CSNB, RP | OS | Yes | 78 |
| PGK1 | O | OS | Yes | 79 |
| PROM1 | O | OS | Yes | 80 |
| R9AP | O | OS | Yes | 81 |
| RDS | MD, RP | OS | Yes | 82 |
| RGS9 | O | OS | Yes | 83 |
| RHO | CSNB, RP | OS | Yes | |
| ROM1 | RP | OS | Yes | 84 |
| RS1 | O | IS, OS/matrix, Müller cells | Yes | 85 |
| SAG | CSNB, RP | OS | Yes | 86 |
| USH1C | USH | OS, IS, synapse | No | 87 |
| PSC complex-cytoskeleton | ||||
| CEP290 | LCA, SYND | TZ | Yes | 88 |
| IQCB1 | SLSN | TZ | Yes | 89 |
| GPR98 | USH | TZ | Yes | 87 |
| MYO7A | USH | TZ, RPE | Yes | 55 |
| NPHP4 | SLSN | TZ | Yes | 90 |
| RP1 | RP | Axoneme | Yes | 25 |
| RPGR | CD, CSNB, RP | TZ | Yes | 91 |
| RPGRIP1 | LCA | TZ, axoneme | Yes | 91 |
| TTC8 | BBS | TZ, centrosome, BB | Yes | 92 |
| TULP1 | LCA, RP | IS, TZ | Yes | 93 |
| USH2A | RP, USH | TZ, synapse | Yes | 87 |
Diseases: CD, cone dystrophy; CSNB, congenital stationary night blindness; MD, macular degeneration; O, other; SLSN, Senior-Loken syndrome; SYND, syndromic; USH, Usher syndrome.
Locations: BB, basal body; NR, not reported; TZ, transition zone.
Inner and Outer Segment Components of the PSC Complex
As described above, we used PSC complexes from rootletin KO mice to distinguish inner (PSC-IS) and outer segment (PSC-OS) components of the PSC complex (Fig. 1). The rootletin KO PSC complex preparation contained 1209 proteins detected by ≥3 peptides (Fig. 3C and Supplemental Table S4). Of these, 98% (1185 proteins) are shared with the wild-type PSC complex proteome, confirming the location of these proteins in the PSC-OS (Fig. 3D and Supplemental Table S1B). Conversely the finding that only 2% (or 24) of the proteins in the rootletin KO PSC complex were not detected in the wild-type PSC complex or cytoskeleton preparations demonstrates excellent reproducibility of the PSC complex preparations and the MS/MS analyses used in this study (Supplemental Table S1D). The specificity of the PSC-OS group of 1185 proteins is demonstrated by the finding that all 44 proteins with functional annotations related to phototransduction are present in this group (Fig. 4A). Furthermore all 36 proteins produced by retinal degeneration disease genes that were detected in the wild-type PSC complex are in the PSC-OS group (Table II). Identification of these PSC-OS proteins will facilitate investigations into cilia biology and vision and pathogenesis of cilia disorders, including retinal degenerations.
Fig. 4. Functional groups and protein domains in the PSC complex proteome and subgroups.
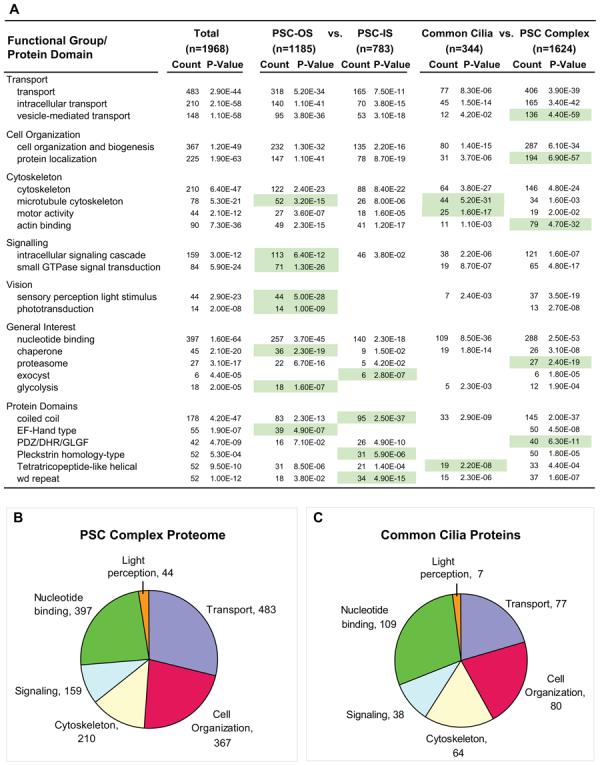
A, results from DAVID and EASE analyses of PSC complex proteome and subgroups. Annotation terms are derived from the Gene Ontology, Protein Information Resource, and Interpro databases. Count, number of proteome or subgroup members within the indicated annotation group. The p value is the Fisher exact test measurement of protein enrichment in annotation terms in PSC complex proteome or subgroup with respect to the total annotated mouse protein set. Green shading highlights enrichment in the indicated subgroups of the PSC complex proteome. B and C, charts showing proportions of proteins in major enriched functional groups for the complete PSC complex proteome (B) and the common cilia proteins (shared with CiliaProteome dataset) (C). Data are from A.
The 783 proteins in the wild-type PSC complex proteome that were not detected in the rootletin KO PSC constitute interesting classes of proteins (Fig. 3D and Supplemental Table S1C). These are PSC-IS proteins that could have activities related to the cilium cytoskeleton, such as transporting proteins to the basal bodies in preparation for export into the cilium (8, 36). It is also possible that some PSC-IS proteins are lower abundance outer segment proteins that were only detected in the PSC complex-cytoskeleton preparation, such as the IFT proteins described above. To estimate the proportion of PSC-IS proteins specifically associated with the cilium cytoskeleton, we used antibodies to 18 PSC-IS group proteins to probe frozen sections of retina and isolated wild-type PSC complexes. Fifteen of the 18 antibodies (83%) stained PSC complex components (Fig. 5 and Supplemental Table S5). We thus estimate that ∼650 of the 783 proteins in the PSC-IS group are specifically associated with the inner segment portion of the PSC complex.
Fig. 5. Locations of novel proteins in mouse PSC complexes.
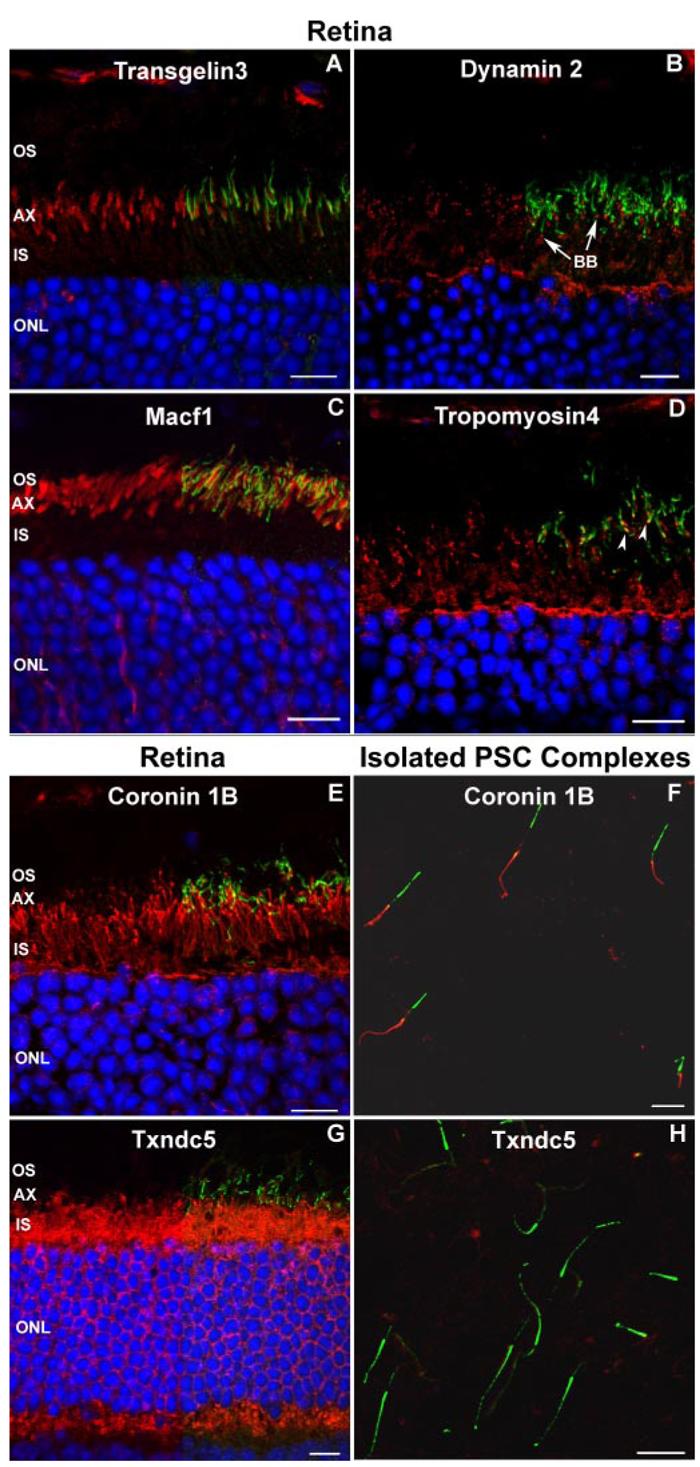
A–D, examples of validation of novel PSC complex proteins. Frozen sections of retina from adult mice were probed with antibodies against the putative PSC complex proteins identified in this study (red). The sections were also probed with antibodies to Rp1 to visualize the photoreceptor axoneme (green). The left half of the image shows the red signal, and the right half of the image shows the merged red and green signals. Nuclei were detected with Hoechst dye (blue). A, Transgelin 3 was detected in the proximal outer segment (OS) adjacent to the axoneme. B, antibodies to Dynamin 2 demonstrated punctate staining at the junction of the inner segment (IS) and OS consistent with basal bodies (BB; arrows). C, Macf1 was located in the proximal OS, closely matching the extent of the axoneme/Rp1 signal. D, Tropomyosin 4 was located in the proximal portion of the axoneme (arrowheads) and inner segments. E–H, example of validation of PSC-IS proteins. Antibodies to coronin 1b and Txndc5 were used to probe frozen sections of mouse retina (E and G) and isolated wild-type PSC complexes (F and H). Coronin 1b was detected in the inner segments on the frozen section (E) and the cytoskeleton of the isolated PSC complex (F) and thus was scored as part of the PSC complex structure. Txndc5 was detected in the inner segments of the frozen section (G) but not in the cytoskeleton of the isolated PSC complex (H) and was scored as not part of the PSC complex. AX, axoneme; ONL, outer nuclear layer. Bars = 10 μm.
Candidate Genes Identified by the PSC Complex Proteome
The genes that encode proteins in the PSC complex proteome are good candidate genes for inherited retinal de-generations and other cilia-related disorders. Consistent with this, during preparation of this report, we became aware of two newly discovered retinal disease genes that produce proteins in the PSC complex proteome.3 To further evaluate the PSC complex proteome for candidate genes, we screened the human homologs of the genes that encode the proteins in the PSC complex proteome to identify those that map into known genetic loci for cilia-related diseases. A total of 60 genes were found to map within the critical intervals for 23 inherited cilia-related diseases (Supplemental Table S6). This list includes 25 candidate genes for eight retinal disorders and eight candidates for six forms of inherited deafness.
Comparisons with Other Cilia Proteomes
To identify conserved and novel cilia proteins in the PSC complex, we compared our proteome with other cilia datasets. Proteomics analyses of axonemes isolated from cultured human respiratory epithelial cells and centrosomes isolated from human lymphoblastic cells have been reported (9, 37). This latter report is relevant to the current analysis because during interphase centrioles function as basal bodies for cilia. Our PSC complex proteome contains 66% of the axoneme and 54% of the centrosome proteins. This demonstrates that the PSC, a specialized sensory cilium, shares many components with other cilia. A summary of comparisons of the PSC complex proteome with 12 previously reported datasets is shown in Table III. The PSC complex proteome has less overlap with cilia datasets generated from bioinformatics or gene expression analyses (21–37%; average, 28%) than with other proteomics analyses (25–87.5%; average, 50%).
Table III. Comparisons of the PSC complex proteome with other cilia datasets.
NR, non-reciprocal; SAGE, serial analysis of gene expression.
| Study | Description | Proteins/genes reporteda |
Proteins/genes comparedb |
Shared with PSC complexc |
|---|---|---|---|---|
| Meta-analyses | ||||
| CiliaProteome V2 (16) | Non-redundant proteins in 10 previous studies | 1226 human (NRe−10) | 1224 mouse (NRe−10) | 344 (28%) |
| Ciliome (17) | Proteins found in ≥3 out of 9 cilia datasets | 196 (48 novel) | 164 (48 novel) | 78 (48%) (17 novel) |
| Proteomics | ||||
| Andersen et al. (37) | Human lymphoblast centrosomes | 112 | 112 | 61 (54%) |
| Broadhead et al. (12) | Trypanosome flagella | 331 | 166 | 42 (25%) |
| Keller et al. (13) | Chlamydomonas centrioles | 129 | 76 | 40 (53%) |
| Ostrowski et al. (9) | Human respiratory epithelial axonemes | 214 | 86 | 57 (66%) |
| Pazour et al. (10) | Chlamydomonas flagella | 652 | 333 | 137 (41%) |
| Smith et al. (11) | Tetrahymena cilia | 223 | 56 | 14 (25%) |
| Schmitt and Wolfrum (94) | Photoreceptor proteins (immunoscreen) | 24 | 24 | 21 (87.5%) |
| Bioinformatics | ||||
| Avidor-Reiss et al. (14) | Genomes of ciliated vs. non-ciliated organisms | 187 | 184 | 47 (26%) |
| Efimenko et al. (95) | Xbx genes C. elegans | 758 | 366 | 99 (27%) |
| Li et al. (15) | Genomes of ciliated vs. non-ciliated organisms | 688 | 427 | 88 (21%) |
| Gene expression | ||||
| Blacque et al. (96) | Xbx genes and SAGE analyses C. elegans cilia | 411 | 339 | 93 (27%) |
| Stolc et al. (97) | Gene expression Chlamydomonas flagella | 220 | 137 | 51 (37%) |
Number of genes/proteins reported is the number of published identifications.
Number of genes/proteins compared is the number of non-redundant proteins or genes for which human or mouse orthologs were identified.
Percentages are of the number of genes/proteins compared.
Proteomes of cilia and flagella from three unicellular eukaryotes, Chlamydomonas reinhardtii, Trypanosoma brucei, and Tetrahymena thermophila, have been reported in the past year (10-12). These proteome data and data from several bioinformatics analyses of cilia genes have recently been collected into databases by two groups (16, 17). In the Cilia-Proteome 1226 non-redundant mammalian orthologs of putative cilia-related proteins were identified by meta-analysis of 10 cilia and basal body datasets reported to date (16). The PSC complex proteome contains 344 of the 1224 (28%) CiliaProteome proteins for which mouse orthologs have been identified (Supplemental Table S1). An additional 153 CiliaProteome proteins were either detected by one to two peptides or were excluded as contaminants (e.g. mitochondrial proteins) in our study. As the majority of the cilia-related proteins in the CiliaProteome database were identified in unicellular organisms or from bioinformatics analyses, this is significant conservation and suggests that these 344 proteins represent a common cilia protein set. Analysis shows that these common cilia proteins include proteins associated with cilia construction and maintenance, such as cytoskeletal proteins, motor proteins, and components of the IFT system (Fig. 4A and Supplemental Table S1).
In this group of 344 common cilia proteins, there are many proteins whose location in photoreceptor cells has not been described previously. This includes a group of 13 proteins produced by genes in which mutations cause diseases related to cilia dysfunction, including Bardet-Biedl syndrome, Senior-Loken syndrome, and Alström syndrome (Table IV). Immunofluorescence analysis of Bbs2 and Bbs7 confirmed our MS/MS identifications with Bbs1 located in basal bodies and Bbs7 located in axonemes of PSCs (not shown; Supplemental Table S5). Similarly many of the IFT proteins have been studied in Chlamydomonas and Caenorhabditis elegans but have not been detected previously in mammals or in the PSC complex. All 12 of the characterized IFT proteins were detected in the PSC complex, including seven that have not been described previously in mammalian cilia (38, 39). All five components of the kinesin and dynein motor proteins involved in IFT were also detected, including Kifap3 whose location in the retina has not been reported previously (Table V).
Table IV.
Novel cilium disease proteins detected in the PSC complex
| Protein/gene | Diseasea | Protein locationb | Ref. |
|---|---|---|---|
| ALMS1 | SYND | Centrosomes, BB | 98 |
| ARL6 (BBS3) | BBS | Cilia C. elegans | 99 |
| BBS1 | BBS | Cilia C. elegans | 92 |
| BBS2 | BBS | Cilia C. elegans | 92 |
| BBS4 | BBS | Centrosomes, BB | 100 |
| BBS5 | BBS | BB other cells | 15 |
| BBS7 | BBS | Cilia C. elegans | 92 |
| KCNV2 | CD | ONL (ISH) | 101 |
| MKS1 | SYND | BB other cells | 102 |
| PTHB1 (BBS9) | BBS | NR | 103 |
| NPHP1 | SLSN | Primary cilia | 104 |
| TRIM32 (BBS11)c | BBS | NR | 105 |
| WFS1 | SYND | ER, vestibular hair cells | 106 |
Diseases: CD, cone dystrophy; SLSN, Senior-Loken Syndrome; SYND, syndromic.
Locations: BB, basal body; ER, endoplasmic reticulum; ONL, outer nuclear layer; ISH, in situ hybridization.
TRIM32 was detected by a single peptide.
Table V.
Intraflagellar transport proteins detected in the PSC complex
| Protein/gene | Protein locationa | First reportb | Ref. |
|---|---|---|---|
| IFT20 | IS, BB, TZ | 38 | |
| IFT52 | IS, BB, TZ | 38 | |
| IFT57 | IS, BB, TZ | 38 | |
| IFT74 | NR | Yes | |
| IFT80 | NR | Yes | |
| IFT81 | NR | Yes | |
| IFT88 | IS, BB, TZ | 38 | |
| IFT122 | NR | Yes | |
| IFT140 | NR | Yes | |
| IFT172 | NR | Yes | |
| WDR19 (IFT139) | NR | 39 | |
| WDR35 (IFT144) | NR | Yes | |
| KIF3A | IS, axoneme | 107 | |
| KIF3B | IS, axoneme | 107 | |
| KIFAP3 | NR | Yes | |
| DYNC2H1 | IS, TZ | 108 | |
| DYNC2LI1 | IS, TZ | 108 |
Locations: BB, basal body; NR, not reported; TZ, transition zone.
First report of protein location in retina or mammalian cilium.
Novel Proteins in the PSC Complex
The PSC complex proteome contains ∼1500 proteins that have not been identified or investigated previously in photoreceptor or other cilia. This number is estimated by subtracting the ∼100 known photoreceptor and phototransduction proteins from the 1624 PSC complex proteins that are not shared with the CiliaProteome. As described above, immunofluorescence analyses validated the location of 45 of 51 proteins for which antibodies were available and whose location in photoreceptor cells had not been reported previously. This includes 15 of 18 proteins from the PSC-IS and 30 of 33 PSC-OS proteins (Supplemental Table S5). Examples of these results are shown in Fig. 5 with Dynamin 2 detected in the basal bodies and Tropomyosin 4 detected in the proximal axonemes. Microtubule-actin cross-linking factor 1 (Macf1) and Transgelin 3 were detected in the proximal outer segment of photoreceptor cells adjacent to the axonemes. The detection of each of these 45 proteins in the PSC complex provides valuable opportunities for investigation of their roles in photoreceptor biology. For example, 10 microtubule plus-end-binding proteins (+TIPs) and related proteins that are thought to help selectively control stabilization of microtubules were detected in the PSC complex (Supplemental Table S1) (40). Antibodies to Clasp 1 and Dynamin 2 confirmed the association of these proteins with the cytoskeleton of the PSC complex (Fig. 5 and Supplemental Table S2). The other six antibodies stained cells in the inner retina, such as Müller cells, suggesting a low level of contamination of the PSC complex preparations with components of other cells, such as Müller cell processes.
It is of interest that 105 of the proteins in the PSC complex proteome are hypothetical, including 60 in the PSC-OS group. The detection of peptides derived from these proteins is thus the first direct evidence that they may have functional roles in cilia or vision. Analysis of these hypothetical proteins shows 14 contain tetratricopeptide repeat, helical, or coiled-coil motifs, which are common in the PSC complex proteome and IFT proteins (41). We have named these proteins Psc100 to Psc204 (Supplemental Table S1E).
Functional Analyses of the PSC Complex Proteome
Several functional groups of proteins are statistically overrepresented in the PSC complex proteome and subgroups (Fig. 4A). For example, almost ¼ (483) of the proteins detected participate in transport, consistent with the polarized nature of the PSC complex. This includes 148 proteins associated with vesicle-mediated transport, including 62 with Rab GTPase domains. Proteins involved in signaling are prominent and concentrated in the PSC-OS group, consistent with other reports that found signaling proteins to be concentrated in cilia (3, 4). Proteins that form rigid scaffolds or bundles, such as those with coiled-coil and WD repeat domains are especially enriched in the PSC-IS group. The WD repeat units form a circularized β-propeller structure that serves as a rigid scaffold for protein interactions. Many IFT proteins contain WD and tetratricopeptide repeats (41). Analysis of the proteins in the portion of the PSC complex proteome that is not shared with other cilia datasets identified enrichment of several functional categories of interest, including transport proteins, actin-binding proteins, and ubiquitin-proteasome system (UPS) proteins (Fig. 4A).
Estimation of PSC-OS Protein Abundance
The long history of quantitative photoreceptor biochemistry provides an opportunity to estimate the relative abundances of the entire set of proteins in our PSC-OS proteome (26, 28, 42). Considering the abundance of each detected peptide as an independent estimate of the number of copies of the parent protein in the sample, we divided the total spectral counts from each protein by the number of observable tryptic peptides (42), a statistic we will identify as spectra per predicted peptide, “S/PP.” Evidence that this quantitative approach is meaningful and reliable comes from a comparison of the S/PP data from wild-type and rootletin KO PSC complexes (Fig. 6A). Remarkably although the isolation procedures were performed in different laboratories and the samples differ in complexity, S/PP values of the 1130 common proteins are close to identical (the unit slope line), indicating that nearly the same number of each of these proteins was present in the PSC complexes of mice of the two genotypes. About 80% of the shared proteins have relative abundances within a factor of 3 (Fig. 6A, gray region) of the identical value (red line) with somewhat greater variability for lower abundance proteins (S/PP < 2).
Fig. 6. Quantitative analyses.
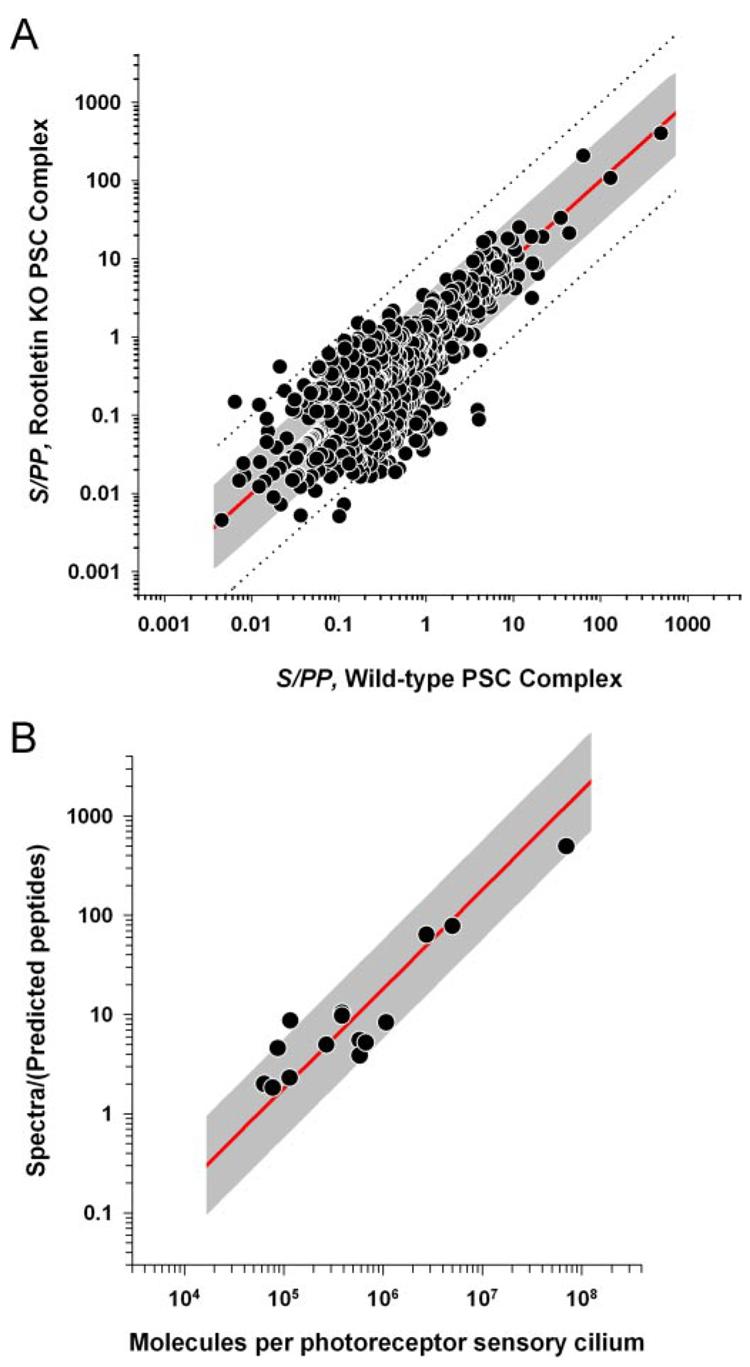
A, S/PP for wild-type and rootletin KO PSC complexes plotted against each other for 1130 shared proteins. The red line indicates unit slope where the S/PP values are equal for proteins in the two different preparations. The dashed lines and gray shading indicate regions within a factor of ±10- and ±3-fold of the unit slope, respectively. B, relationship between MS/MS spectral counts and the copy numbers of 15 different proteins in the mouse photoreceptor outer segment. Details regarding the 15 standard proteins are provided in Supplemental Table S7. The ordinate of each point is the total number of spectra from the protein divided by the predicted number of observable tryptic peptides (S/PP); the abscissa value gives the independently determined NC of the protein in the rod outer segment. In all cases the protein copy numbers were determined relative to rhodopsin whose copy number in the mouse rod outer segment is 7 ×107. The gray line expresses the hypothesis of a linear relationship between copy number and recorded spectra, i.e. S/PP = k × NC. The line was estimated by least squares regression in log/log coordinates with k = 1.805 ×10−5.
To estimate the protein copy numbers in each photoreceptor outer segment, we compared the reported amounts of 15 photoreceptor outer segment proteins with their spectral count data from the wild-type PSC complexes (Supplemental Table S7). An absolute scale is provided by rhodopsin of which 7 × 107 molecules are present in a mouse rod outer segment (29). The relationship between S/PP and the reported number of copies (NC) for the 15 outer segment proteins is linear (Fig. 6B): thus, the gray line expresses the hypothesis S/PP = k × NC and provides a reasonable description over a nearly 10,000-fold protein concentration range. Using the estimated value of k, the copy number of each PSC-OS protein identified in this study was determined (Supplemental Table S8). The copy numbers range from 7 × 107 molecules for rhodopsin to 300 for Herc2, a large protein thought to play a role in vesicular trafficking (43).
DISCUSSION
We report here a comprehensive proteomics analysis of the mouse PSC complex. The PSC complex proteome determined in this study contains 1968 proteins detected by three or more unique peptides, making it one of the largest organelle proteomes reported to date (44). Our PSC complex proteome contains many novel cilia and photoreceptor proteins. This dataset has the potential to open new avenues for research directed at fundamental questions about cilia biology, such as how these highly polarized structures are built and maintained and how these processes are disrupted in human disease.
The PSC complex proteome contains many cilia proteins not identified previously in photoreceptors. This includes 13 proteins produced by genes that harbor mutations that cause cilia disease (Table IV). The detection of these proteins in photoreceptors is consistent with the hypothesis that defects in these proteins cause disease by disrupting cilia structure or function and focuses future investigation of disease pathogenesis (2). Our PSC complex proteome also contains seven IFT proteins not described previously in mammalian cilia or photoreceptors (Table V). These data confirm the importance of IFT in mammalian cilia and the high degree of conservation of this axonemal transport system. These transport processes are especially important in photoreceptor cilia, where large numbers of proteins are required to maintain the outer segment, which are completely renewed every 10 days (45).
The PSC complex proteome also contains ∼1500 proteins not identified previously in cilia. This includes novel candidate disease genes. The identification of 60 proteins encoded by genes that map within the critical intervals for 23 inherited cilia-related disorders will help prioritize genes in these linkage intervals for mutation screening (Supplemental Table S6). This is especially true for the 34 genes in the list that produce proteins in the PSC-OS group. For example, the gene for transmembrane protein 27 maps within the critical region for retinitis pigmentosa 23 (RP23) (46). Expression of the protein produced by this gene, which is also called collectrin, has been reported previously only in the kidney and pancreas, both organs in which primary cilia play an important role (47). Furthermore it is likely that other PSC complex proteins are encoded by genes located in unidentified disease loci as evidenced by the recent detection of two novel disease proteins in the PSC complex proteome.3
The novel cilia proteins detected in the PSC complex proteome may also be informative about photoreceptor biology. For example, 27 components of the UPS were detected in the PSC complex but are not shared with other cilia (Fig. 4A). The UPS plays an important role in several cellular pathways via the coordinated degradation of their protein components (48). Although proteasomes are typically concentrated at the centrosome, the role of the UPS in cilia and photoreceptor function has not been evaluated in detail (49). It has been suggested that the UPS may play a role in setting the levels of phototransduction proteins, which are tightly regulated (50). The UPS also has an important role in removing damaged proteins from the cell (51). Dysfunction of this process is associated with several neurodegenerative disorders, including Parkinson disease (48). PSC complexes are subjected to high levels of oxidative stress resulting from obligatory exposure of the retina to light (52). The finding of UPS components in the PSC complex suggests that local recognition and clearance of oxidatively modified proteins within the PSC complex by the UPS may be one of the mechanisms by which photoreceptor cells cope with chronic light stress. Consistent with these hypotheses, ubiquitin is abundant in the PSC complex and was detected at multiple molecular weights, indicating conjugation to PSC complex proteins.
The results of the mass spectrometric analyses show that the PSC complex proteome is relatively comprehensive and has only limited contamination by non-PSC proteins. For example, the 98% agreement between the analyses of the rootletin KO and wild-type PSC complex preparations shows excellent reproducibility of the LC-MS/MS analyses (Figs. 3 and 6A). In addition, the agreement between the two datasets demonstrates good depth of analysis provided by separating the protein preparations into 30 fractions by electrophoresis and using long LC gradients with a linear ion trap mass spectrometer. Furthermore 36 of 38 proteins produced by retinal degeneration disease genes that are known to be located in the PSC complex were detected (Table II). The two proteins that were not detected in our analyses are harmonin and the β3 subunit of the cone cyclic nucleotide-gated channel (Cngb3). Evidence suggests that harmonin is a soluble protein (53); thus it is possible it was lost during the isolation of the PSC complexes. Membrane proteins such as the cone cyclic nucleotide-gated channel β3 are difficult to detect using mass spectrometry in part because their membrane-spanning regions are very hydrophobic and thus lack the arginine and lysine residues required for cleavage by trypsin. Most tryptic peptides derived from channel proteins are thus outside the size and hydrophobicity ranges detected by the mass spectrometric methods used (54). Because only 3% of photoreceptors in the mouse retina are cones, it is likely that the combination of having six membrane-spanning regions and being present only in cones prevented detection of Cngb3.
As another assessment of the specificity of the PSC proteome, we performed immunofluorescence analyses of mouse retina using antibodies to 51 proteome members that were not described previously in PSC complexes. All but six of these antibodies confirmed the mass spectrometric data (Supplemental Table S5). The other six antibodies stained cells in the inner retina, such as Müller cells, suggesting a low level of contamination of the PSC complex preparations with components of other cells, such as Müller cell processes. Further analysis of the proteins to which these antibodies are directed is warranted as they may also be present at low abundance in PSCs. There are several examples of PSC complex proteins that require immunoelectron microscopy for detection in retinal sections, such as myosin VIIa (55). Myosin VIIa was readily detected in the PSC complex proteome (Supplemental Table S1; eight peptides), illustrating the power of proteomics analysis to reveal low abundance proteins. Proteins from mitochondria and ribosomes, which can segregate with PSC complexes due to their high abundance in photoreceptor inner segments, accounted for less than 15% of the total proteins detected; this is comparable to the levels of mitochondrial and ribosomal proteins detected in other cilia and basal body proteomics analyses (9, 13). In sum, these analyses support the conclusion that the majority of the proteins in the proteome are truly present in the PSC complex.
Acceleration of the Pace of Discovery
Analysis of the PSC complex proteome has the potential to accelerate the pace of discovery regarding cilia biology. For example, an insight derived from our analyses is that the PSC complex as a functional organelle includes components in both the inner and outer segments of photoreceptor cells (Fig. 1 and Supplemental Table S1). The detection of ∼650 proteins predicted to be functionally part of the cilium structure that extends into the inner segment of photoreceptor cells emphasizes the essential connection between the inner and outer segment portions of the PSC complex. Further study of the PSC-IS proteins detected in our analyses may help identify additional proteins required for PSC complex function. For instance, almost all of the components of the exocyst complex were detected in the PSC-IS group of proteins. The exocyst complex has been shown to be located in primary cilia in renal tubule epithelial cells and plays a central role in the development of renal tubules and cysts (56). There is recent evidence that the exocyst also plays an important role in vesicle-mediated transport of proteins to the Drosophila rhabdomere, but the role of the exocyst in mammalian photoreceptors has not yet been investigated (57).
The link between the inner and outer segment components of the PSC complex is also highlighted by the recent recognition that the movement of signal transduction proteins between the inner and outer segments of photoreceptor cells is an important mechanism by which photoreceptor cells adapt to changes in light intensity (30). Stimulus-dependent redistribution of signaling proteins into and out of cilia also mediates transduction of external signals by primary cilia, including Hedgehog signaling and sensation of urine flow, both of which ultimately lead to modulation of transcription of target genes (3). Protein movement may thus be a general mechanism by which cilia transduce information about the external environments of their cells. The PSC-IS group proteins detected in the PSC complex are candidates to participate in the movement of proteins in response to light or other stimuli in the PSC and other cilia.
Conservation of the Mammalian PSC
Based on the analysis of PSC complexes from rootletin KO mice, our data show that the PSC complex proteome contains 1185 proteins confirmed to be part of the PSC-OS (Fig. 3). Comparison of the PSC-OS proteome with other cilia datasets shows that the outer segment contains 2–5 times more proteins than reported for cilia from unicellular organisms, whose proteomes range from 223 in Tetrahymena to 652 in Chlamydomonas (Table III).
Although a portion of the additional proteins in the PSC complex are associated with visual function, including 44 proteins associated with phototransduction, detection of these additional proteins likely reflects a general increase in complexity of mammalian cilia. This conclusion is supported by the observation that many protein classes in cilia from unicellular organisms are augmented in the PSC complex proteome. For example, of the 11 kinesin components and 14 microtubule-associated proteins found in the PSC, only five and three are present in flagella of Chlamydomonas, respectively (10). The increased number of proteins in the PSC complex reflects, at least in part, the increased complexity of the mouse genome as the percentage of genes devoted to encoding cilia proteins is similar in the mouse, Chlamydomonas, and Trypanosoma genomes; this is not true of the Tetrahymena genome (Table VI). The increased complexity of mammalian cilia also suggests that additional members of other protein classes will be found in mammalian cilia.
Table VI.
Cilia proteomes and genome sizes
| Species | Cilia proteome | Transcriptomea | Ref. | |
|---|---|---|---|---|
| % | ||||
| Mus musculus | 1,185 (PSC-OS) | 32,661 | 3.6 | Ensembl (www.ensembl.org/Mus_musculus /index.html) |
| C. reinhardtii | 652 | 15,256 | 4.3 | United States Department of Energy Joint Genome Institute (genome.jgi-psf.org/Chlre3/Chlre3.info.html) |
| T. brucei | 331 | 9,068 | 3.7 | 109 |
| T. thermophila | 223 | 27,000 | 0.83 | 110 |
Reported number of transcriptional units or gene transcripts.
It is also evident from the comparisons that there is notable conservation of cilia proteins. At least 344 proteins are shared between the non-redundant cilia-related proteins in the Cilia-Proteome (16) and the PSC complex proteome and represent a set of common cilia proteins, emphasizing the preservation of the basic components of cilia structure throughout evolution. For example, the PSC complex shares 137 proteins with flagella from Chlamydomonas, a unicellular green alga estimated to have emerged as a species 700 million years ago (58). Furthermore the relative proportions of the major enriched functional protein groups are similar in the common cilia proteins and the PSC complex proteome (Fig. 3, B and C).
Implications of Quantification of the PSC Complex Proteome Components
Precise regulation of protein levels is a major theme in cell biology. In photoreceptor cells over- or underexpression of specific proteins can lead to cell dysfunction and death with alterations in rhodopsin levels being the classic example (59, 60). It is difficult, however, to measure the levels of individual proteins on a large scale especially of proteins for which activity assays or quantitative immunoassays are not available. We found that total spectral counts normalized to total expected peptides (S/PP) within the analyzed mass range correlated well with the known amounts of 15 outer segment proteins (Fig. 6B). Specifically 14 of the benchmark points lie within a factor of 3 of the best fitting line. This is consistent with prior reports that the number of spectra detected for a given protein in MS/MS analyses can be used to estimate the relative abundance of that protein (26, 27).
The value S/PP is an estimate of protein abundance and is not corrected for inherent variations in peptide “detectability.” For example, as indicated above peptides from membrane proteins are less efficiently detected in LC-MS/MS experiments due to their hydrophobicity (54). Rhodopsin, which has seven transmembrane segments, is a relevant example. Because the majority of tryptic peptides from rhodopsin are too large to be detected using the LC-MS/MS parameters applied in our analysis, we used the hydrophilic peptide from the C terminus of rhodopsin for the analysis in Fig. 6B. Even this peptide appears to be less “detectable” than the peptides in the majority of the other benchmark outer segment proteins as the point for rhodopsin (most abundant protein) falls below the regression line (Fig. 6B). That said, it is evident from Fig. 6B that S/PP provides a good estimate of relative protein abundance. This may in part be because the S/PP calculation is an average for all peptides detected from a given protein and thus “averages over” some of the inherent variation in peptide detectability. The S/PP calculation can be refined in the future as the reasons for the variation in peptide detectability are better defined (61).
The estimated copy number of each protein in our PSC-OS proteome (Fig. 6 and Supplemental Table S8) provides a basis for predicting the stoichiometries of protein complexes in outer segments and could be useful for functional studies of these proteins and their interactions and roles in the pathogenesis of diseases caused by cilia dysfunction. Knowledge of the amounts of specific proteins in photoreceptor outer segments may also be important for directing the development of accurate gene augmentation therapies for inherited retinal degenerations and other cilia disorders, some of which are nearing clinical application (62). For example, mutations in CEP290 have recently been reported to cause the cilia disorders Joubert syndrome and Leber congenital amaurosis (LCA), a form of congenital blindness (63-65). The proteome data indicate that ∼9000 copies of Cep290 protein are present in each mouse outer segment. In contrast, ∼580,000 copies of Rds, whose mutation causes several forms of retinal degeneration, are present (Supplemental Table S8). These examples reveal that different delivery and expression strategies will be needed to effect successful therapy for different types of inherited retinal degenerations and other cilia disorders.
Supplementary Material
Acknowledgments
We thank Tom Beer, Tony Chang, and Kaye Speicher in the Wistar Proteomics Core Facility for performing the LC-MS/MS analyses and assistance with data analysis. We also thank Li Fu and Alexei Saveliev for technical assistance and Donita Garland for comments on the manuscript.
Footnotes
The abbreviations used are: PSC, photoreceptor sensory cilium; IFT, intraflagellar transport; PSC-IS, inner segment portion of the PSC complex; PSC-OS, outer segment portion of the PSC complex; RP, retinitis pigmentosa; RPE, retinal pigment epithelium; UPS, ubiquitinproteasome system; BBS, Bardet-Biedl syndrome; KO, knock-out; Pipes, 1,4-piperazinediethanesulfonic acid; bis-Tris, 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol; DAVID, Database for Annotation, Visualization, and Integrated Discovery; EASE, Expression Analysis Systematic Explorer; S/PP, spectra per predicted peptide; NC, number of copies; IS, inner segment; OS, outer segment; LCA, Leber congenital amaurosis.
H.-Y. Tang, G. Tan, and D. W. Speicher, manuscript in preparation..
C. Inglehearn, personal communication.
This work was supported by National Institutes of Health Grants EY12910, CA10815, and CA77048 and by the F. M. Kirby Foundation, Foundation Fighting Blindness, Research to Prevent Blindness, Rosanne Silbermann Foundation, Mackall Foundation Trust, and Commonwealth Universal Research Enhancement Program.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
REFERENCES
- 1.Pazour GJ, Witman GB. The vertebrate primary cilium is a sensory organelle. Curr. Opin. Cell Biol. 2003;15:105–110. doi: 10.1016/s0955-0674(02)00012-1. [DOI] [PubMed] [Google Scholar]
- 2.Pan J, Wang Q, Snell WJ. Cilium-generated signaling and cilia-related disorders. Lab. Investig. 2005;85:452–463. doi: 10.1038/labinvest.3700253. [DOI] [PubMed] [Google Scholar]
- 3.Singla V, Reiter JF. The primary cilium as the cell's antenna: signaling at a sensory organelle. Science. 2006;313:629–633. doi: 10.1126/science.1124534. [DOI] [PubMed] [Google Scholar]
- 4.Davis EE, Brueckner M, Katsanis N. The emerging complexity of the vertebrate cilium: new functional roles for an ancient organelle. Dev. Cell. 2006;11:9–19. doi: 10.1016/j.devcel.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 5.Beisson J, Wright M. Basal body/centriole assembly and continuity. Curr. Opin. Cell Biol. 2003;15:96–104. doi: 10.1016/s0955-0674(02)00017-0. [DOI] [PubMed] [Google Scholar]
- 6.Yang J, Gao J, Adamian M, Wen XH, Pawlyk B, Zhang L, Sanderson MJ, Zuo J, Makino CL, Li T. The ciliary rootlet maintains long-term stability of sensory cilia. Mol. Cell. Biol. 2005;25:4129–4137. doi: 10.1128/MCB.25.10.4129-4137.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horst CJ, Johnson LV, Besharse JC. Transmembrane assemblage of the photoreceptor connecting cilium and motile cilium transition zone contain a common immunologic epitope. Cell Motil. Cytoskelet. 1990;17:329–344. doi: 10.1002/cm.970170408. [DOI] [PubMed] [Google Scholar]
- 8.Yang J, Li T. The ciliary rootlet interacts with kinesin light chains and may provide a scaffold for kinesin-1 vesicular cargos. Exp. Cell Res. 2005;309:379–389. doi: 10.1016/j.yexcr.2005.05.026. [DOI] [PubMed] [Google Scholar]
- 9.Ostrowski LE, Blackburn K, Radde KM, Moyer MB, Schlatzer DM, Moseley A, Boucher RC. A proteomic analysis of human cilia: identification of novel components. Mol. Cell. Proteomics. 2002;1:451–465. doi: 10.1074/mcp.m200037-mcp200. [DOI] [PubMed] [Google Scholar]
- 10.Pazour GJ, Agrin N, Leszyk J, Witman GB. Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 2005;170:103–113. doi: 10.1083/jcb.200504008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith JC, Northey JG, Garg J, Pearlman RE, Siu KW. Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. J. Proteome Res. 2005;4:909–919. doi: 10.1021/pr050013h. [DOI] [PubMed] [Google Scholar]
- 12.Broadhead R, Dawe HR, Farr H, Griffiths S, Hart SR, Portman N, Shaw MK, Ginger ML, Gaskell SJ, McKean PG, Gull K. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 2006;440:224–227. doi: 10.1038/nature04541. [DOI] [PubMed] [Google Scholar]
- 13.Keller LC, Romijn EP, Zamora I, Yates JR, III, Marshall WF. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 14.Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 2004;117:527–539. doi: 10.1016/s0092-8674(04)00412-x. [DOI] [PubMed] [Google Scholar]
- 15.Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 2004;117:541–552. doi: 10.1016/s0092-8674(04)00450-7. [DOI] [PubMed] [Google Scholar]
- 16.Gherman A, Davis EE, Katsanis N. The ciliary proteome database: an integrated community resource for the genetic and functional dissection of cilia. Nat. Genet. 2006;38:961–962. doi: 10.1038/ng0906-961. [DOI] [PubMed] [Google Scholar]
- 17.Inglis PN, Boroevich KA, Leroux MR. Piecing together a ciliome. Trends Genet. 2006;22:491–500. doi: 10.1016/j.tig.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 18.Papermaster DS, Dreyer WJ. Rhodopsin content in the outer segment membranes of bovine and frog retinal rods. Biochemistry. 1974;13:2438–2444. doi: 10.1021/bi00708a031. [DOI] [PubMed] [Google Scholar]
- 19.Liu Q, Zuo J, Pierce EA. The retinitis pigmentosa 1 protein is a photoreceptor microtubule-associated protein. J. Neurosci. 2004;24:6427–6436. doi: 10.1523/JNEUROSCI.1335-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fleischman D, Denisevich M, Raveed D, Pannbacker RG. Association of guanylate cyclase with the axoneme of retinal rods. Biochim. Biophys. Acta. 1980;630:176–186. doi: 10.1016/0304-4165(80)90419-5. [DOI] [PubMed] [Google Scholar]
- 21.Speicher KD, Kolbas O, Harper S, Speicher DW. Systematic analysis of peptide recoveries from in-gel digestions for femtomole protein identifications in proteome studies. J. Biomol. Tech. 2000;11:74–86. [PMC free article] [PubMed] [Google Scholar]
- 22.Tabb DL, McDonald WH, Yates JR., III DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McConnell DG. The isolation of retinal outer segment fragments. J. Cell Biol. 1965;27:459–473. doi: 10.1083/jcb.27.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat. Genet. 2006;38:576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- 25.Liu Q, Zhou J, Daiger SP, Farber DB, Heckenlively JR, Smith JE, Sullivan LS, Zuo J, Milam AH, Pierce EA. Identification and subcellular localization of the RP1 protein in human and mouse photoreceptors. Investig. Ophthalmol. Vis. Sci. 2002;43:22–32. [PMC free article] [PubMed] [Google Scholar]
- 26.Old WM, Meyer-Arendt K, Aveline-Wolf L, Pierce KG, Mendoza A, Sevinsky JR, Resing KA, Ahn NG. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteomics. 2005;4:1487–1502. doi: 10.1074/mcp.M500084-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of relative abundance ratios derived from peptide ion chromatograms and spectrum counting for quantitative proteomic analysis using stable isotope labeling. Anal. Chem. 2005;77:6218–6224. doi: 10.1021/ac050846r. [DOI] [PubMed] [Google Scholar]
- 28.Ishihama Y, Oda Y, Tabata T, Sato T, Nagasu T, Rappsilber J, Mann M. Exponentially modified protein abundance index (em-PAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Lyubarsky AL, Daniele LL, Pugh EN., Jr. From candelas to photoisomerizations in the mouse eye by rhodopsin bleaching in situ and the light-rearing dependence of the major components of the mouse ERG. Vision Res. 2004;44:3235–3251. doi: 10.1016/j.visres.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 30.Calvert PD, Strissel KJ, Schiesser WE, Pugh EN, Jr., Arshavsky VY. Light-driven translocation of signaling proteins in vertebrate photoreceptors. Trends Cell Biol. 2006;16:560–568. doi: 10.1016/j.tcb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 31.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 32.Hosack DA, Dennis G, Jr., Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang J, Liu X, Yue G, Adamian M, Bulgakov O, Li T. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. J. Cell Biol. 2002;159:431–440. doi: 10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat. Rev. Mol. Cell. Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 35.van Wijk E, van der Zwaag B, Peters T, Zimmerman U, Te BH, Kersten FF, Marker T, Aller E, Hoefsloot LH, Cremers CW, Cremers FP, Wolfrum U, Knipper M, Roepman R, Kremer H. The DFNB31 gene product whirlin connects to the Usher protein network in the cochlea and retina by direct association with USH2A and VLGR1. Hum. Mol. Genet. 2006;15:761–765. doi: 10.1093/hmg/ddi490. [DOI] [PubMed] [Google Scholar]
- 36.Deretic D. Post-Golgi trafficking of rhodopsin in retinal photoreceptors. Eye. 1998;12:526–530. doi: 10.1038/eye.1998.141. [DOI] [PubMed] [Google Scholar]
- 37.Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 38.Pazour GJ, Baker SA, Deane JA, Cole DG, Dickert BL, Rosenbaum JL, Witman GB, Besharse JC. The intraflagellar transport protein, IFT88, is essential for vertebrate photoreceptor assembly and maintenance. J. Cell Biol. 2002;157:103–113. doi: 10.1083/jcb.200107108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Efimenko E, Blacque OE, Ou G, Haycraft CJ, Yoder BK, Scholey JM, Leroux MR, Swoboda P. Caenorhabditis elegans DYF-2, an ortholog of human WDR19, is a component of the IFT machinery in sensory cilia. Mol. Biol. Cell. 2006;17:4801–4811. doi: 10.1091/mbc.E06-04-0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr. Opin. Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 41.Jekely G, Arendt D. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. BioEssays. 2006;28:191–198. doi: 10.1002/bies.20369. [DOI] [PubMed] [Google Scholar]
- 42.Rappsilber J, Ryder U, Lamond AI, Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walkowicz M, Ji Y, Ren X, Horsthemke B, Russell LB, Johnson D, Rinchik EM, Nicholls RD, Stubbs L. Molecular characterization of radiation- and chemically induced mutations associated with neuromuscular tremors, runting, juvenile lethality, and sperm defects in jdf2 mice. Mamm. Genome. 1999;10:870–878. doi: 10.1007/s003359901106. [DOI] [PubMed] [Google Scholar]
- 44.Yates JR, III, Gilchrist A, Howell KE, Bergeron JJ. Proteomics of organelles and large cellular structures. Nat. Rev. Mol. Cell. Biol. 2005;6:702–714. doi: 10.1038/nrm1711. [DOI] [PubMed] [Google Scholar]
- 45.Young RW. The renewal of photoreceptor cell outer segments. J. Cell Biol. 1967;33:61–72. doi: 10.1083/jcb.33.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hardcastle AJ, Thiselton DL, Zito I, Ebenezer N, Mah TS, Gorin MB, Bhattacharya SS. Evidence for a new locus for X-linked retinitis pigmentosa (RP23) Investig. Ophthalmol. Vis. Sci. 2000;41:2080–2086. [PubMed] [Google Scholar]
- 47.Akpinar P, Kuwajima S, Krutzfeldt J, Stoffel M. Tmem27: a cleaved and shed plasma membrane protein that stimulates pancreatic beta cell proliferation. Cell Metab. 2005;2:385–397. doi: 10.1016/j.cmet.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Ross CA, Pickart CM. The ubiquitin-proteasome pathway in Parkinson's disease and other neurodegenerative diseases. Trends Cell Biol. 2004;14:703–711. doi: 10.1016/j.tcb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 49.Badano JL, Teslovich TM, Katsanis N. The centrosome in human genetic disease. Nat. Rev. Genet. 2005;6:194–205. doi: 10.1038/nrg1557. [DOI] [PubMed] [Google Scholar]
- 50.Obin M, Lee BY, Meinke G, Bohm A, Lee RH, Gaudet R, Hopp JA, Arshavsky VY, Willardson BM, Taylor A. Ubiquitylation of the transducin βγ subunit complex. Regulation by phosducin. J. Biol. Chem. 2002;277:44566–44575. doi: 10.1074/jbc.M205308200. [DOI] [PubMed] [Google Scholar]
- 51.Poppek D, Grune T. Proteasomal defense of oxidative protein modifications. Antioxid. Redox Signal. 2006;8:173–184. doi: 10.1089/ars.2006.8.173. [DOI] [PubMed] [Google Scholar]
- 52.Boulton M, Rozanowska M, Rozanowski B. Retinal photodamage. J. Photochem. Photobiol. B Biol. 2001;64:144–161. doi: 10.1016/s1011-1344(01)00227-5. [DOI] [PubMed] [Google Scholar]
- 53.Reiners J, Marker T, Jurgens K, Reidel B, Wolfrum U. Photoreceptor expression of the Usher syndrome type 1 protein protocadherin 15 (USH1F) and its interaction with the scaffold protein harmonin (USH1C) Mol. Vis. 2005;11:347–355. [PubMed] [Google Scholar]
- 54.Wu CC, Yates JR., III The application of mass spectrometry to membrane proteomics. Nat. Biotechnol. 2003;21:262–267. doi: 10.1038/nbt0303-262. [DOI] [PubMed] [Google Scholar]
- 55.Liu X, Vansant G, Udovichenko IP, Wolfrum U, Williams DS. Myosin VIIa, the product of the Usher 1B syndrome gene, is concentrated in the connecting cilia of photoreceptor cells. Cell Motil. Cytoskelet. 1997;37:240–252. doi: 10.1002/(SICI)1097-0169(1997)37:3<240::AID-CM6>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 56.Rogers KK, Wilson PD, Snyder RW, Zhang X, Guo W, Burrow CR, Lipschutz JH. The exocyst localizes to the primary cilium in MDCK cells. Biochem. Biophys. Res. Commun. 2004;319:138–143. doi: 10.1016/j.bbrc.2004.04.165. [DOI] [PubMed] [Google Scholar]
- 57.Beronja S, Laprise P, Papoulas O, Pellikka M, Sisson J, Tepass U. Essential function of Drosophila Sec6 in apical exocytosis of epithelial photoreceptor cells. J. Cell Biol. 2005;169:635–646. doi: 10.1083/jcb.200410081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amati BB, Goldschmidt-Clermont M, Wallace CJ, Rochaix JD. cDNA and deduced amino acid sequences of cytochrome c from Chlamydomonas reinhardtii: unexpected functional and phylogenetic implications. J. Mol. Evol. 1988;28:151–160. doi: 10.1007/BF02143507. [DOI] [PubMed] [Google Scholar]
- 59.Olsson JE, Gordon JW, Pawlyk BS, Roof DJ, Hayes A, Molday RS, Mukai S, Cowley GS, Berson EL, Dryja TP. Transgenic mice with a rhodopsin mutation (Pro23His): a mouse model of autosomal dominant retinitis pigmentosa. Neuron. 1992;9:815–830. doi: 10.1016/0896-6273(92)90236-7. [DOI] [PubMed] [Google Scholar]
- 60.Rosenfeld PJ, Cowley GS, McGee TL, Sandberg MA, Berson EL, Dryja TP. A null mutation in the rhodopsin gene causes rod photoreceptor dysfunction and autosomal recessive retinitis pigmentosa. Nat. Genet. 1992;1:209–213. doi: 10.1038/ng0692-209. [DOI] [PubMed] [Google Scholar]
- 61.Mallick P, Schirle M, Chen SS, Flory MR, Lee H, Martin D, Ranish J, Raught B, Schmitt R, Werner T, Kuster B, Aebersold R. Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 62.Acland GM, Aguirre GD, Bennett J, Aleman TS, Cideciyan AV, Bennicelli J, Dejneka NS, Pearce-Kelling SE, Maguire AM, Palczewski K, Hauswirth WW, Jacobson SG. Long-term restoration of rod and cone vision by single dose rAAV-mediated gene transfer to the retina in a canine model of childhood blindness. Mol. Ther. 2005;12:1072–1082. doi: 10.1016/j.ymthe.2005.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sayer JA, Otto EA, O'Toole JF, Nurnberg G, Kennedy MA, Becker C, Hennies HC, Helou J, Attanasio M, Fausett BV, Utsch B, Khanna H, Liu Y, Drummond I, Kawakami I, Kusakabe T, Tsuda M, Ma L, Lee H, Larson RG, Allen SJ, Wilkinson CJ, Nigg EA, Shou C, Lillo C, Williams DS, Hoppe B, Kemper MJ, Neuhaus T, Parisi MA, Glass IA, Petry M, Kispert A, Gloy J, Ganner A, Walz G, Zhu X, Goldman D, Nurnberg P, Swaroop A, Leroux MR, Hildebrandt F. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat. Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 64.Valente EM, Silhavy JL, Brancati F, Barrano G, Krishnaswami SR, Castori M, Lancaster MA, Boltshauser E, Boccone L, Al-Gazali L, Fazzi E, Signorini S, Louie CM, Bellacchio E, Bertini E, Dallapiccola B, Gleeson JG. Mutations in CEP290, which encodes a centrosomal protein, cause pleiotropic forms of Joubert syndrome. Nat. Genet. 2006;38:623–625. doi: 10.1038/ng1805. [DOI] [PubMed] [Google Scholar]
- 65.den Hollander AI, Koenekoop RK, Yzer S, Lopez I, Arends ML, Voesenek KE, Zonneveld MN, Strom TM, Meitinger T, Brunner HG, Hoyng CB, van den Born LI, Rohrschneider K, Cremers FP. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am. J. Hum. Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun H, Nathans J. Stargardt's ABCR is localized to the disc membrane of retinal rod outer segments. Nat. Genet. 1997;17:15–16. doi: 10.1038/ng0997-15. [DOI] [PubMed] [Google Scholar]
- 67.Akey DT, Zhu X, Dyer M, Li A, Sorensen A, Blackshaw S, Fukuda-Kamitani T, Daiger SP, Craft CM, Kamitani T, Sohocki MM. The inherited blindness associated protein AIPL1 interacts with the cell cycle regulator protein NUB1. Hum. Mol. Genet. 2002;11:2723–2733. doi: 10.1093/hmg/11.22.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cook NJ, Molday LL, Reid D, Kaupp UB, Molday RS. The cGMP-gated channel of bovine rod photoreceptors is localized exclusively in the plasma membrane. J. Biol. Chem. 1989;264:6996–6999. [PubMed] [Google Scholar]
- 69.Hirano AA, Hack I, Wassle H, Duvoisin RM. Cloning and immunocytochemical localization of a cyclic nucleotide-gated channel alpha-subunit to all cone photoreceptors in the mouse retina. J. Comp Neurol. 2000;421:80–94. doi: 10.1002/(sici)1096-9861(20000522)421:1<80::aid-cne5>3.0.co;2-o. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kohl S, Baumann B, Broghammer M, Jagle H, Sieving P, Kellner U, Spegal R, Anastasi M, Zrenner E, Sharpe LT, Wissinger B. Mutations in the CNGB3 gene encoding the β-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum. Mol. Genet. 2000;9:2107–2116. doi: 10.1093/hmg/9.14.2107. [DOI] [PubMed] [Google Scholar]
- 71.Takemoto DJ, Cunnick JM. Visual transduction in rod outer segments. Cell. Signal. 1990;2:99–104. doi: 10.1016/0898-6568(90)90013-z. [DOI] [PubMed] [Google Scholar]
- 72.Lerea CL, Somers DE, Hurley JB, Klock IB, Bunt-Milam AH. Identification of specific transducin α subunits in retinal rod and cone photoreceptors. Science. 1986;234:77–80. doi: 10.1126/science.3529395. [DOI] [PubMed] [Google Scholar]
- 73.Palczewski K, McDowell JH, Hargrave PA. Purification and characterization of rhodopsin kinase. J. Biol. Chem. 1988;263:14067–14073. [PubMed] [Google Scholar]
- 74.Kachi S, Nishizawa Y, Olshevskaya E, Yamazaki A, Miyake Y, Wakabayashi T, Dizhoor A, Usukura J. Detailed localization of photoreceptor guanylate cyclase activating protein-1 and -2 in mammalian retinas using light and electron microscopy. Exp. Eye Res. 1999;68:465–473. doi: 10.1006/exer.1998.0629. [DOI] [PubMed] [Google Scholar]
- 75.Cuenca N, Lopez S, Howes K, Kolb H. The localization of guanylyl cyclase-activating proteins in the mammalian retina. Investig. Ophthalmol. Vis. Sci. 1998;39:1243–1250. [PubMed] [Google Scholar]
- 76.Hallett MA, Delaat JL, Arikawa K, Schlamp CL, Kong F, Williams DS. Distribution of guanylate cyclase within photoreceptor outer segments. J. Cell Sci. 1996;109:1803–1812. doi: 10.1242/jcs.109.7.1803. [DOI] [PubMed] [Google Scholar]
- 77.Applebury ML, Antoch MP, Baxter LC, Chun LL, Falk JD, Farhangfar F, Kage K, Krzystolik MG, Lyass LA, Robbins JT. The murine cone photoreceptor: a single cone type expresses both S and M opsins with retinal spatial patterning. Neuron. 2000;27:513–523. doi: 10.1016/s0896-6273(00)00062-3. [DOI] [PubMed] [Google Scholar]
- 78.Farber DB. From mice to men: the cyclic GMP phosphodiesterase gene in vision and disease. The Proctor Lecture. Investig. Ophthalmol. Vis. Sci. 1995;36:263–275. [PubMed] [Google Scholar]
- 79.Hsu SC, Molday RS. Glycolytic enzymes and a GLUT-1 glucose transporter in the outer segments of rod and cone photoreceptor cells. J. Biol. Chem. 1991;266:21745–21752. [PubMed] [Google Scholar]
- 80.Maw MA, Corbeil D, Koch J, Hellwig A, Wilson-Wheeler JC, Bridges RJ, Kumaramanickavel G, John S, Nancarrow D, Roper K, Weigmann, Huttner WB, Denton MJ. A frameshift mutation in prominin (mouse)-like 1 causes human retinal degeneration. Hum. Mol. Genet. 2000;9:27–34. doi: 10.1093/hmg/9.1.27. [DOI] [PubMed] [Google Scholar]
- 81.Hu G, Wensel TG. R9AP, a membrane anchor for the photoreceptor GTPase accelerating protein, RGS9–1. Proc. Natl. Acad. Sci. U. S. A. 2002;99:9755–9760. doi: 10.1073/pnas.152094799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Molday RS, Hicks D, Molday L. Peripherin. A rim-specific membrane protein of rod outer segment discs. Investig. Ophthalmol. Vis. Sci. 1987;28:50–61. [PubMed] [Google Scholar]
- 83.He W, Cowan CW, Wensel TG. RGS9, a GTPase accelerator for phototransduction. Neuron. 1998;20:95–102. doi: 10.1016/s0896-6273(00)80437-7. [DOI] [PubMed] [Google Scholar]
- 84.Bascom RA, Manara S, Collins L, Molday RS, Kalnins VI, McInnes RR. Cloning of the cDNA for a novel photoreceptor membrane protein (rom-1) identifies a disk rim protein family implicated in human retinopathies. Neuron. 1992;8:1171–1184. doi: 10.1016/0896-6273(92)90137-3. [DOI] [PubMed] [Google Scholar]
- 85.Molday LL, Hicks D, Sauer CG, Weber BH, Molday RS. Expression of X-linked retinoschisis protein RS1 in photoreceptor and bipolar cells. Investig. Ophthalmol. Vis. Sci. 2001;42:816–825. [PubMed] [Google Scholar]
- 86.Broekhuyse RM, Leunissen JL, Verkley AJ. Ultrastructural localization of S-antigen in retinal structures. Curr. Eye Res. 1985;4:73–77. doi: 10.3109/02713688508999970. [DOI] [PubMed] [Google Scholar]
- 87.Reiners J, van Wijk E, Marker T, Zimmermann U, Jurgens K, Te BH, Overlack N, Roepman R, Knipper M, Kremer H, Wolfrum U. Scaffold protein harmonin (USH1C) provides molecular links between Usher syndrome type 1 and type 2. Hum. Mol. Genet. 2005;14:3933–3943. doi: 10.1093/hmg/ddi417. [DOI] [PubMed] [Google Scholar]
- 88.Chang B, Khanna H, Hawes N, Jimeno D, He S, Lillo C, Parapuram SK, Cheng H, Scott A, Hurd RE, Sayer JA, Otto EA, Attanasio M, O'Toole JF, Jin G, Shou C, Hildebrandt F, Williams DS, Heckenlively JR, Swaroop A. In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet. 2006;15:1847–1857. doi: 10.1093/hmg/ddl107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Otto EA, Loeys B, Khanna H, Hellemans J, Sudbrak R, Fan S, Muerb U, O'Toole JF, Helou J, Attanasio M, Utsch B, Sayer JA, Lillo C, Jimeno D, Coucke P, De PA, Reinhardt R, Klages S, Tsuda M, Kawakami I, Kusakabe T, Omran H, Imm A, Tippens M, Raymond PA, Hill J, Beales P, He S, Kispert A, Margolis B, Williams DS, Swaroop A, Hildebrandt F. Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior-Loken syndrome and interacts with RPGR and calmodulin. Nat. Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 90.Roepman R, Letteboer SJ, Arts HH, van Beersum SE, Lu X, Krieger E, Ferreira PA, Cremers FP. Interaction of nephrocystin-4 and RPGRIP1 is disrupted by nephronophthisis or Leber congenital amaurosis-associated mutations. Proc. Natl. Acad. Sci. U. S. A. 2005;102:18520–18525. doi: 10.1073/pnas.0505774102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hong DH, Yue G, Adamian M, Li T. Retinitis pigmentosa GTPase regulator (RPGRr)-interacting protein is stably associated with the photoreceptor ciliary axoneme and anchors RPGR to the connecting cilium. J. Biol. Chem. 2001;276:12091–12099. doi: 10.1074/jbc.M009351200. [DOI] [PubMed] [Google Scholar]
- 92.Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, Mah AK, Johnsen RC, Cavender JC, Lewis RA, Leroux MR, Beales PL, Katsanis N. Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature. 2003;425:628–633. doi: 10.1038/nature02030. [DOI] [PubMed] [Google Scholar]
- 93.Hagstrom SA, Duyao M, North MA, Li T. Retinal degeneration in tulp1−/− mice: vesicular accumulation in the interphotoreceptor matrix. Investig. Ophthalmol. Vis. Sci. 1999;40:2795–2802. [PubMed] [Google Scholar]
- 94.Schmitt A, Wolfrum U. Identification of novel molecular components of the photoreceptor connecting cilium by immunoscreens. Exp. Eye Res. 2001;73:837–849. doi: 10.1006/exer.2001.1086. [DOI] [PubMed] [Google Scholar]
- 95.Efimenko E, Bubb K, Mak HY, Holzman T, Leroux MR, Ruvkun G, Thomas JH, Swoboda P. Analysis of xbx genes in C. elegans. Development. 2005;132:1923–1934. doi: 10.1242/dev.01775. [DOI] [PubMed] [Google Scholar]
- 96.Blacque OE, Perens EA, Boroevich KA, Inglis PN, Li C, Warner A, Khattra J, Holt RA, Ou G, Mah AK, McKay SJ, Huang P, Swoboda P, Jones SJ, Marra MA, Baillie DL, Moerman DG, Shaham S, Leroux MR. Functional genomics of the cilium, a sensory organelle. Curr. Biol. 2005;15:935–941. doi: 10.1016/j.cub.2005.04.059. [DOI] [PubMed] [Google Scholar]
- 97.Stolc V, Samanta MP, Tongprasit W, Marshall WF. Genome-wide transcriptional analysis of flagellar regeneration in Chlamydomonas reinhardtii identifies orthologs of ciliary disease genes. Proc. Natl. Acad. Sci. U. S. A. 2005;102:3703–3707. doi: 10.1073/pnas.0408358102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hearn T, Spalluto C, Phillips VJ, Renforth GL, Copin N, Hanley NA, Wilson DI. Subcellular localization of ALMS1 supports involvement of centrosome and basal body dysfunction in the pathogenesis of obesity, insulin resistance, and type 2 diabetes. Diabetes. 2005;54:1581–1587. doi: 10.2337/diabetes.54.5.1581. [DOI] [PubMed] [Google Scholar]
- 99.Fan Y, Esmail MA, Ansley SJ, Blacque OE, Boroevich K, Ross AJ, Moore SJ, Badano JL, May-Simera H, Compton DS, Green JS, Lewis RA, van Haelst MM, Parfrey PS, Baillie DL, Beales PL, Katsanis N, Davidson WS, Leroux MR. Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat. Genet. 2004;36:989–993. doi: 10.1038/ng1414. [DOI] [PubMed] [Google Scholar]
- 100.Kim JC, Badano JL, Sibold S, Esmail MA, Hill J, Hoskins BE, Leitch CC, Venner K, Ansley SJ, Ross AJ, Leroux MR, Katsanis N, Beales PL. The Bardet-Biedl protein BBS4 targets cargo to the pericentriolar region and is required for microtubule anchoring and cell cycle progression. Nat. Genet. 2004;36:462–470. doi: 10.1038/ng1352. [DOI] [PubMed] [Google Scholar]
- 101.Wu H, Cowing JA, Michaelides M, Wilkie SE, Jeffery G, Jenkins SA, Mester V, Bird AC, Robson AG, Holder GE, Moore AT, Hunt DM, Webster AR. Mutations in the gene KCNV2 encoding a voltage-gated potassium channel subunit cause “cone dystrophy with supernormal rod electroretinogram” in humans. Am. J. Hum. Genet. 2006;79:574–579. doi: 10.1086/507568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kyttala M, Tallila J, Salonen R, Kopra O, Kohlschmidt N, Paavola-Sakki P, Peltonen L, Kestila M. MKS1, encoding a component of the flagellar apparatus basal body proteome, is mutated in Meckel syndrome. Nat. Genet. 2006;38:155–157. doi: 10.1038/ng1714. [DOI] [PubMed] [Google Scholar]
- 103.Nishimura DY, Swiderski RE, Searby CC, Berg EM, Ferguson AL, Hennekam R, Merin S, Weleber RG, Biesecker LG, Stone EM, Sheffield VC. Comparative genomics and gene expression analysis identifies BBS9, a new Bardet-Biedl syndrome gene. Am. J. Hum. Genet. 2005;77:1021–1033. doi: 10.1086/498323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Otto EA, Schermer B, Obara T, O'Toole JF, Hiller KS, Mueller AM, Ruf RG, Hoefele J, Beekmann F, Landau D, Foreman JW, Goodship JA, Strachan T, Kispert A, Wolf MT, Gagnadoux MF, Nivet H, Antignac C, Walz G, Drummond IA, Benzing T, Hildebrandt F. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat. Genet. 2003;34:413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chiang AP, Beck JS, Yen HJ, Tayeh MK, Scheetz TE, Swiderski RE, Nishimura DY, Braun TA, Kim KY, Huang J, Elbedour K, Carmi R, Slusarski DC, Casavant TL, Stone EM, Sheffield VC. Homozygosity mapping with SNP arrays identifies TRIM32, an E3 ubiquitin ligase, as a Bardet-Biedl syndrome gene (BBS11) Proc. Natl. Acad. Sci. U. S. A. 2006;103:6287–6292. doi: 10.1073/pnas.0600158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cryns K, Thys S, Van Laer L, Oka Y, Pfister M, Van Nassauw L, Smith RJ, Timmermans JP, Van Camp G. The WFS1 gene, responsible for low frequency sensorineural hearing loss and Wolfram syndrome, is expressed in a variety of inner ear cells. Histochem. Cell Biol. 2003;119:247–256. doi: 10.1007/s00418-003-0495-6. [DOI] [PubMed] [Google Scholar]
- 107.Whitehead JL, Wang SY, Bost-Usinger L, Hoang E, Frazer KA, Burnside B. Photoreceptor localization of the KIF3A and KIF3B subunits of the heterotrimeric microtubule motor kinesin II in vertebrate retina. Exp. Eye Res. 1999;69:491–503. doi: 10.1006/exer.1999.0724. [DOI] [PubMed] [Google Scholar]
- 108.Mikami A, Tynan SH, Hama T, Luby-Phelps K, Saito T, Crandall JE, Besharse JC, Vallee RB. Molecular structure of cytoplasmic dynein 2 and its distribution in neuronal and ciliated cells. J. Cell Sci. 2002;115:4801–4808. doi: 10.1242/jcs.00168. [DOI] [PubMed] [Google Scholar]
- 109.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, Bohme U, Hannick L, Aslett MA, Shallom J, Marcello L, Hou L, Wickstead B, Alsmark UC, Arrowsmith C, Atkin RJ, Barron AJ, Bringaud F, Brooks K, Carrington M, Cherevach I, Chillingworth TJ, Churcher C, Clark LN, Corton CH, Cronin A, Davies RM, Doggett J, Djikeng A, Feldblyum T, Field MC, Fraser A, Goodhead I, Hance Z, Harper D, Harris BR, Hauser H, Hostetler J, Ivens A, Jagels K, Johnson D, Johnson J, Jones K, Kerhornou AX, Koo H, Larke N, Landfear S, Larkin C, Leech V, Line A, Lord A, MacLeod A, Mooney PJ, Moule S, Martin DM, Morgan GW, Mungall K, Norbertczak H, Ormond D, Pai G, Peacock CS, Peterson J, Quail MA, Rabbinowitsch E, Rajandream MA, Reitter C, Salzberg SL, Sanders M, Schobel S, Sharp S, Simmonds M, Simpson AJ, Tallon L, Turner CM, Tait A, Tivey AR, Van Aken S, Walker D, Wanless D, Wang S, White B, White O, Whitehead S, Woodward J, Wortman J, Adams MD, Embley TM, Gull K, Ullu E, Barry JD, Fairlamb AH, Opperdoes F, Barrell BG, Donelson JE, Hall N, Fraser CM, Melville SE, El-Sayed NM. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 110.Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Jr., Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA, Krieger CJ, del Toro C, Ryder HF, Williamson SC, Barbeau RA, Hamilton EP, Orias E. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.