Abstract
Purpose
The TP53 tumor suppressor gene encodes a sequence-specific transcription factor that is able to transactivate several sets of genes, the promoters of which include appropriate response elements. Although human cancers frequently contain mutated p53, the alleles as well as the clinical expression are often heterogeneous. Germ line mutations of TP53 result in cancer proneness syndromes known as Li-Fraumeni, Li-Fraumeni – like, and nonsyndromic predisposition with or without family history. p53 mutants can be classified as partial deficiency alleles or severe deficiency alleles depending on their ability to transactivate a set of human target sequences, as measured using a standardized yeast-based assay (see http://www.umd.be:2072/index.html).We have investigated the extent to which the functional features of p53 mutant alleles determine clinical features in patients who have inherited these alleles and have developed cancer.
Experimental Design
We retrieved clinical data from the IARC database (see http://www.p53.iarc.fr/Germline.html) for all cancer patients with germline p53 mutations and applied stringent statistical evaluations to compare the functional classification of p53 alleles with clinical phenotypes.
Results
Our analyses reveal that partial deficiency alleles are associated with a milder family history (P = 0.007), a lower numbers of tumors (P = 0.007), and a delayed disease onset (median, 31 versus 15 years; P = 0.007) which could be related to distinct tumor spectra.
Conclusions
These findings establish for the first time significant correlations between the residual transactivation function of individual TP53 alleles and clinical variables in patients with inherited p53 mutations who develop cancer.
The TP53 tumor suppressor gene (chromosome 17p13; OMIM no. 191170) encodes a protein involved in many pathways that control cellular responses to various stress signals (1–3). The p53 protein is a sequence-specific transcription factor constitutively expressed in most cell types and is activated by genotoxic stress, mainly at a posttranslational level, to transactivate effector genes from target response elements (REs). The disruption of the p53 pathway is found in almost all tumor types, predominantly through mutation of the TP53 gene itself. In contrast with other tumor suppressor genes, TP53 mutations are usually of the missense type in one allele, generally but not necessarily followed by loss of the second allele during tumor progression. About 50% of all tumor types harbor p53 mutations (4, 5). The high number of somatic missense mutations found in the DNA-binding domain in tumors, and the high number of different single amino acid changes which they produce (~ 1,300), suggests that p53 function is extremely sensitive to perturbation and that there is selection for cells expressing a mutant protein by virtue of its specific functionality. The latter possibility could reflect either a dominant-negative effect produced by an abnormal p53 allele due to the fact that p53 is a tetramer (1), or a gain of novel functional properties (6, 7). Many cancer-associated p53 mutations do not result in complete loss of function. Instead, there is considerable heterogeneity, as individual mutant proteins may have lost some wild-type functions while still retaining (or acquiring) others (6–17).
Germ line p53 mutations have provided formal proof that p53 has a major role in the development of cancer. Based on the clinical expression of p53 mutations in heterozygotes, the following cancer proneness syndromes (in order of decreasing severity) have been identified: Li-Fraumeni syndrome (LFS), Li-Fraumeni–like syndrome (LFL), and nonsyndromic predispositions with or without family history (FH and noFH, respectively; refs. 18, 19). The spectrum of p53 germ line mutations is wide, with 92 different missense alleles described in the IARC database (5).5 Based on their ability to transactivate a set of human target sequences, these missense p53 mutants can be classified as partial deficiency (PD) alleles or severe deficiency (SD) alleles (see details in Materials and Methods and in Results). In addition, 52 p53 mutants could be classified as obligate SD (O-SD) alleles by virtue of nonsense mutations, frameshifts, or other mutations that give rise to a truncated protein.
Several studies have indicated that specific mutations in p53 may affect the type of tumor and the age of onset, and that these effects may correlate with structure-based groupings of p53 mutations (19–21). On the other hand, the effect of the functional heterogeneity of p53 mutations on the severity of associated diseases has not been assessed. In contrast with other inherited cancer syndromes, which are predominantly characterized by site-specific cancers, LFS presents with a variety of tumor types (19). The seven most frequent tumor types (described below) account for ~ 72% of the reported cases.
We have investigated the extent to which a systematic functional classification of all germ line p53 alleles can predict clinical features in patients with inherited p53 mutations who develop cancer. Clinical data were retrieved from the IARC database (5).5 Our results, based on stringent statistical analyses, show a significant correlation between the residual transactivation function of individual p53 alleles and the development of cancer.
Materials and Methods
Data set of p53 transactivation capacities
We retrieved functional data from the literature.6 The data set contains a summary of transactivation capabilities towards eight different p53 REs (p21/WAF1, MDM2, BAX, 14-3-3σ, p53AIP1, GADD45, NOXA, and p53R2) that are upstream of a common reporter. The EGFP or DsRed reporter genes provided quantitative analyses of the transcriptional capability for each mutant. We were given access to an early version of the database, in which transcriptional activity towards each RE was divided into four classes: class I >75%; class II ≥50% and <75%; class III ≥25% and <50%; class IV <25%; (see Acknowledgement). According to these results, we have classified a mutant as SD if there was <25% of wild-type activity on every RE. The PD alleles were defined as those that showed ≥25% of wild-type activity toward at least one RE. A summary functional score for each of the 92 alleles on eight REs (or for just the three apoptotic REs) is provided in Supplementary Table S1A (SD alleles) and B (PD alleles). For some analyses, PD alleles were further divided based on the number of REs towards which they showed ≥25% of wild-type transactivation activity (Supplementary Table S2A-C).
Clinical definitions
Classic LFS is defined as a proband with a sarcoma before the age of 45 years and a first-degree relative with any cancer before the age of 45 years plus an additional first- or second-degree relative in the same lineage with any cancer before the age of 45 years or a sarcoma at any age (18). This definition has been relaxed to define LFL cases (22). LFL syndrome is defined as a proband with any childhood cancer, or a sarcoma, brain tumor, or adrenocortical tumor before the age of 45 years, plus a first- or second-degree relative in the same lineage with a typical LFS tumor at any age, plus an additional first- or second-degree relative in the same lineage with any cancer before the age of 60 years. Twenty to 40% of LFL families harbor mutations in the TP53 gene (22, 23). FH refers to family history of cancer that does not fulfill LFS or any of the LFL definitions, and noFH refers to no family history of cancer.
IARC database
This relational database contains information on families with LFS/LFL syndromes and those that do not fulfill the clinical definitions of LFS/LFL (i.e., FH and noFH), although they carry a germ line mutation in the TP53 gene. Data are available on family members with cancer that are either TP53 mutation carriers or that have not been examined for their p53 allele status, as well as on family structure, tumor samples, details on the germ line mutation, mutation detection method, and the publication in which the family is described. Details of annotations can be found at the IARC web site.5 Clinical data were downloaded from the database without additional curating, with the exceptions noted in Supplementary Table S1A and B.
Statistical tests
Statistical comparisons were done with nonparametric tests (Fisher exact test) and with the within-cluster resampling approach (24). The P values are given in the text.
In the analysis of the data from the IARC database, it is necessary to take into account two key features of the data. The first is the lack of statistical independence among tumors (because individuals may have multiple tumors) and among individuals (because families may have multiple individuals). Because observations accrue to the database by voluntary submission, not by some defined probability-based sampling plan, subtle and unknown biases may be introduced that cannot be fully accounted for in any statistical analysis. One potential source of bias that we wanted to be especially cautious about, however, was the possibility that the number of affected members in a family or the number of distinct cancers affecting an individual (which could influence the chance that a family or individual might enter the database) might be associated with one of the clinical outcomes of interest such as tumor type or age at diagnosis. In the statistical literature, this potential source of bias is called “informative cluster size” (here, a family is a cluster).
We regarded families as statistically independent. For testing hypotheses about characteristics of families (e.g., clinical class, proportion of families in which at least one individual had multiple tumors), we used Fisher exact test which is appropriate when families are independent. To avoid dependence among tumors, we analyzed each individual’s first tumor only. For testing hypotheses about the characteristics of individuals (e.g., age at diagnosis and tissue site of first tumor), we used the within-cluster resampling approach (24), which uses data from every individual but gives equal weight to each family. This approach appropriately adjusts for within-family correlations and, in addition, protects against biases that might accrue from informative cluster size. The basic idea is to sample one individual from each family at random and compute the desired estimate (proportion, mean, etc.). The process is repeated many times allowing different individuals in the families represented by multiple individuals to all enter the analysis but in different repeated samples; individuals from families represented by only one individual enter in every repeated sample. The final estimate is the mean of the estimates from each repeated sample. The variance of this estimate is computed as previously described (24). This method can be adapted for estimation and testing with continuous variables like age or with categorical variables like tumor site. Test statistics were computed from within-cluster resampling estimates of differences in outcome (proportion, mean, etc.) between functional classes divided by the corresponding estimated standard error, and compared with the normal distribution for assessing P values.
Results
The functionality of p53 mutants has been extensively investigated using transactivation assays based in yeast, in which p53 REs were placed upstream of reporter genes (12, 13, 17, 25, 26). Overall, there was good agreement between the results obtained in yeast and in human cells for a large proportion of p53 alleles (27). The largest yeast-based systematic analysis examined >2,000 mutations, including all the germ line p53 alleles, using eight REs and quantitative fluorescent reporters (16, 28).6 Based on that data set, we classified the germ line p53 mutants as PD alleles if trans-activation function was retained towards at least one target sequence. The remaining p53 mutants were classified as SD or O-SD alleles if the mutations resulted in truncations. According to this derived functional classification, 43 of the alleles were PD and 49 were SD. The 52 O-SD alleles were a control group in the sense that they were assumed to have a complete loss of transactivation activity.
p53 functionality was used to query clinical data including the site of the tumor, occurrence of multiple tumors in the same individual, and confirmed inheritance of a germ line p53 mutation in the public IARC germ line p53 mutant database.5 We limited the data set to the seven most frequent tumor types, i.e., soft tissue (connective) sarcomas and osteosarcomas (bones), breast cancer, brain tumors, hematopoietic tumors (leukemia + lymphoma), adrenocortical carcinoma, and bronchus/lung cancer. These account for ~ 72% of the reported cases. The remaining cases were heterogeneous with very few observed tumors in more than 20 different tissues and/or tumor types. This restriction on tissue targets excluded two PD p53 alleles (N210Y and S227T) from consideration as they were associated with only five rare tumors according to the IARC database. We have also excluded from the analyses individuals with inherited R337H, a unique PD allele, which is unusually frequent in the Brazilian population and seems to predispose mainly to adrenocortical carcinomas in children. A complete summary of the data retrieved from the IARC database for SD and PD alleles is available in Supplementary Table S1A and B, respectively.
First, we examined whether p53 functional status correlates with the distribution of clinical classes. In FH families, the frequency of PD p53 alleles was much higher than that of SD alleles (18 of 58 versus 15 of 119; P = 0.007, Fisher exact test). The opposite was found for families with full-blown LFS (13 of 58 versus 51 of 119; P = 0.009; Table 1A). The patterns were similar when the data were presented in terms of affected individuals and tumors. Because the PD group is heterogeneous, in that it comprises p53 alleles retaining some function toward at least one and up to eight REs, we also considered more homogeneous PD subgroups (see Supplementary Table S2A-C). PD alleles with lower functionality were more similar to the SD group of alleles, whereas PD alleles with higher residual functionality were enriched in the less severe clinical classes.
Table 1.
Effect of different functional subsets of p53 mutant alleles on severity of cancer risk
| (A) Alleles classified with respect to transactivation of all target genes | |||||
|---|---|---|---|---|---|
| Functional class
|
PD
|
SD
|
O-SD
|
||
| No. of p53 alleles
|
40* |
49
|
52
|
||
| Clinical classes | Families† | ||||
| Not available‡ | 2 (3.4) | 5 (4.2) | 1 (1.5) | ||
| noFH‡ | 8 (13.8) | 19 (16.0) | 9 (13.2) | ||
| FH§ | 18 (31.0) | 15 (12.6) | 9 (13.2) | ||
| LFL‡ | 17 (29.3) | 29 (24.4) | 20 (29.4) | ||
| LFS|| | 13 (22.4) | 51 (42.9) | 29 (42.6) | ||
| Total no. of families | 58 (100) | 119 (100) | 68 (100) | ||
| Clinical classes | Individuals† | ||||
|
| |||||
| Not available | 2 (1.2) | 5 (1.6) | 1 (0.4) | ||
| noFH | 8 (4.6) | 19 (6.2) | 9 (3.5) | ||
| FH | 31 (17.9) | 35 (11.4) | 19 (7.4) | ||
| LFL | 60 (34.7) | 80 (26.0) | 76 (29.6) | ||
| LFS | 72 (41.6) | 169 (54.9) | 152 (59.1) | ||
| Total no. of individuals | 173 (100) | 308 (100) | 257 (100) | ||
| Clinical classes | Tumors† | ||||
|
| |||||
| Not available | 4 (2.0) | 8 (1.9) | 1 (0.3) | ||
| noFH | 10 (5.1) | 33 (8.0) | 16 (5.1) | ||
| FH | 37 (18.6) | 41 (9.9) | 20 (6.3) | ||
| LFL | 70 (35.2) | 110 (26.5) | 89 (28.3) | ||
| LFS | 78 (39.2) | 223 (53.7) | 189 (60.0) | ||
| Total no. of tumors | 199 (100) | 415 (100) | 315 (100) | ||
| (B) Alleles classified with respect to transactivation of apoptotic genes only | |||||
|
| |||||
| Functional class | PD for apoptotic REs | SD for apoptotic REs | |||
|
| |||||
| No. of p53 alleles | 32 | 57 | |||
|
| |||||
| Clinical classes | Families¶ | ||||
|
| |||||
| Not available** | 2 (5.4) | 5 (3.6) | |||
| noFH** | 7 (18.9) | 20 (14.3) | |||
| FH†† | 13 (35.1) | 20 (14.3) | |||
| LFL** | 11 (29.7) | 35 (25.0) | |||
| LFS‡‡ | 4 (10.8) | 60 (42.9) | |||
| Total no. | 37 (100) | 140 (100) | |||
| Clinical classes | Individuals¶ | ||||
|
| |||||
| Not available | 2 (2.5) | 5 (1.3) | |||
| noFH | 7 (8.6) | 20 (5.0) | |||
| FH | 23 (28.4) | 43 (10.8) | |||
| LFL | 39 (48.1) | 101 (25.2) | |||
| LFS | 10 (12.3) | 231 (57.8) | |||
| Total no. | 81 (100) | 400 (100) | |||
| Clinical classes | Tumors¶ | ||||
|
| |||||
| Not available | 4 (4.1) | 8 (1.6) | |||
| noFH | 9 (9.2) | 34 (6.6) | |||
| FH | 27 (27.6) | 51 (9.9) | |||
| LFL | 47 (48.0) | 133 (25.8) | |||
| LFS | 11 (11.2) | 290 (56.2) | |||
| Total no. | 98 (100) | 516 (100) | |||
| (C) Risk of multiple tumors | |||||
|
| |||||
| Functional class (no.) | PD | SD | O-SD | PDa§§ | SDa§§ |
|
| |||||
| Families | 58 | 119 | 68 | 37 | 140 |
| Families in which at least one individual has multiple tumors|| || | 19 | 66 | 28 | 12 | 73 |
| Individuals | 173 | 308 | 257 | 81 | 400 |
| Individuals with multiple tumors¶¶ | 20 | 84 | 45 | 13 | 91 |
Of the 43 PD alleles, three were excluded because they were associated with only five rare tumors (N210Y, S227T) or seemed to predispose only to pediatric adrenocortical carcinomas (R337H).
Numbers (and percentages) of families, individuals, and tumors carrying germ line PD, SD, and O-SD p53 alleles. Rounding off may prevent percentages from summing exactly to a value of 1. Clinical class is a characteristic of families in that the class definition may depend in part on the number of affected family members and every affected individual in a family is assigned the same clinical class. Consequently, families are the appropriate units for statistical analyses of how the distribution of the clinical classes may depend on mutation functional class. We report P values only for analyses based on families, but display the distributions for individuals and for tumors for completeness.
Percentage of families in clinical classes NA, noFH, and LFL did not differ significantly among those carrying PD, SD, or O-SD alleles (all P ≥ 0.42).
A significantly higher percentage of FH families carried PD alleles than carried SD or O-SD alleles (P = 0.007 versus SD; P = 0.02 versus O-SD; Fisher’s exact test) but the percentage of FH families that carried SD and O-SD alleles did not differ significantly (P =1.0).
A significantly lower percentage of LFS families carried PD alleles than carried SD or O-SD alleles (P = 0.009 versus SD; P = 0.03 versus O-SD; Fisher’s exact test) but the percentage of LFS families that carried SD and O-SD alleles do not differ significantly (P = 1.0).
Numbers (and percentage) of families, individuals, and tumors carrying germ line PD and SD p53 alleles. Rounding off may result in percentages not summing precisely to a value of 1. Clinical class is a characteristic of families in that the class definition may depend in part on the number of affected family members and every affected individual in a family is assigned the same clinical class. Consequently, families are the appropriate units for statistical analysis of how the distribution of the clinical classes may depend on mutation functional class. We report P values only for analyses based on families, but display the distributions for individuals and for tumors for completeness.
Percentage of families in clinical classes NA, noFH, and LFL did not differ significantly between those carrying PD alleles for apoptotic REs and those carrying SD alleles for apoptotic REs (all P ≥ 0.61).
A significantly higher percentage of FH families carried PD alleles for apoptotic REs than carried SD alleles for apoptotic REs (P = 0.008).
A significantly lower percentage of LFS families carried PD alleles for apoptotic REs than carried SD alleles for apoptotic REs (P = 0.0003).
PD or SD alleles for apoptotic REs.
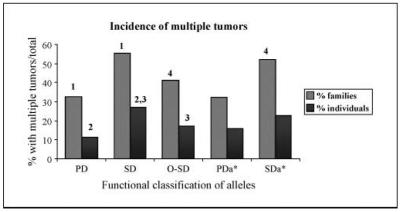
Families in which at least one individual had multiple tumors more commonly carried SD than PD alleles (P = 0.007) and more commonly carried SD alleles for apoptotic REs than PD alleles for apoptotic REs (P = 0.05; Fisher’s exact test). Comparisons between O-SD and either PD (P = 0.36) or SD (P = 0.07) were not statistically significant.
Individuals with multiple tumors more commonly carried SD than PD or O-SD alleles (P = 0.002 and 0.0004, respectively; within-cluster resampling test; ref. 24). The comparisons between PD and O-SD alleles (P = 0.89) and between SD alleles for apoptotic REs and PD alleles for apoptotic REs (P = 0.12) were not statistically significant.
Next, we inquired whether clinical expression correlates with the target-specific transactivating functions of p53. Because the induction of apoptosis is one of the key roles of p53 (1), we grouped alleles based on their activity on apoptotic REs (PDa and SDa; Table 1B). The resulting PDa group was enriched for highly functional PD p53 alleles (see “score” column in Supplementary Table S1B), whereas the SDa group contained weak PD alleles in addition to the SD alleles. Nevertheless, the previously observed differences in the distribution of clinical classes were maintained, particularly with respect to LFS.
Consistent with these findings, we observed that the incidence of individuals and their families that developed multiple tumors was lower with PD alleles than with SD alleles (Table 1C; Fig. 1). The difference was even more striking when the analysis was limited only to confirmed heterozygotes (designated as “carriers” in the IARC database; Supplementary Table S3; P = 0.002). Similar differences were also observed with the subgroups of PD alleles stratified according to the functional score (Supplementary Table S2C).
Fig. 1.
Effect of different functional subsets of p53 mutant alleles on the risk of multiple tumors. Percentage of families with at least one member that has developed multiple tumors over the total number of families (hatched columns); percentage of individuals that developed multiple tumors over the total number of individuals (solid columns) for each of the indicated p53 mutant classes (see the footnote to Table 1C for details). PDa* and SDa*, classification based on results with apoptotic REs: 1, P = 0.007; 2, P = 0.002; 3, P = 0.004; 4, P = 0.05.
One of the features that most dramatically illustrates the cancer risk of subjects who have inherited a TP53 mutation is the early age at which cancer appears, and this feature is also reflected in mouse models of LFS (20, 21). In our analysis (Table 2; Fig. 2) we found a marked difference between patients carrying PD and SD alleles in terms of the age at which their first cancer was diagnosed (median, 31 versus 15 years; P = 0.0007). This result was confirmed with the various subgroups of PD alleles compared with the SD group (data not shown). Patients with O-SD alleles are intermediate between these two groups. If we further focus the analysis on p53 function as it affects proapoptotic genes, a similar pattern is found (Table 2).
Table 2.
Effect of p53 functional status on age at diagnosis in confirmed carriers considering only the first tumor
| p53 functional status
|
|||||
|---|---|---|---|---|---|
| PD | SD | O-SD | PDa* | SDa* | |
| No. | |||||
| Individuals | 80 | 151 | 104 | 40 | 191 |
| Families | 48 | 106 | 57 | 30 | 124 |
| Mean | |||||
| All individuals | 29.8 | 19.0 | 23.1 | 30.2 | 21.2 |
| Adjusted for family† | 26.9 | 18.2 | 21.8 | 26.1 | 19.7 |
| 95% confidence limits† | 21.6–32.3 | 15.6–20.9 | 18.3–25.4 | 18.0–34.1 | 17.2–22.1 |
| Median | |||||
| All individuals | 31 | 15 | 25 | 31.5 | 22 |
| Adjusted for family† | 29 | 14 | 24 | 27 | 17 |
| 95% confidence limits† | 21.5–36.4 | 9.9–18.1 | 15.3–32.7 | 13.0–41.0 | 11.8–22.2 |
| P values† (above diagonal for comparison of means and below for comparison of medians) | |||||
| PD | — | 0.004 | 0.12 | — | — |
| SD | 0.0007 | — | 0.11 | 0.07 | — |
| O-SD | 0.36 | 0.07 | — | 0.34 | 0.33 |
| PDa | — | 0.10 | 0.81 | — | 0.14 |
| SDa | — | — | 0.18 | 0.19 | — |
NOTE: Age at diagnosis was available for only a subset of individuals in the IARC database and the missing data could bias these comparisons.
For apoptotic REs.
Calculated using the within-cluster resampling method (24).
Fig. 2.
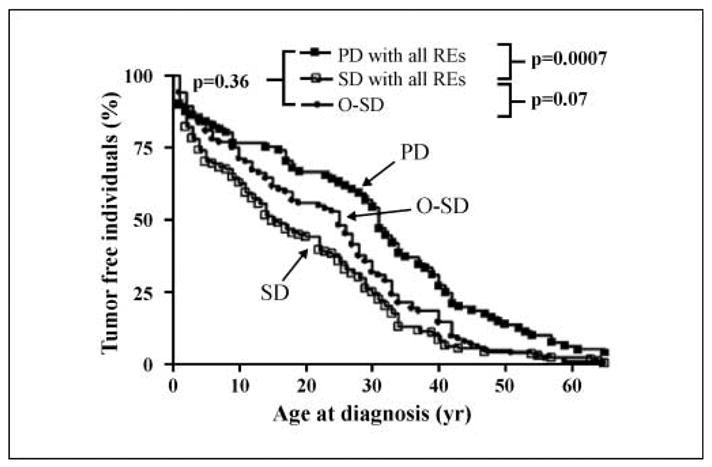
p53 functionality and clinical variables: age at diagnosis for the overall tumor spectrum. The percentage of tumor-free individuals is plotted as a function of age up to age 65 (Kaplan-Meier method). The reported P values are the results of a within-cluster resampling analysis (ref. 24; see Materials and Methods). The analyses were restricted to confirmed germline carriers whose age at diagnosis was recorded in the database. For patients with multiple tumors, only the first tumor was considered because secondary malignancies might be influenced by the nature of the tissue, history, and therapeutic interventions on the first tumor. All p53 REs were considered. The number of individuals and families represented in the figure are provided in Table 2.
A higher incidence of breast cancer (P = 0.05, within-cluster resampling; ref. 24) and lower incidence of connective tissue tumors (P = 0.0006, within-cluster resampling; ref. 24) and bone tumors (P = 0.07, within-cluster resampling; ref. 24) were observed with PD alleles (Fig. 3). This change in the tumor spectrum could be related to the differences in the age at diagnosis.
Fig. 3.
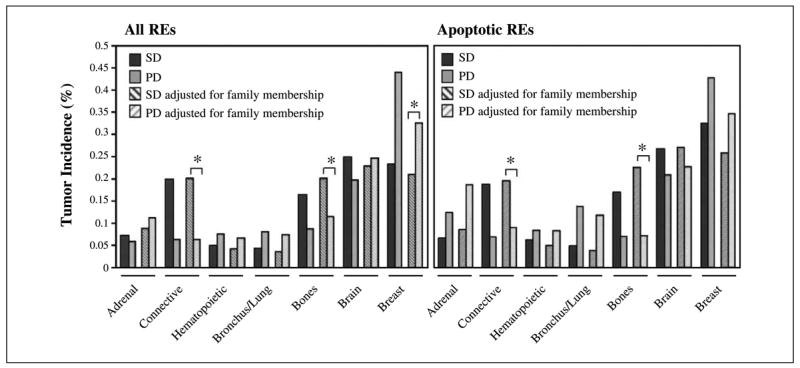
p53 functionality and variation in tissue specificity. Variation in tissue specificity in relation to p53 functional status versus all, or versus the apoptotic REs. p53 mutant alleles were divided into two groups (PD and SD) and the distribution (%) of tumors in the seven most frequent tissue targets is shown (solid columns). For each tissue, estimates of tumor frequencies that are adjusted for family membership are also presented (hatched columns). These estimates give equal weight to each family. The family-based estimates can be larger or smaller than the individual-based estimates (first two columns). Statistical significance is provided only for comparisons of family-based estimates because those tests properly account for possible correlations between family members. Considering all p53 REs and family-based estimates, partial functionality is associated with higher incidence of breast cancer (*, P = 0.05) and with lower incidence of connective (*, P = 0.0006) and bone (*, P = 0.07) tumors (within-cluster resampling; ref. 24). These differences were confirmed considering apoptotic REs for connective (*, P = 0.05) and bone (*, P = 0.05) tumors but not for breast cancer (P = 0.27). A nonsignificant tendency for higher incidence of bronchus/lung tumors with partial functionality, observed with all REs (P = 0.18), approached statistical significance when apoptotic REs were considered (P = 0.08). Adrenal, brain, and hematopoietic tumors showed little evidence of differences across functional classes (minimum, P = 0.13).
Discussion
There are 92 different p53 missense germ line alleles in the IARC database. These mutations are associated with clinically distinct syndromic and nonsyndromic cancer proneness. No clear-cut association between pattern of mutations and clinical manifestations have been established thus far. In fact, mutations in different protein domains can result in the same clinically defined grouping; whereas in different families, the same mutation can give different clinical outcomes (see Supplementary Table S1A and B).6 For example, the Arg175His allele is present in 14 families, of which 11 are LFS/LFL and 3 are FH/noFH. Similar results are seen with Arg248Trp (of 10 families, 9 are LFS/LFL). This may be explained by the fact that multiple genes and biological pathways as well as complex gene/environment interactions are involved in cancer development. Furthermore, within a particular pedigree, the time (or generations) after the appearance of a p53 germ line mutation could influence disease expression (29). Functional polymorphisms both in coding and in regulatory sequences of the p53 gene itself, of p53 target genes and of genes involved in modulating p53 activity (e.g., MDM2) could also modify p53-related responses (30–33).
In seeking possible correlations between p53 mutations affecting different structural domains and clinical features in LFS families, the IARC database curators (19) have shown that in individuals with a p53 mutation, brain tumors were associated with missense p53 mutations located in the DNA-binding loop that contact the minor groove of DNA (P = 0.01), whereas adrenal gland carcinomas were associated with missense mutations located in the loops opposite to the protein-DNA contact surface (P = 0.003).
Our approach has been to consider the functional heterogeneity of p53 alleles, determined by standardized functional assays, as a means of addressing genotype-phenotype correlations in familial cancers. We used experimental functionality data available on all 92 reported missense germ line p53 alleles (16, 28)6 and simply divided mutants into two categories (PD and SD) using criteria described in Materials and Methods. The PD and SD alleles were distributed, with different frequencies, in all structural regions of the DNA-binding domains (Supplementary Fig. S1). We found that p53 mutant functionality identifies groups of familial cancer patients with different clinical features and outcomes. Overall, the PD p53 alleles are preferentially associated with a milder family history of cancer (P = 0.007, Fisher exact test; Table 1A), a lower risk of developing multiple tumors (Table 1C; Fig. 1), a tendency towards delayed disease onset (Table 2; Fig. 2), and a higher relative risk for breast cancer (P = 0.05, within-cluster resampling; ref. 24; Fig. 3). Within the PD group, alleles with a nearly total loss of function behave almost like SD alleles, whereas alleles with higher residual functions were associated with a milder family history (Table 1B; Supplementary Table S2B and C).
Two mouse models of LFS have been recently reported (20, 21). One group (21) engineered p53R172H/+ and p53R270H/+ mice (corresponding to the SD human hotspots R175H and R273H, respectively). Interestingly, these two groups of animals developed allele-specific tumor spectra which were different from that seen in heterozygous p53+/− mice. Allele-specific effects were also observed in experiments with derived primary cells. These differences cannot be attributed merely to the loss of transactivation (21). In contrast, in a different strain of mice, there was no difference in the tumor spectrum between p53R172H/R172H and p53−/− mice (20), indicating that the phenotype is highly dependent on the overall genetic background. The same must be true in humans because families with identical germ line mutations can present different clinical syndromes (e.g., LFS, LFL, FH, and noFH). The importance of p53 mutant functionality in determining clinical features can be inferred when the results with the p53R172P/R172P knock-in mice are considered (34). In contrast with R172H, the R172P mutant is a PD allele. Using the survival of p53−/− mice as a reference, p53R172P/R172P mice showed a much higher survival due to reduced tumor burden [see Fig. 4A in ref. 34] than p53R172H/R172H [see Fig. 2A in ref. 20]. Thus, the combined results obtained using the “knock-in” mice models are clearly supportive of our observations on different factors, including the intrinsic functional heterogeneity of p53 mutants, modulating clinical outcomes in p53 germ line carriers.
The comparison between the clinical features of individuals and families having SD and O-SD p53 alleles may also help to clarify whether p53 mutants merely act as tumor suppressor genes. Our analyses revealed that SD and O-SD alleles were similarly distributed in the different clinical classes (P = 1; Table 1A). However, individuals (and families) with O-SD alleles tend to show a lower incidence of multiple tumors (for individuals, P = 0.0004; for families, P = 0.07; Table 1C) and a trend for a delayed disease onset (median, 25 versus 15 years; P = 0.07; Table 2; Fig. 2) with respect to those with SD alleles. These observations could be explained simply by the level of residual functional p53 tetramers (haploinsufficiency); however, they do not exclude the possibility that at least some p53 alleles may behave as oncogenes (7).
Compared with other inherited disorders, those associated with p53 mutations have an added level of complexity because somatic mutations must occur for the disease phenotype to develop. This work shows that, despite this complexity, genotype-phenotype correlations can be pinpointed, particularly if the functional features of mutant alleles are appropriately analyzed. Indeed, the functional characteristics of p53 germinal mutations seem to be a predictor of disease expression in terms of age of onset and number of tumors. These findings have clinical implications because it is clear that regardless of their initial syndromic classification, subjects with SD alleles are at greater risk, suggesting a more cautious approach to clinical management.
Supplementary Material
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
Acknowledgments
We thank Dr. Thierry Soussi for granting access to an early version of the p53 database.
This work is dedicated to Olga Cattaneo Fronza with love.
Grant support: Associazione Italiana per la Ricerca sul Cancro, Allenza Contro il Cancro; FIRB, the Intramural Research Program of the NIH, and NIEHS. Yari Ciribilli was working under an FIRC fellowship. The research of M.A. Resnick and J. Jordan was supported by intramural research funds from the National Institute of Environmental Health Sciences, NIH.
Footnotes
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Soussi T, Lozano G. p53 mutation heterogeneity in cancer. Biochem Biophys Res Commun. 2005;331:834–42. doi: 10.1016/j.bbrc.2005.03.190. [DOI] [PubMed] [Google Scholar]
- 3.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 4.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–78. [PubMed] [Google Scholar]
- 5.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–14. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 6.Bossi G, Lapi E, Strano S, Rinaldo C, Blandino G, Sacchi A. Mutant p53 gain of function: reduction of tumor malignancy of human cancer cell lines through abrogation of mutant p53 expression. Oncogene. 2006;25:304–9. doi: 10.1038/sj.onc.1209026. [DOI] [PubMed] [Google Scholar]
- 7.Di Agostino S, Strano S, Emiliozzi V, et al. Gain of function of mutant p53: the mutant p53/NF-Yprotein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 8.Rowan S, Ludwig RL, Haupt Y, et al. Specific loss of apoptotic but not cell-cycle arrest function in a human tumor derived p53 mutant. EMBOJ. 1996;15:827–38. [PMC free article] [PubMed] [Google Scholar]
- 9.Ludwig RL, Bates S, Vousden KH. Differential activation of target cellular promoters by p53 mutants with impaired apoptotic function. Mol Cell Biol. 1996;16:4952–60. doi: 10.1128/mcb.16.9.4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monti P, Campomenosi P, Ciribilli Y, et al. Tumour p53 mutations exhibit promoter selective dominance over wild type p53. Oncogene. 2002;21:1641–8. doi: 10.1038/sj.onc.1205250. [DOI] [PubMed] [Google Scholar]
- 11.Monti P, Campomenosi P, Ciribilli Y, et al. Characterization of the p53 mutants ability to inhibit p73 β transactivation using a yeast-based functional assay. Oncogene. 2003;22:5252–60. doi: 10.1038/sj.onc.1206511. [DOI] [PubMed] [Google Scholar]
- 12.Campomenosi P, Monti P, Aprile A, et al. p53 mutants can often transactivate promoters containing a p21 but not Bax or PIG3 responsive elements. Oncogene. 2001;20:3573–9. doi: 10.1038/sj.onc.1204468. [DOI] [PubMed] [Google Scholar]
- 13.Inga A, Monti P, Fronza G, Darden T, Resnick MA. p53 mutants exhibiting enhanced transcriptional activation and altered promoter selectivity are revealed using a sensitive, yeast-based functional assay. Oncogene. 2001;20:501–13. doi: 10.1038/sj.onc.1204116. [DOI] [PubMed] [Google Scholar]
- 14.Inga A, Resnick MA. Novel human p53 mutations that are toxic to yeast can enhance transactivation of specific promoters and reactivate tumor p53 mutants. Oncogene. 2001;20:3409–19. doi: 10.1038/sj.onc.1204457. [DOI] [PubMed] [Google Scholar]
- 15.Inga A, Storici F, Darden TA, Resnick MA. Differential transactivation by the p53 transcription factor is highly dependent on p53 level and promoter target sequence. Mol Cell Biol. 2002;22:8612–25. doi: 10.1128/MCB.22.24.8612-8625.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kato S, Han SY, Liu W, et al. Understanding the function-structure and function-mutation relationships of p53 tumor suppressor protein by high-resolution missense mutation analysis. Proc Natl Acad Sci U S A. 2003;100:8424–9. doi: 10.1073/pnas.1431692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Resnick MA, Inga A. Functional mutants of the sequence-specific transcription factor p53 and implications for master genes of diversity. Proc Natl Acad Sci US A. 2003;100:9934–9. doi: 10.1073/pnas.1633803100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li FP, Fraumeni JF, Jr, Mulvihill JJ, et al. A cancer family syndrome in twenty-four kindreds. Cancer Res. 1988;48:5358–62. [PubMed] [Google Scholar]
- 19.Olivier M, Goldgar DE, Sodha N, et al. Li-Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer Res. 2003;63:6643–50. [PubMed] [Google Scholar]
- 20.Lang GA, Iwakuma T, Suh YA, et al. Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell. 2004;119:861–72. doi: 10.1016/j.cell.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 21.Olive KP, Tuveson DA, Ruhe ZC, et al. Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell. 2004;119:847–60. doi: 10.1016/j.cell.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 22.Birch JM, Hartley AL, Tricker KJ, et al. Prevalence and diversity of constitutional mutations in the p53 gene among 21 Li-Fraumeni families. Cancer Res. 1994;54:1298–304. [PubMed] [Google Scholar]
- 23.Evans DG, Birch JM, Thorneycroft M, McGown G, Lalloo F, Varley JM. Low rate of TP53 germline mutations in breast cancer/sarcoma families not fulfilling classical criteria for Li-Fraumeni syndrome. J Med Genet. 2002;39:941–4. doi: 10.1136/jmg.39.12.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hoffman EB, Sen PK, Winberg CR. Within-cluster resampling. Biometrika. 2001;88:1121–34. [Google Scholar]
- 25.Flaman JM, Robert V, Lenglet S, Moreau V, Iggo R, Frebourg T. Identification of human p53 mutations with differential effects on the bax and p21 promoters using functional assays in yeast. Oncogene. 1998;16:1369–72. doi: 10.1038/sj.onc.1201889. [DOI] [PubMed] [Google Scholar]
- 26.Di Como CJ, Prives C. Human tumor-derived p53 proteins exhibit binding site selectivity and temperature sensitivity for transactivation in a yeast-based assay. Oncogene. 1998;16:2527–39. doi: 10.1038/sj.onc.1202041. [DOI] [PubMed] [Google Scholar]
- 27.Kakudo Y, Shibata H, Otsuka K, Kato S, Ishioka C. Lack of correlation between p53-dependent transcriptional activity and the ability to induce apoptosis among 179 mutant p53s. Cancer Res. 2005;65:2108–14. doi: 10.1158/0008-5472.CAN-04-2935. [DOI] [PubMed] [Google Scholar]
- 28.Hamroun D, Kato S, Ishioka C, Claustres M, Beroud C, Soussi T. The UMD TP53 database and website: update and revisions. Hum Mutat. 2006;27:14–20. doi: 10.1002/humu.20269. [DOI] [PubMed] [Google Scholar]
- 29.Brown BW, Costello TJ, Hwang SJ, Strong LC. Generation or birth cohort effect on cancer risk in Li-Fraumeni syndrome. Hum Genet. 2005;118:489–98. doi: 10.1007/s00439-005-0016-x. [DOI] [PubMed] [Google Scholar]
- 30.Tomso DJ, Inga A, Menendez D, et al. Functionally distinct polymorphic sequences in the human genome that are targets for p53 transactivation. Proc Natl Acad Sci US A. 2005;102:6431–6. doi: 10.1073/pnas.0501721102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bond GL, Hu W, Bond EE, et al. A single nucleotide polymorphism in the MDM2 promoter attenuates the p53 tumor suppressor pathway and accelerates tumor formation in humans. Cell. 2004;119:591–602. doi: 10.1016/j.cell.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 32.Menendez D, Krysiak O, Inga A, Krysiak B, Resnick MA, Schonfelder G. A SNP in the flt-1 promoter integrates the VEGF system into the p53 transcriptional network. Proc Natl Acad Sci U S A. 2006;103:1406–11. doi: 10.1073/pnas.0508103103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bougeard G, Baert-Desurmont S, Tournier I, et al. Impact of the MDM2 SNP309 and p53 Arg72Pro polymorphism on age of tumour onset in Li-Fraumeni syndrome. J Med Genet. 2006;43:531–3. doi: 10.1136/jmg.2005.037952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu G, Parant JM, Lang G, et al. Chromosome stability, in the absence of apoptosis, is critical for suppression of tumorigenesis in Trp53 mutant mice. Nat Genet. 2004;36:63–8. doi: 10.1038/ng1282. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).