Abstract
Numerous plant RNA viruses have associated with them satellite (sat) RNAs that have little or no nucleotide sequence similarity to either the viral or host genomes but are completely dependent on the helper virus for replication. We report here on the discovery of a 682-nt circular DNA satellite associated with tomato leaf curl geminivirus (TLCV) infection in northern Australia. This is the first demonstration that satellite molecules are not limited to RNA viral systems. The DNA satellite (TLCV sat-DNA) is strictly dependent for replication on the helper virus replication-associated protein and is encapsidated by TLCV coat protein. It has no significant open reading frames, and it shows no significant sequence similarity to the 2766-nt helper-virus genome except for two short motifs present in separate putative stem–loop structures: TAATATTAC, which is universally conserved in all geminiviruses, and AATCGGTGTC, which is identical to a putative replication-associated protein binding motif in TLCV. Replication of TLCV sat-DNA is also supported by other taxonomically distinct geminiviruses, including tomato yellow leaf curl virus, African cassava mosaic virus, and beet curly top virus. Therefore, this unique DNA satellite does not appear to strictly conform with the requirements that dictate the specificity of interaction of geminiviral replication-associated proteins with their cognate origins as predicted by the current model of geminivirus replication.
Keywords: geminivirus, tomato leaf curl virus, DNA satellite
Subviral agents, viroids, and satellites are well known in infectious RNA systems (1–4). Apart from viroids, which are autonomously replicating, numerous plant RNA viruses (1, 2) have associated with them RNA satellites that show little or no nucleotide sequence similarity to the viral or host genomes, but which are completely dependent on the helper virus for replication. Unlike satellite viruses, which encode the genetic information for their own coat protein, RNA satellites are packaged in coat protein encoded by the helper virus. Thus, particles containing RNA satellites are antigenically indistinguishable from those of the helper virus.
Both linear and circular satellites are found to be associated with plant RNA viruses (4). Some satellites have been found to exacerbate viral symptoms or induce symptoms quite distinct from the those induced by the helper virus alone (2). However, a number of them interfere with helper virus replication and ameliorate disease expression, which has led to considerable interest into the investigation of their potential as sources of viral resistance (5).
More than 30 RNA viruses are currently recognized as having one or more satellites associated with them (2). However, to date, similar DNA species associated with DNA viruses have not been recorded. We now report on the isolation and characterization of a small, circular DNA satellite associated with a geminivirus. Geminiviruses are a group of plant viruses characterized by their twin-shaped isometric particles; these viruses are responsible for major crop losses worldwide. Their genomes consist of either one or two circular single-stranded DNA components of approximately 2.5–3.0 kb in size (6). Tomato leaf curl virus (TLCV) belongs to geminivirus subgroup III, the members of which are characterized by either monopartite or bipartite genomes, transmission by the whitefly Bemisia tabaci, and by the infection of dicotyledonous hosts.
Small subgenomic DNA species are often associated with geminivirus infection (7). These DNAs are derived by a partial deletion of the wild-type viral genome and, as such, show a high degree of sequence homology to the helper virus. In contrast, the small circular DNA molecule found to be associated with TLCV infection (which we describe here) is not derived from the helper virus genome, and it displays characteristics analogous to the viral RNA satellites. This type of molecule has not been found to be associated with any other geminivirus or, indeed, any other plant, animal, or bacterial DNA virus.
MATERIALS AND METHODS
Construction of Virus Clones.
Construction of infectious clones of the geminiviruses TLCV, TLCV-D1 strain, tomato yellow leaf curl virus (TYLCV)-Sardinian strain, African cassava mosaic virus (ACMV) (A and B), and beet curly top virus (BCTV) have been described previously (8–12). TLCV mutant constructs were produced as described in Rigden et al. (13).
Cloning and Manipulation of TLCV sat-DNA (DNA-2).
DNA-2 was purified from TLCV-infected tomato material as described previously for TLCV (8) but included a treatment with RNase A. Purified DNA-2 was treated with Klenow fragment in the presence of random decamers (Bresatec, Adelaide, Australia) to convert any single-stranded DNA (ssDNA) to double-stranded DNA (dsDNA) and digested with RsaI. The two resultant fragments (150 and 532 nt) were cloned into EcoRV-cut Bluescript SK+ (Strategene) and sequenced. A unique NcoI site was identified within the sequence and was used to reclone the same DNA. Sequence comparison of the NcoI clone with the RsaI clones confirmed it to be a full length circular monomer of DNA-2.
Site-directed mutagenesis of TLCV sat-DNA was carried out using an Altered Sites kit (Promega). Wild-type and mutated TLCV sat-DNA constructs were cloned into pBin19 as either NcoI dimers or NcoI/SpeI 1.3-mers and then introduced into Agrobacterium tumefaciens (strain C58) by electroporation.
Whole Plant Infectivity Assays.
Agroinoculation was performed with an overnight culture of A. tumefaciens containing tandem–repeat constructs in pBin19 (8). Bacterial cultures bearing TLCV- and TLCV sat-DNA- encoding plasmids were mixed in equal proportions for coagroinoculation experiments. New developing leaves were sampled 21–28 days postinoculation, and the presence of replicative DNA forms was detected either by dot–blot analysis (14) or Southern blot analysis as described below.
Leaf Strip Transient Assay.
The transient replication assay was modified as described in ref. 15. Leaf strips (1–2 mm) cut from in vitro grown tobacco plants (Nicotiana tabacum cv. Samsun) were cocultivated with A. tumefaciens on 0.7% bacto-agar plates containing culture medium (0.43% Murashige and Skoog basal salt mixture, 3% sucrose, 0.01% Gamborg vitamins, 0.0001% benzyladenine, 0.00001% naphthaleneacetic acid) in the dark at 25°C. After 48 h, strips were transferred to 25 ml of culture medium supplemented with 0.025% kanamycin and 0.05% cefotaxime and incubated for 6 days at 25°C in normal room light with gentle agitation at 50 rpm. Strips (150 mg) were blotted dry and frozen in liquid N2 prior to DNA extraction.
Extraction and Analysis of DNA.
Total nucleic acid was extracted as described previously (8) but included a treatment with RNase A. Inoculation sites were excised from the stem as described (16). DNA species were separated (5 μg/lane) in 1.8% agarose gels containing 0.5 μg⋅ml−1 ethidium bromide to facilitate separation of the ssDNA and dsDNA forms. Unless otherwise stated, DNA was blotted and hybridized with 32P-labeled DNA probes as described (13).
Immunocapture PCR.
Datura (Datura stramonium L.) leaves were extracted in 10 vol of 50 mM Tris⋅HCl (pH 8.0), 5 mM EDTA, 2% polyvinylpyrrolidone, and 0.05% Tween 20 by grinding and centrifugation at 13,000 × g for 15 min at 4°C. TLCV particles were captured using a polyclonal antibody against the coat protein of a serologically related geminivirus, ACMV (17). DNA was released from each microtitre well (as described in ref. 18) in a total volume of 20 μl; 1 μl of which was used for 30 cycles of PCR amplification (19) using TLCV-specific primers (nt positions 605 to 632 and 1626 to 1605) or TLCV sat-DNA-specific primers (nt positions 504 to 485 and 505 to 522), which produce PCR products of 1021 and 682 bp, respectively.
Sequence Analysis.
Sequence alignments were carried out using the programs GAP, BESTFIT and DOTPLOT from the University of Wisconsin Genetics Computer Group (20).
RESULTS AND DISCUSSION
Isolation and Characterization of a DNA Satellite of TLCV.
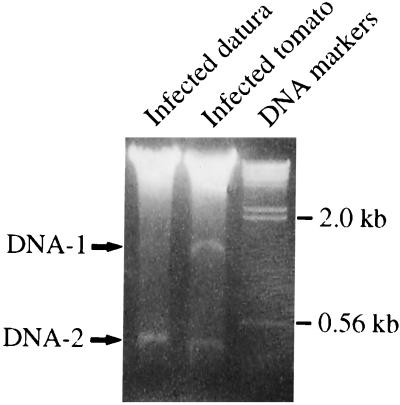
Leaf curl virus-infected tomato plants were collected from a field near Darwin, Australia and grafted onto tomato (Lycopersicon esculentum Mill. cv. Grosse Lisse) and Datura stocks. Leaf curl symptoms were observed on host plants 4–6 weeks after grafting. Electrophoretic separation of DNA extracts of infected leaf tissue revealed the presence of two distinct bands designated DNA-1 and DNA-2 (Fig. 1) in addition to the plant genomic DNA. We have previously shown DNA-1 to be the 2766-nt circular DNA genome of TLCV (8). Southern blot analysis with a TLCV probe did not reveal any cross-hybridization with DNA-2 (data not shown), indicating that DNA-2 was not derived from the TLCV genome but may represent a previously undescribed DNA species associated with TLCV infection.
Figure 1.
Isolation and characterization of a circular 682-nt DNA molecule associated with TLCV infection. Ethidium bromide-stained 1.2% agarose gel showing the presence of two DNA species (arrows) in TLCV-infected plant material.
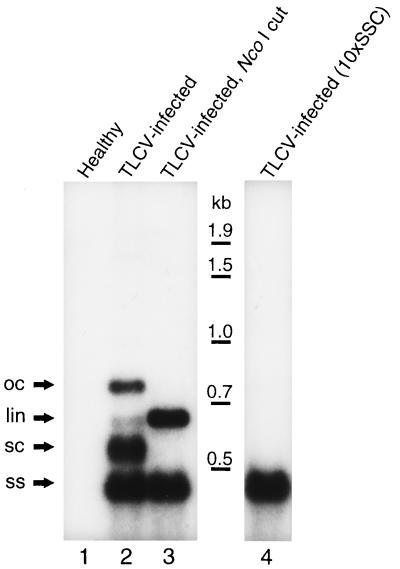
The cloning and sequencing of DNA-2 (purified from TLCV-infected tomato tissue) revealed it to be a circular molecule of 682 nt. This clone detected replicative forms of DNA-2, but not TLCV, in TLCV-infected tomato tissues (Fig. 2, lane 2). No hybridization was observed with this clone against total nucleic acid extracted from healthy plants (Fig. 2, lane 1). Both double-stranded (ds) and single-stranded (ss) forms of DNA-2 were found to be present in infected tissues (Fig. 2 compare lanes 2–4).
Figure 2.
Detection of DNA-2 replicative forms in TLCV-infected tomato. Identification of open circular dsDNA (oc), linear dsDNA (lin), supercoiled dsDNA (sc) and ssDNA (ss) forms was confirmed by digestion of total nucleic acid extracts with NcoI that linearized the oc and sc forms but had no effect on the ssDNA (compare lanes 2 and 3), and blotting of samples under nondenaturing conditions with 10 × SSC resulting in specific detection of ssDNA but not dsDNA (compare lanes 2 and 4).
The complete sequence of the 682-nt DNA-2 molecule is shown in Fig. 3. Global sequence alignment of DNA-2 with TLCV using the program GAP did not reveal any extensive similarity, confirming that it is not a subgenomic DNA species of TLCV. Examination of the DNA-2 sequence did, however, reveal the presence of a putative stem–loop structure formed by 11-nt GC-rich inverted repeats between nt 670-680 and nt 10-20 containing the loop sequence TAATATTAC. This nonanucleotide sequence element, within a putative stem–loop structure, is universally conserved in geminiviruses (6) and strongly suggested that DNA-2 may be a geminivirus-associated molecule.
Figure 3.
Nucleotide sequence of DNA-2. Numbering begins with the first base (T) of the geminivirus conserved nonanucleotide sequence element (boldface). Inverted repeats forming a putative stem–loop structure are shown with arrows.
Searches of the nonredundant nucleotide database with the DNA-2 sequence did not reveal any significant matches with any existing sequences. Analysis of open reading frames (ORF) indicated the presence of a number of small ORFs (i.e., 30–70 aa), but RNA Polymerase II promoter elements required to initiate transcription appear to be absent. This suggested that DNA-2 is not transcriptionally active, and it may rely on geminiviral and/or host factors for replication and systemic spread.
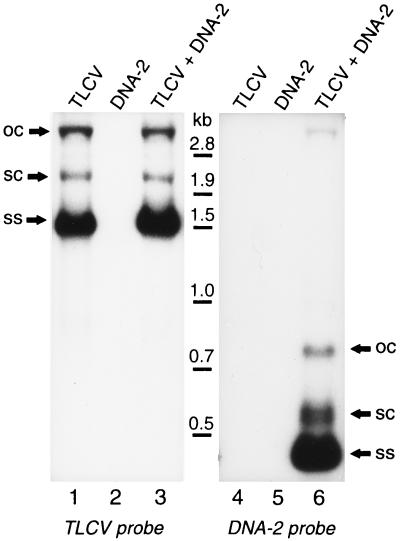
To test this hypothesis, a dimeric clone of DNA-2 was constructed and introduced into the binary vector pBin19 for agroinoculation of Datura plants in the presence and absence of an infectious dimeric clone of TLCV (8). Plants agroinoculated with TLCV or TLCV + DNA-2 displayed typical severe symptoms of leaf curl infection after 21 days, whereas plants inoculated with DNA-2 alone remained healthy. Southern blot analysis revealed that plants inoculated with TLCV alone or with TLCV + DNA-2 contained ds and ss circular forms of TLCV DNA (Fig. 4, lanes 1, 3). However, replicative intermediates of DNA-2 were only detected in the leaves of plants agroinoculated with DNA-2 in the presence of TLCV(Fig. 4, lane 6). Also, there was no evidence of DNA-2 replication in tissue at the inoculation sites of plants inoculated with DNA-2 alone (data not shown). These results demonstrate that replication and systemic spread of DNA-2 is absolutely dependent on the presence of the helper virus TLCV. This, together with the lack of sequence similarity between DNA-2 and the helper virus, shows strong parallels between the properties of DNA-2 and the RNA satellites (1). On the basis of this evidence, we propose DNA-2 to be a DNA satellite of TLCV and have therefore named it TLCV sat-DNA.
Figure 4.
Analysis of replicating DNA species in plants agroinoculated with cloned DNA-2 in the presence and absence of TLCV. Datura plants were agroinoculated with either a dimeric TLCV clone (pTLCVBIN1; 8), a dimeric clone of DNA-2, or both as described in the text. Replicative DNA forms are labeled as shown in Fig. 2.
Although many viral RNA satellites have been identified and characterized (1, 2), no comparable DNA satellite has previously been described. Bacteriophage P4 has been classified as a DNA satellite (4), but in direct contrast to the characteristics of RNA satellites, it is a large (11.6 kb) self-replicating phage containing up to 13 functional genes, which is capable of maintaining itself as a multicopy plasmid in infected host cells, and only requires a helper phage for lytic growth (21). Thus, TLCV sat-DNA represents the first DNA satellite to be described, for any viral system, that displays the fundamental characteristics of the RNA satellites of plant viruses.
TLCV Gene Products Required for Replication and Encapsidation of TLCV sat-DNA.
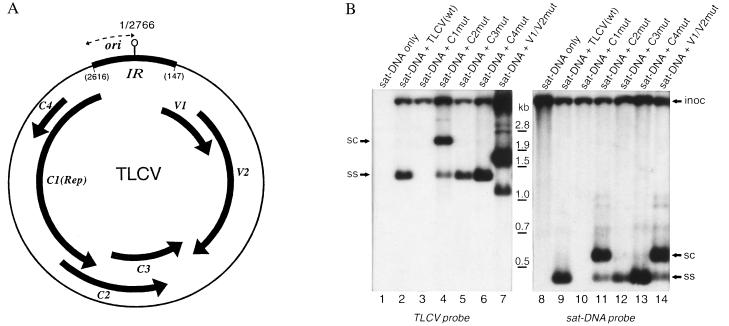
The TLCV genome contains six functional genes (Fig. 5A) involved in viral infection. Of these, only the C1 gene encoding the replication-associated protein is absolutely required for viral replication (13). To examine the interaction between each of these viral gene products and the satellite, a transient replication assay using a tobacco leaf strip explant system was developed. This assay examined satellite replication and encapsidation with a range of TLCV mutants in which specific viral gene products had been disrupted (13). No satellite replication was observed in the absence of TLCV (Fig. 5B, lane 8) or in the presence of a C1(Rep)-TLCV mutant (Fig. 5B, lane 10), confirming the absolute dependence of satellite replication on TLCV replication. TLCV sat-DNA replication was not, however, dependent on the presence of any of the other five TLCV gene products (Fig. 5B, lanes 11–14), although it can be seen that changes in the relative levels of ss and ds forms of satellite DNA mirror those of the helper virus in response to mutation of specific TLCV gene products. Thus, inhibition of virion-sense gene products (i.e., V1 & V2, Fig. 5A), either by direct mutation (Fig. 5B, lanes 7 and 14) or indirectly through mutation of the C2 gene product (Fig. 5B, lanes 4 and 11) that is thought to regulate virion-sense gene expression (22), leads to a reduction in the level of ssDNA and an increase in the level of dsDNA replicative forms of both TLCV and the satellite. This result suggests that satellite DNA may be encapsidated by TLCV coat protein, and is in agreement with vector transmission studies showing cotransmission of TLCV and TLCV sat-DNA by B. tabaci (data not shown) that involves recognition of the viral capsid protein.
Figure 5.
Role of TLCV gene products in replication and encapsidation of TLCV sat-DNA. A, Genome organization of TLCV. Functional ORFs are displayed as thick arrows. The position of the conserved stem–loop structure (|○), intergenic region (IR), and the putative ori (↔) are marked. B, Southern blot of total nucleic acid extracts of tobacco leaf strips coagroinoculated with a dimeric TLCV sat-DNA construct and various TLCV ORF mutant constructs. Replicative DNA forms are labeled as in Fig. 2. High Mr DNA detected at the top of each blot represents inoculum (inoc) DNA isolated from residual A. tumefaciens on the leaf strips at the time of extraction. Note the TLCV V1/V2 mutant involves a 510-nt deletion resulting in the smaller sc and ssDNA forms observed in lane 7.
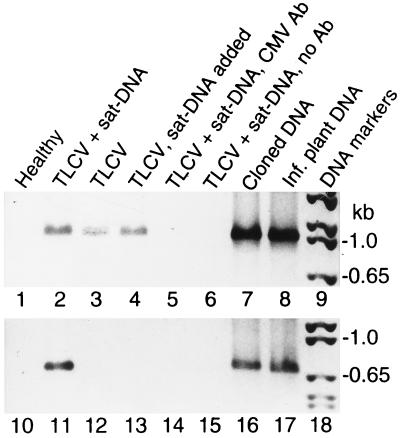
Direct evidence of encapsidation of satellite DNA by TLCV coat protein was obtained with an immuno-capture PCR technique (18) using a polyclonal antibody directed against TLCV coat protein. Both TLCV and satellite DNA were found to be present in particles immunocaptured from extracts of infected plants (Fig. 6, lanes 2 and 11). No satellite-specific PCR product was detected in extracts from plants inoculated with TLCV alone (lane 12) or in the same extracts that had been spiked with satellite DNA (lane 13), demonstrating the specificity of the immuno-capture step for encapsidated nucleic acid. These results confirm encapsidation of satellite DNA by TLCV coat protein.
Figure 6.
Encapsidation of TLCV sat-DNA by helper virus coat protein. TLCV and TLCV sat-DNA associated with encapsidated viral particles was detected by immunocapture PCR. PCR was carried out with TLCV-specific (lanes 1–8) or TLCV sat-DNA-specific primers (lanes 10–17). The negative photographic image of the ethidium-stained PCR products are shown. Lanes 1–6 and 10–15 show PCR products obtained with template immunocaptured from extracts of Datura plants agroinoculated with TLCV or TLCV+TLCV sat-DNA, respectively. Extract shown in lanes 4 and 13 was spiked prior to immunocapture with total nucleic acid containing TLCV sat-DNA extracted from twice the weight of tissue used for immunocapture. Negative controls consisted of extract immunocaptured with an unrelated antibody against cucumber mosaic virus coat protein (lanes 5 and 14) or with no antibody (lanes 6 and 15). Control PCR reactions were carried out with using either cloned DNA templates (lanes 7 and 16) or DNA isolated from TLCV-infected plants as described in Fig. 1 (lanes 8 and 17).
TLCV sat-DNA Contains a TLCV Rep-Binding Motif.
The geminiviral origin of replication (ori) has been mapped to the left side of the intergenic region (Fig. 5A), and it is thought to be composed of at least three functional modules (23). These include a putative stem–loop structure containing the conserved TAATATTAC motif that is the site of Rep-mediated cleavage (24), a high-affinity Rep-binding site that determines viral-specific recognition of its cognate origin, and at least one additional element, located between the Rep-binding and cleavage sites, that contributes to specific origin recognition by viral trans-acting factors. In the case of bipartite geminiviruses, which are composed of two distinct genomic components (A and B), replication of both components by the same Rep protein is achieved through the presence of a ∼200-nt “common” region comprising the intergenic region (6).
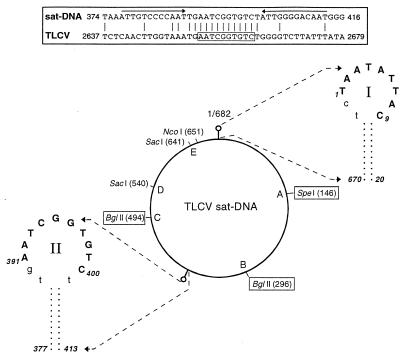
It is clear from the lack of sequence similarity between TLCV sat-DNA and TLCV that the satellite does not contain a TLCV-like intergenic region analogous to the common region of the bipartite geminiviruses, yet TLCV is clearly capable of supporting high levels of satellite replication (Fig. 4). To examine this further, a more sensitive sequence comparison between the complete TLCV sat-DNA genome and the TLCV intergenic region was carried out using the program DOTPLOT (20). This analysis revealed that, in addition to the presence of the conserved geminiviral stem–loop structure (Fig. 7, stem–loop I), TLCV sat-DNA contains a second, small stretch of 13 nucleotides between nt 389 and 401 that is identical to a stretch of sequence within the TLCV intergenic region (Fig. 7, Top). Significantly, this conserved sequence contains the motif AATCGGTGTC, which has been postulated to act as a specific Rep-binding or recognition domain within the ori of TLCV (25). This motif may also be involved in recognition and binding of TLCV Rep to the satellite.
Figure 7.
Identification of putative sequences involved in TLCV-supported satellite replication. Putative stem–loop structures (|○) are shown in expanded form to show the conserved geminiviral sequence motif in stem–loop I (nt 1–9, boldface) and the putative TLCV Rep-binding motif in stem–loop II (nt 391–400, boldface). Introduced restriction sites generated by site-directed mutagensis are boxed. Letters A–E refer to regions deleted or inverted in mutant satellite constructs as described in the text. Inset (Top) shows a region of sequence conservation between TLCV sat-DNA and the TLCV as detected by DOTPLOT analysis. Inverted repeats forming a putative stem–loop structure are shown with arrows. The putative Rep binding motif (25) within the TLCV intergenic region is boxed.
Geminiviruses infecting dicotyledonous plants, including TLCV, typically contain adjacent direct repeats of Rep-binding motifs within a discrete region ∼20 to 150 nt upstream of the conserved stem–loop structure (23, 25), and both repeats appear to be required for efficient high-affinity Rep binding (26). In contrast, the satellite appears to contain only a single putative TLCV Rep-binding motif; this is located ∼280 nt upstream of the Rep cleavage site within stem–loop I (Fig. 7). Furthermore, this motif is part of a second putative, highly stable (ΔG = −21.8 kcal mol−1) stem–loop structure within the satellite (Fig. 7, stem–loop II), which is absent in TLCV. The significance of the stem–loop II structure, in terms of Rep-binding and satellite replication, has yet to be determined. Local melting of dsDNA at the Rep-binding site, postulated to occur during viral replication (24), may induce extrusion and/or stabilization of stem–loop II, and this may act to bring the replication-associated protein into close proximity to the Rep cleavage site in stem–loop I.
A preliminary series of mutagenesis experiments were carried out to examine the role of other sequence elements within the satellite genome that may be important in TLCV-mediated satellite replication. Introduction of single-base mutations at positions A, B, and C (Fig. 7) were found to have no effect on satellite infectivity (i.e., 6 of 6 plants inoculated with each mutant were found to be systemically infected with TLCV sat-DNA in the presence of TLCV). Satellite replication was also maintained in constructs in which the A-B region had been deleted (5 of 5 plants infected). However, manipulation of sequences in the B-E region (nt 296–641), i.e. B-C deletion, B-C inversion, or D-E deletion, were found to completely inhibit satellite infectivity (0 of 6 plants infected for each separate mutant tested). The lack of TLCV-mediated replication of satellite constructs with mutations in the B-E region was confirmed using the leaf strip explant assay (data not shown), thus demonstrating that the lack of infectivity of these mutants in whole plant assays was due to a inhibition of replication, and not by an effect on satellite movement. These results strongly support a distributed organization of sequence elements important in replication of TLCV sat-DNA in contrast to the apparently compact organization of such elements in geminivirus genomic DNA molecules.
Rep-ori Specificity, TLCV sat-DNA Defies the Rule.
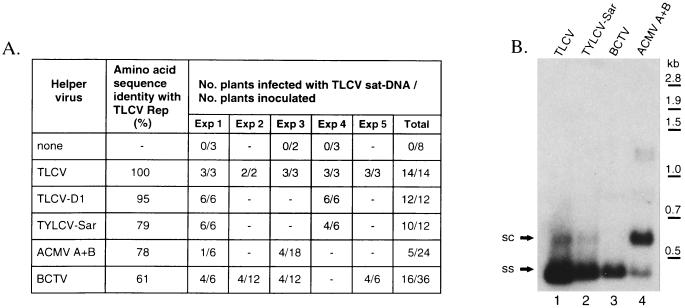
Given the marked differences in the organization of the TLCV and satellite oris, we examined whether satellite replication, unlike that of geminiviruses, could be supported by heterologous replication-associated proteins. Datura plants were co-agroinoculated with TLCV sat-DNA and each of a set of three dicot-infecting geminiviruses with replication-associated proteins of varying similarity to TLCV. Satellite replication was supported by each of the helper viruses tested (Fig. 8A), but it appeared to have little effect on the expression of symptoms associated with each geminivirus (data not shown). Although all agroinoculated plants developed virus symptoms, satellite inoculation efficiency varied markedly depending on the helper virus, ranging from 100% with the D1 isolate of TLCV down to 21% with ACMV (Fig. 8A). There was also a significant difference in the level of satellite DNA supported by the different viruses (Fig. 8B). Plants inoculated with the geminiviruses most closely related to TLCV, i.e. the TLCV-D1 strain (data not shown) and the TYLCV-Sardinian strain (Fig. 8B, lane 2), supported satellite DNA replication at levels approaching that observed in the presence of wild-type TLCV (Fig. 8B, lane 1). However, satellite levels were significantly lower in BCTV-infected plants (Fig. 8B, lane 3), ranging from 1 to 20% of TLCV controls, whereas ACMV (Fig. 8B, lane 4) was only capable of supporting satellite replication at levels approximating 1% or less of control plants. These results demonstrate that the replication of TLCV sat-DNA can be supported by heterologous replication-associated proteins from different geminiviruses.
Figure 8.
Replication of TLCV sat-DNA is supported by different geminiviruses encoding heterologous replication-associated proteins. Datura plants were agroinoculated with a dimeric clone of TLCV sat-DNA alone or together with infectious constructs of different helper geminiviruses: TLCV, TLCV-D1 strain, TYLCV-Sardinian strain, ACMV A + B or BCTV. All plants inoculated with these viruses developed disease symptoms. A, Summary of satellite infectivity experiments as determined by dot–blot analysis using a TLCV sat-DNA probe. Amino acid sequence identity between the TLCV Rep and the protein homologue of the other geminiviruses was calculated using GAP. B, Southern blot of total nucleic acid extracts of plants identified as positive for satellite DNA. Replicative forms of satellite DNA were detected with a TLCV sat-DNA probe. Due to the large variation in satellite DNA levels obtained with the different helper viruses, the relative loadings of total nucleic acid per lane were adjusted as follows: lanes 1 and 2 (0.75 μg); lane 3 (0.31 μg); lane 4 (5 μg) representing relative loadings of 1:1:16:66 for lanes 1–4, respectively. TLCV sat-DNA forms are labeled as in Fig. 2.
It is interesting to note that ratio of satellite dsDNA to ssDNA is markedly increased in ACMV-infected tissue (Fig. 8B, lane 4) compared with the other helper viruses (Fig. 8B, lanes 1–3). This increased ratio is similar to that observed in the presence of a TLCV V1/V2 mutant (Fig. 5B, lane 14) that does not support systemic spread (16), suggesting that movement of satellite DNA may not take place efficiently with ACMV as a helper virus. Unlike monopartite geminiviruses that require the coat protein for systemic spread (6), bipartite geminiviruses such as ACMV utilize specific movement proteins encoded on the B component for systemic spread. Such movement proteins may not support the efficient movement of the satellite DNA molecule, which has adapted to spread with the coat protein of a monopartite geminivirus.
Trans-replication of a single ori by a such a diverse group of heterologous replication-associated proteins has not been previously observed with any geminivirus system. Pseudorecombinants between different bipartite geminiviruses involving A component-supported replication of a heterologous B component have been observed, but only between strains of the same virus or between very closely related viruses with highly homologous common regions and identical Rep-binding motifs (26). The only exception to this has been a report of the generation of an infectious pseudorecombinant between two distinct bipartite geminiviruses, bean dwarf mosaic virus and tomato mottle virus (27). However, even in this case, a high degree of homology exists between the common regions of these two geminiviruses (27), and the predicted Rep-binding motifs of each virus diverge at only one nucleotide position (26).
It is tempting to speculate that the presence of a direct repeat of the sequence element TGGTGGT at nucleotide positions 653–659 and 662–668 (Fig. 3) may have a role in the capacity of TLCV sat-DNA to interact with a diverse group of heterologous replication-associated proteins. This repeated element shows a high degree of similarity to the conserved core sequence of putative Rep-binding motifs of a range of dicot-infecting geminiviruses, i.e., TGG(T/G/A)G including TYLCV-Sar, BCTV, and ACMV. The location of this GG-rich repeat immediately upstream of the conserved stem–loop structure also shows a high degree of similarity to the structural organization of geminiviral origins (23, 26). Although it is possible that this motif may be involved in binding TLCV Rep to the satellite, the results of the initial mutagenesis experiments clearly indicate that sequences located more distant from the conserved stem–loop structure (i.e., in the B-C region, Fig. 7) have a significant role in TLCV-mediated satellite replication.
The origin of this unique DNA satellite remains unknown. The capacity of TLCV sat-DNA to be replicated by a number of different geminiviruses demonstrates a degree of commonality that suggests an ancient origin for this subviral DNA species, more so than a recent evolutionary event specifically associated with TLCV infection in northern Australia. Helper viruses such as BCTV, for example, which clearly support TLCV sat-DNA replication, have never been detected in this country. If the satellite did arise early in evolution, it raises the question whether other, as yet undiscovered, satellite DNA-like molecules are associated with geminiviruses in other parts of the world.
Acknowledgments
We thank John Stanley for infectious clones of ACMV and BCTV, Bryan Harrison for the ACMV antibody, and Terri King for excellent technical assistance. This research was supported in part by Gene Shears Pty.
ABBREVIATIONS
- TLCV
tomato leaf curl virus
- TYLCV
tomato yellow leaf curl virus: ACMV, African cassava mosaic virus
- BCTV
beet curly top virus
- ds
double-stranded
- ss
single-stranded
- Rep
replication-associated protein
- ORF
open reading frame
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank data base (accession no: U74627).
References
- 1.Francki R. Annu Rev Microbiol. 1985;39:151–174. doi: 10.1146/annurev.mi.39.100185.001055. [DOI] [PubMed] [Google Scholar]
- 2.Liu Y Y, Cooper J I. Rev Plant Pathol. 1994;73:371–387. [Google Scholar]
- 3.Taylor J M. Annu Rev Microbiol. 1992;46:253–276. doi: 10.1146/annurev.mi.46.100192.001345. [DOI] [PubMed] [Google Scholar]
- 4.Mayo M A, Berns K I, Fritsch C, Kaper J M, Jackson A O, Leibowitz M J, Taylor J M. In: Virus taxonomy. Classification and nomenclature of viruses. Sixth Report of the International Committee on Taxonomy of Viruses. Fauquet A M F, editor. New York: Springer; 1995. pp. 487–492. [Google Scholar]
- 5.Tien P, Wu G. Adv Virus Res. 1991;39:321–339. doi: 10.1016/s0065-3527(08)60799-x. [DOI] [PubMed] [Google Scholar]
- 6.Lazarowitz S G. Crit Rev Plant Sci. 1992;11:327–349. [Google Scholar]
- 7.Stenger D C, Stevenson M C, Hormuzdi S G, Bisaro D M. J Gen Virol. 1992;73:237–242. doi: 10.1099/0022-1317-73-2-237. [DOI] [PubMed] [Google Scholar]
- 8.Dry I B, Rigden J E, Krake L R, Mullineaux P M, Rezaian M A. J Gen Virol. 1993;74:147–151. doi: 10.1099/0022-1317-74-1-147. [DOI] [PubMed] [Google Scholar]
- 9.Behjatnia S A A, Dry I B, Krake L R, Conde B D, Connelly M I R, Randles J W, Rezaian M A. Phytopathology. 1996;86:880–886. [Google Scholar]
- 10.Kheyr-Pour A, Bendahmane M, Matzeit V, Accotto G P, Crespi S, Gronenborn B. Nucleic Acids Res. 1991;19:6763–6769. doi: 10.1093/nar/19.24.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klinkenberg F A, Ellwood S, Stanley J. J Gen Virol. 1989;70:1837–1844. [Google Scholar]
- 12.Briddon R W, Watts J, Markham P G, Stanley J. Virology. 1989;172:628–633. doi: 10.1016/0042-6822(89)90205-5. [DOI] [PubMed] [Google Scholar]
- 13.Rigden J E, Dry I B, Krake L R, Rezaian M A. Proc Natl Acad Sci USA. 1996;93:10280–10284. doi: 10.1073/pnas.93.19.10280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maule A J, Hull R, Donson J. J Virol Methods. 1983;6:215–224. doi: 10.1016/0166-0934(83)90048-4. [DOI] [PubMed] [Google Scholar]
- 15.Elmer J S, Brand L, Sunter G, Gardiner W E, Bisaro D M, Rogers S G. Nucleic Acids Res. 1988;16:7043–7060. doi: 10.1093/nar/16.14.7043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rigden J E, Dry I B, Mullineaux P M, Rezaian M A. Virology. 1993;193:1001–1005. doi: 10.1006/viro.1993.1215. [DOI] [PubMed] [Google Scholar]
- 17.Thomas J E, Massalski P R, Harrison B D. J Gen Virol. 1986;67:2739–2748. [Google Scholar]
- 18.Wetzel T, Candresse T, Macquaire G, Ravelonandro M, Dunez J. J Virol Methods. 1992;39:27–37. doi: 10.1016/0166-0934(92)90122-t. [DOI] [PubMed] [Google Scholar]
- 19.Rezaian M A, Krake L R, Golino D A. Intervirology. 1992;34:38–43. doi: 10.1159/000150261. [DOI] [PubMed] [Google Scholar]
- 20.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christie G E, Calendar R. Annu Rev Genet. 1990;24:465–490. doi: 10.1146/annurev.ge.24.120190.002341. [DOI] [PubMed] [Google Scholar]
- 22.Sunter G, Bisaro D M. Virology. 1991;180:416–419. doi: 10.1016/0042-6822(91)90049-h. [DOI] [PubMed] [Google Scholar]
- 23.Fontes E P B, Gladfelter H J, Schaffer R L, Petty I T D, Hanley-Bowdoin L. Plant Cell. 1994;6:405–416. doi: 10.1105/tpc.6.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Orozco B M, Hanley-Bowdoin L. J Microbiol. 1996;70:148–158. doi: 10.1128/jvi.70.1.148-158.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arguello-Astorga G R, Guevara-Gonzalez R G, Herrera-Estrella L R, Rivera-Bustamante R F. Virology. 1994;203:90–100. doi: 10.1006/viro.1994.1458. [DOI] [PubMed] [Google Scholar]
- 26.Fontes E P B, Eagle P A, Sipe P S, Luckow V A, Hanley-Bowdoin L. J Biol Chem. 1994;269:8459–8465. [PubMed] [Google Scholar]
- 27.Gilbertson R L, Hidayat S H, Paplomatas E J, Rojas M R, Hou Y M, Maxwell D P. J Gen Virol. 1993;74:23–31. doi: 10.1099/0022-1317-74-1-23. [DOI] [PubMed] [Google Scholar]