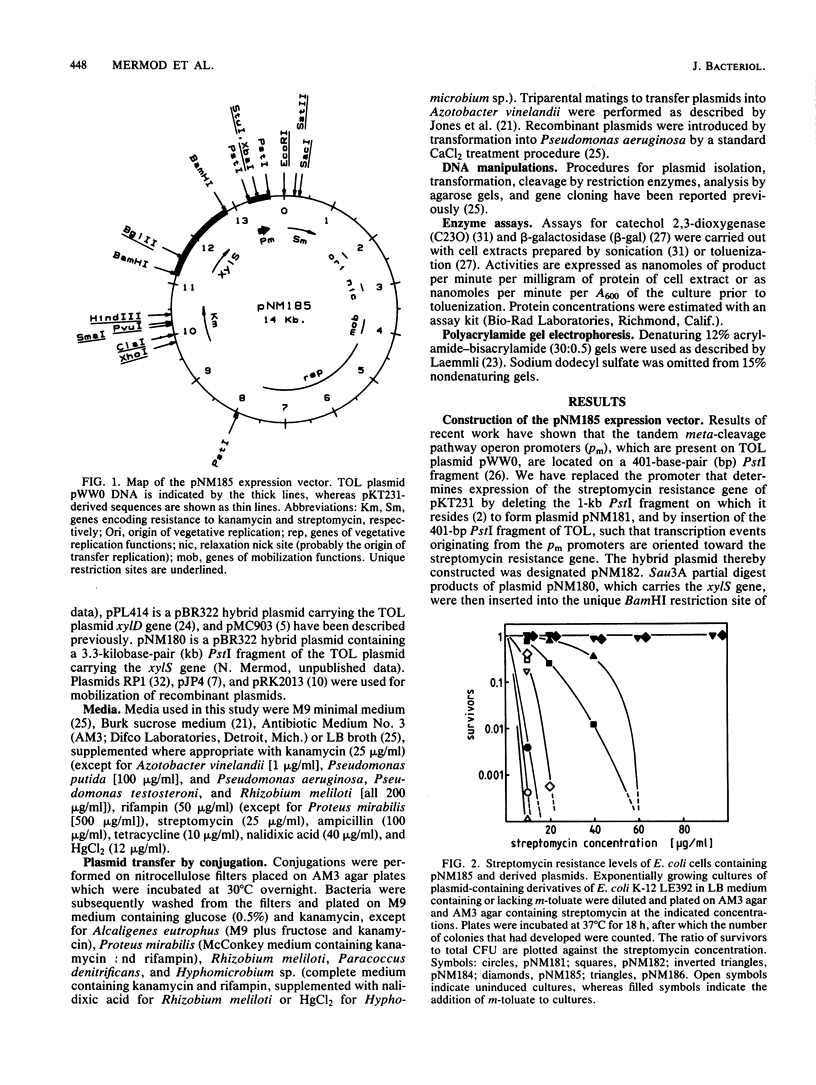
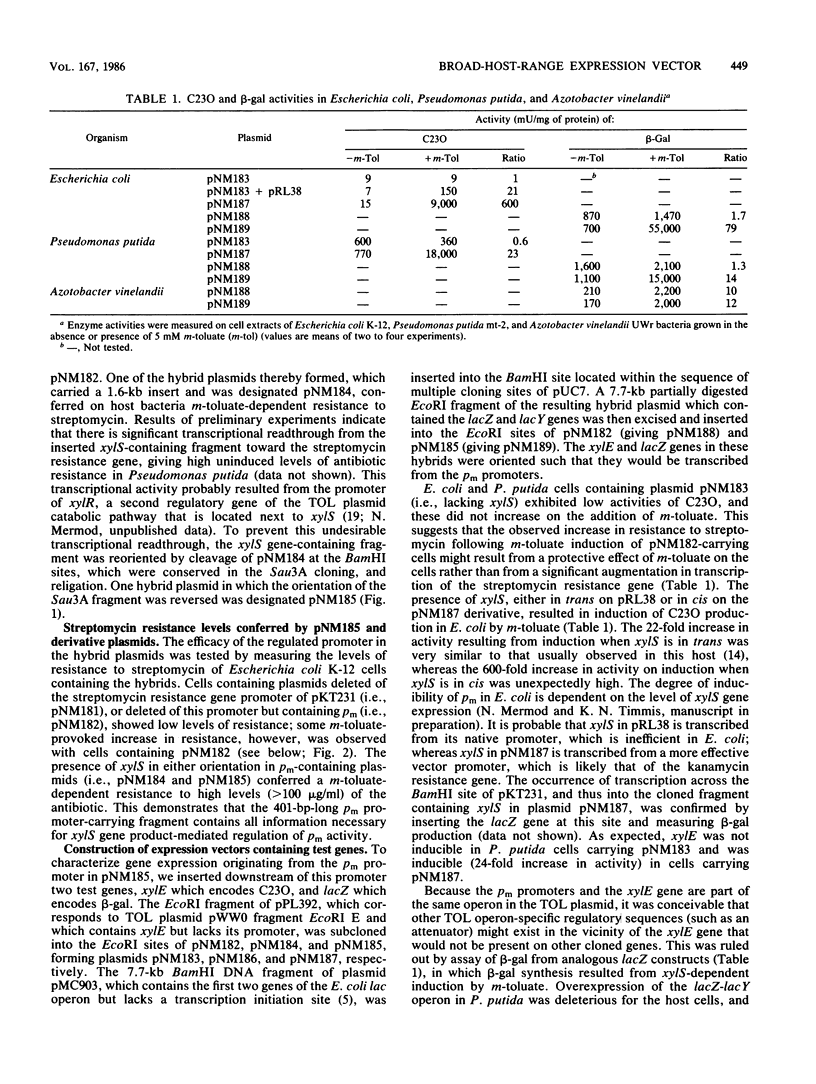
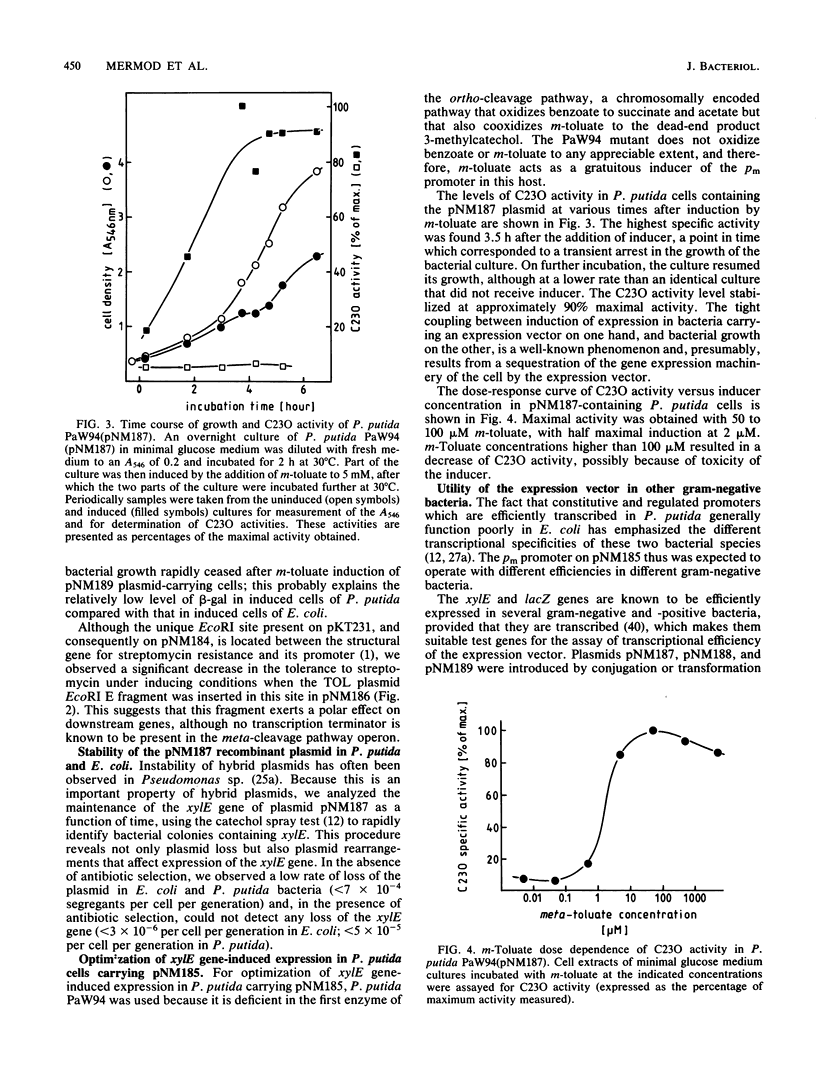
Abstract
A pKT231-based broad-host-range plasmid vector was constructed which enabled regulation of expression of cloned genes in a wide range of gram-negative bacteria. This vector, pNM185, contained upstream of its EcoRI, SstI, and SstII cloning sites the positively activated pm twin promoters of the TOL plasmid and xylS, the gene of the positive regulator of these promoters. Expression of cloned genes was induced with micromolar quantities of benzoate or m-toluate, the inexpensive coinducers of the pm promoters. Expression of a test gene, xylE, which specifies catechol 2,3-dioxygenase, cloned in this vector was tested in representative strains of a variety of gram-negative bacteria. Regulated expression of xylE was observed in most strains examined, and induced levels of enzyme representing up to 5% of total cellular protein and ratios of induced:noninduced levels of enzyme up to a factor of 600 were observed. The level of xylE gene expression in different bacteria tended to be correlated with their phylogenetic distance from Pseudomonas putida.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bagdasarian M. M., Amann E., Lurz R., Rückert B., Bagdasarian M. Activity of the hybrid trp-lac (tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range, controlled-expression vectors. Gene. 1983 Dec;26(2-3):273–282. doi: 10.1016/0378-1119(83)90197-x. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M., Lurz R., Rückert B., Franklin F. C., Bagdasarian M. M., Frey J., Timmis K. N. Specific-purpose plasmid cloning vectors. II. Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene. 1981 Dec;16(1-3):237–247. doi: 10.1016/0378-1119(81)90080-9. [DOI] [PubMed] [Google Scholar]
- Bagdasarian M., Timmis K. N. Host: vector systems for gene cloning in Pseudomonas. Curr Top Microbiol Immunol. 1982;96:47–67. doi: 10.1007/978-3-642-68315-2_4. [DOI] [PubMed] [Google Scholar]
- Bishop P. E., Brill W. J. Genetic analysis of Azotobacter vinelandii mutant strains unable to fix nitrogen. J Bacteriol. 1977 May;130(2):954–956. doi: 10.1128/jb.130.2.954-956.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Chou J., Cohen S. N. In vitro gene fusions that join an enzymatically active beta-galactosidase segment to amino-terminal fragments of exogenous proteins: Escherichia coli plasmid vectors for the detection and cloning of translational initiation signals. J Bacteriol. 1980 Aug;143(2):971–980. doi: 10.1128/jb.143.2.971-980.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty T., Montenegro M. A., Sanyal S. C., Helmuth R., Bulling E., Timmis K. N. Cloning of enterotoxin gene from Aeromonas hydrophila provides conclusive evidence of production of a cytotonic enterotoxin. Infect Immun. 1984 Nov;46(2):435–441. doi: 10.1128/iai.46.2.435-441.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Pemberton J. M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981 Feb;145(2):681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Weightman A. J., Knackmuss H. J., Timmis K. N. Transposon mutagenesis and cloning analysis of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134(pJP4). J Bacteriol. 1985 Jan;161(1):85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn N. W., Holloway B. W. Pleiotrophy of p-fluorophenylalanine-resistant and antibiotic hypersensitive mutants of Pseudomonas aeruginosa. Genet Res. 1971 Oct;18(2):185–197. doi: 10.1017/s0016672300012593. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox G. E., Stackebrandt E., Hespell R. B., Gibson J., Maniloff J., Dyer T. A., Wolfe R. S., Balch W. E., Tanner R. S., Magrum L. J. The phylogeny of prokaryotes. Science. 1980 Jul 25;209(4455):457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- Franklin F. C., Bagdasarian M., Bagdasarian M. M., Timmis K. N. Molecular and functional analysis of the TOL plasmid pWWO from Pseudomonas putida and cloning of genes for the entire regulated aromatic ring meta cleavage pathway. Proc Natl Acad Sci U S A. 1981 Dec;78(12):7458–7462. doi: 10.1073/pnas.78.12.7458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Lehrbach P. R., Lurz R., Rueckert B., Bagdasarian M., Timmis K. N. Localization and functional analysis of transposon mutations in regulatory genes of the TOL catabolic pathway. J Bacteriol. 1983 May;154(2):676–685. doi: 10.1128/jb.154.2.676-685.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harayama S., Lehrbach P. R., Timmis K. N. Transposon mutagenesis analysis of meta-cleavage pathway operon genes of the TOL plasmid of Pseudomonas putida mt-2. J Bacteriol. 1984 Oct;160(1):251–255. doi: 10.1128/jb.160.1.251-255.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes E. J., Bayly R. C. Control of catechol meta-cleavage pathway in Alcaligenes eutrophus. J Bacteriol. 1983 Jun;154(3):1363–1370. doi: 10.1128/jb.154.3.1363-1370.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Ebina Y., Nakazawa A., Nakazawa T. Nucleotide sequence surrounding transcription initiation site of xylABC operon on TOL plasmid of Pseudomonas putida. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1688–1691. doi: 10.1073/pnas.81.6.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Determination of the transcription initiation site and identification of the protein product of the regulatory gene xylR for xyl operons on the TOL plasmid. J Bacteriol. 1985 Sep;163(3):863–869. doi: 10.1128/jb.163.3.863-869.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Molecular cloning of gene xylS of the TOL plasmid: evidence for positive regulation of the xylDEGF operon by xylS. J Bacteriol. 1981 Nov;148(2):413–418. doi: 10.1128/jb.148.2.413-418.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inouye S., Nakazawa A., Nakazawa T. Nucleotide sequence of the promoter region of the xylDEGF operon on TOL plasmid of Pseudomonas putida. Gene. 1984 Sep;29(3):323–330. doi: 10.1016/0378-1119(84)90061-1. [DOI] [PubMed] [Google Scholar]
- Jones R., Woodley P., Robson R. Cloning and organisation of some genes for nitrogen fixation from Azotobacter chroococcum and their expression in Klebsiella pneumoniae. Mol Gen Genet. 1984;197(2):318–327. doi: 10.1007/BF00330980. [DOI] [PubMed] [Google Scholar]
- Kunz D. A., Chapman P. J. Catabolism of pseudocumene and 3-ethyltoluene by Pseudomonas putida (arvilla) mt-2: evidence for new functions of the TOL (pWWO) plasmid. J Bacteriol. 1981 Apr;146(1):179–191. doi: 10.1128/jb.146.1.179-191.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lehrbach P. R., Zeyer J., Reineke W., Knackmuss H. J., Timmis K. N. Enzyme recruitment in vitro: use of cloned genes to extend the range of haloaromatics degraded by Pseudomonas sp. strain B13. J Bacteriol. 1984 Jun;158(3):1025–1032. doi: 10.1128/jb.158.3.1025-1032.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod N., Lehrbach P. R., Reineke W., Timmis K. N. Transcription of the TOL plasmid toluate catabolic pathway operon of Pseudomonas putida is determined by a pair of co-ordinately and positively regulated overlapping promoters. EMBO J. 1984 Nov;3(11):2461–2466. doi: 10.1002/j.1460-2075.1984.tb02156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton N. P., Clarke L. E. Identification of the promoter of the Pseudomonas gene coding for carboxypeptidase G2. J Mol Appl Genet. 1985;3(1):26–35. [PubMed] [Google Scholar]
- Nakai C., Kagamiyama H., Nozaki M., Nakazawa T., Inouye S., Ebina Y., Nakazawa A. Complete nucleotide sequence of the metapyrocatechase gene on the TOI plasmid of Pseudomonas putida mt-2. J Biol Chem. 1983 Mar 10;258(5):2923–2928. [PubMed] [Google Scholar]
- Nakazawa T., Inouye S., Nakazawa A. Physical and functional mapping of RP4-TOL plasmid recombinants: analysis of insertion and deletion mutants. J Bacteriol. 1980 Oct;144(1):222–231. doi: 10.1128/jb.144.1.222-231.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Evans W. C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971 Jun 11;20(3):400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- Thomas C. M. Molecular genetics of broad host range plasmid RK2. Plasmid. 1981 Jan;5(1):10–19. doi: 10.1016/0147-619x(81)90074-3. [DOI] [PubMed] [Google Scholar]
- Tybulewicz V. L., Falk G., Walker J. E. Rhodopseudomonas blastica atp operon. Nucleotide sequence and transcription. J Mol Biol. 1984 Oct 25;179(2):185–214. doi: 10.1016/0022-2836(84)90465-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Woese C. R., Stackebrandt E., Weisburg W. G., Paster B. J., Madigan M. T., Fowler V. J., Hahn C. M., Blanz P., Gupta R., Nealson K. H. The phylogeny of purple bacteria: the alpha subdivision. Syst Appl Microbiol. 1984;5:315–326. doi: 10.1016/s0723-2020(84)80034-x. [DOI] [PubMed] [Google Scholar]
- Worsey M. J., Franklin F. C., Williams P. A. Regulation of the degradative pathway enzymes coded for by the TOL plasmid (pWWO) from Pseudomonas putida mt-2. J Bacteriol. 1978 Jun;134(3):757–764. doi: 10.1128/jb.134.3.757-764.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsey M. J., Williams P. A. Metabolism of toluene and xylenes by Pseudomonas (putida (arvilla) mt-2: evidence for a new function of the TOL plasmid. J Bacteriol. 1975 Oct;124(1):7–13. doi: 10.1128/jb.124.1.7-13.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeyer J., Wasserfallen A., Timmis K. N. Microbial mineralization of ring-substituted anilines through an ortho-cleavage pathway. Appl Environ Microbiol. 1985 Aug;50(2):447–453. doi: 10.1128/aem.50.2.447-453.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zukowski M. M., Gaffney D. F., Speck D., Kauffmann M., Findeli A., Wisecup A., Lecocq J. P. Chromogenic identification of genetic regulatory signals in Bacillus subtilis based on expression of a cloned Pseudomonas gene. Proc Natl Acad Sci U S A. 1983 Feb;80(4):1101–1105. doi: 10.1073/pnas.80.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]