Summary
Hypoxia-inducible factor-1, HIF1, transcriptionally activates over 200 genes vital for cell homeostasis and angiogenesis. We developed a computational model to gain a detailed quantitative understanding of how HIF1 acts to sense oxygen and respond to hypoxia. The model consists of kinetic equations describing the intracellular variation of 17 compounds, including HIF1, iron, prolyl hydroxylase, oxygen, ascorbate, 2-oxoglutarate, von Hippel Lindau protein and associated complexes. We tested an existing hypothesis of a switch-like change in HIF1 expression in response to a gradual decrease in O2 concentration. Our model predicts that depending on the molecular environment, such as intracellular iron levels, the hypoxic response varies considerably. We show HIF1-activated cellular responses can be divided into two categories: a steep, switch-like response to O2 and a gradual one. Discovery of this dual response prompted comparison of two therapeutic strategies, ascorbate and iron supplementation, and prolyl hydroxylase targeting, to predict under what microenvironments either effectively increases HIF1α hydroxylation. Results provide crucial insight into the effects of iron and prolyl hydroxylase on oxygen sensing. The model advances quantitative molecular level understanding of HIF1 pathways – an endeavor that will help elucidate the diverse responses to hypoxia found in cancer, ischemia and exercise.
Keywords: Computational modeling, Mathematical modeling, Oxygen sensing, Hypoxic response
Introduction
The transcription factor HIF (hypoxia-inducible factor) plays a crucial role in mammalian response to oxygen (O2) levels. HIF1, the first characterized member of the HIF family, transcriptionally activates hundreds of genes associated with angiogenesis in cancer, exercise and ischemia; energy metabolism; nutrient transport; and cell migration (Semenza, 2004; Wang et al., 1995).
Angiogenesis, the formation of blood vessels from preexisting vessels, is regulated in part by local tissue O2 levels. This regulatory pathway links cell and tissue metabolic demand with vascular oxygen supply. The pathway is intricately governed by HIF1 regulation and HIF1 transcriptional activation of angiogenic factors.
HIF1 is a heterodimer, comprised of subunits HIF1α and HIF1β. The beta subunit is constitutively expressed in cells. Expression of the alpha subunit may be induced by a number of pathways, and its degradation is highly sensitive to O2 levels. Called a ‘master switch for hypoxic gene expression’ (Powell, 2003; Semenza, 2004), intracellular HIF1α in normoxia is experimentally undetectable; during hypoxia, it rapidly accumulates in the cell nucleus, and triggers gene expression. Molecular players involved in this process have come to light over the past six years. In normoxia, enzymes called prolyl hydroxylase domains (PHDs) react with HIF1α (Fig. 1). PHDs hydroxylate HIF1α at Pro402 and Pro564 in the oxygen-dependant degradation domain. The activity of PHDs depends on the amount of oxygen available. Three isoforms of HIF PHDs exist: PHD1, PHD2 and PHD3. Each isoform performs a separate function, with different kinetic properties and primary cellular locations (Appelhoff et al., 2004). PHD2 is the most abundant prolyl hydroxylase isoform in the cell cytoplasm during normoxia, and it has been credited as a controller of steady-state HIF1α concentrations under these conditions in a range of cell types (Berra et al., 2003). Conversely, in normoxia, intracellular PHD1 and PHD3 levels are low, if experimentally detectable (Appelhoff et al., 2004).
Fig. 1.
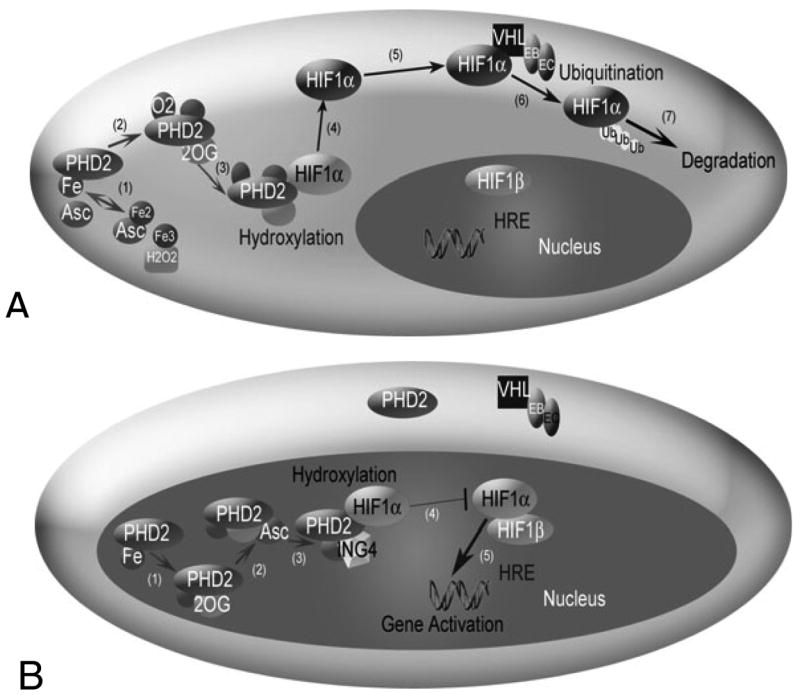
The HIF1 pathway in normoxia (A) and hypoxia (B). (A) HIF1α hydroxylation and degradation in the presence of oxygen involves: (1) the independent oxidation-reduction reactions of ascorbate (Asc) and iron (Fe); (2) and (3) prolyl hydroxlyase 2 (PHD2) binding to Fe, 2-oxoglutarate (2OG), and O2; (4) PHD2 hydroxylation of HIF1α; (5) unbound hydroxylated HIF1α moving in the cell cytoplasm; (6) the von Hippel Lindau (VHL)-Elongin B (EB)-Elongin C (EC) complex ubiquitylating HIF1α; and (7) HIF1α degradation. A change in shading of HIF1α indicates addition of a hydroxyl group. (B) In hypoxia, HIF1α enters the nucleus, where hydroxylation, but no degradation occurs. (1) and (2) PHD2 binding to Fe, 2OG and Asc, but not O2. (3) The protein inhibitor of growth 4 (ING4) binding to PHD2 may regulate HIF1α transcriptional activity and (4) block HIF1α-HIF1β binding. When HIF1α-HIF1β binding occurs, the HIF1 dimer can transcriptionally activate genes at the hypoxia response element (HRE) site.
Following the reaction with PHD, the hydroxylated HIF1α is free to bind to a von Hippel Lindau (VHL) ubiquitin ligase complex, which tags HIF1α for proteasomal destruction. Like PHD2, VHL is found primarily in the cell cytosol in many cell types (Los et al., 1996), though VHL trafficking between nucleus and cytoplasm may be necessary in HIF1α degradation during reoxygenation from hypoxia (Groulx and Lee, 2002). VHL forms a stable complex with proteins including Elongin B, Elongin C, Cul2 and Rbx1 (Kamura et al., 1999). This VHL ligase complex binds to hydroxylated HIF1α and tags the protein with a polyubiquitin tail (Ivan et al., 2001; Kamura et al., 2000). A multi-protein complex, called the proteasome, recognizes this tail and destroys HIF1α.
In hypoxia, HIF1α escapes hydroxylation, accumulates and enters the cell nucleus, where it binds to HIF1β (known also as ARNT) (Fig. 1). The dimer transcriptionally activates a host of genes, including those encoding the angiogenic protein vascular endothelial growth factor (VEGF) and its receptor, Flk-1 or VEGFR2 (Milkiewicz et al., 2003); platelet-derived growth factor (Bos et al., 2005); and erythropoetin (Marti, 2004). By activating these genes, HIF1 contributes to angiogenesis, which provides nutrients to facilitate tumor growth or to extend muscle contraction, for example. As prominent players in the cell response to hypoxia and the onset of angiogenesis, HIF1 and its related pathways are attractive therapeutic targets in cancer and ischemia (Hewitson and Schofield, 2004).
In vitro studies have shown how the hypoxic response varies in tumors, based on their vascular microenvironment (Blouw et al., 2003). A balance of HIF1α levels and HIF1α activity seems necessary to achieve health (Josko and Mazurek, 2004; Koshiji and Huang, 2004). The underlying molecular mechanisms of how this balance is achieved and how the system responds to its microenvironment are not fully understood. One hypothesis is that HIF1α acts literally as a ‘switch’ – an on/off mechanism for the onset of hypoxia-induced angiogenesis when a critical O2 level is reached (Kohn et al., 2004). Fundamental issues include understanding how HIF1α acting as a generic switch would be correlated to the varied hypoxic responses found in tumor cells. Alternatively, if HIF1α and its pathways do not act as a switch, the observed sensitivity to oxygen and the rapid induction of hypoxic genes would need to be otherwise explained. We address these questions, by developing a detailed model of HIF1α degradation, which allows molecular mechanisms to be tested quantitatively. The one known existing computer simulation related to HIF1, a network representation of cell hypoxic response, has included a subset of core HIF1 pathways and tested the hypothetical dependency of gene expression on HIF1α synthesis and degradation rates (Kohn et al., 2004). This model led to the above HIF1α switch hypothesis, which to our knowledge, has not been tested further. No computational model has explored the biochemical kinetics of the HIF1 pathways in detail or in a quantitative relationship to experimental data. The current model is the first molecular level, mechanistic model of HIF1 hydroxylation and degradation. We used the model to study the effects of different intracellular molecular compositions on hypoxic response, where the cellular microenvironment is currently inaccessible in vivo, and is only measurable in isolated instances in vitro.
From the model, we predict several key characteristics of the mechanisms involved in the HIF1 pathway. We show that HIF1-activated cellular responses can be divided into two categories depending on the molecular environment: a switch-like response to O2 levels, and a gradual one. We found conditions where iron and PHD2 are individual sensors of oxygen and determinants of the hypoxic response; and we showed the combined effects of three highly oxygen-sensitive compounds. From these studies, we compare two proposed therapeutic strategies targeting the HIF1 pathway, iron supplementation and PHD2 targeting, and predict under what microenvironments either would most efficiently increase HIF1α hydroxylation. These observations contribute to a better understanding of the hypoxic response at the molecular level and should stimulate further computational and experimental exploration, with particular applications to therapies that target cofactors in HIF1α hydroxylation.
Results
Model validation
Double reciprocal plots of the hydroxylation reaction before binding to HIF1α were consistent with experimental results from the collagen PHDs (Myllyla et al., 1977); this confirmed that the model represents the uncompetitive binding of Fe2+, O2 and 2-oxoglutarate (2-OG) to PHD2 (Fig. 2A shows the example of iron binding). The reaction can proceed without ascorbate (Fig. 2B,C), however, high ascorbate concentrations (>100 μM) significantly increase the reaction rate (Fig. 2B).
Fig. 2.
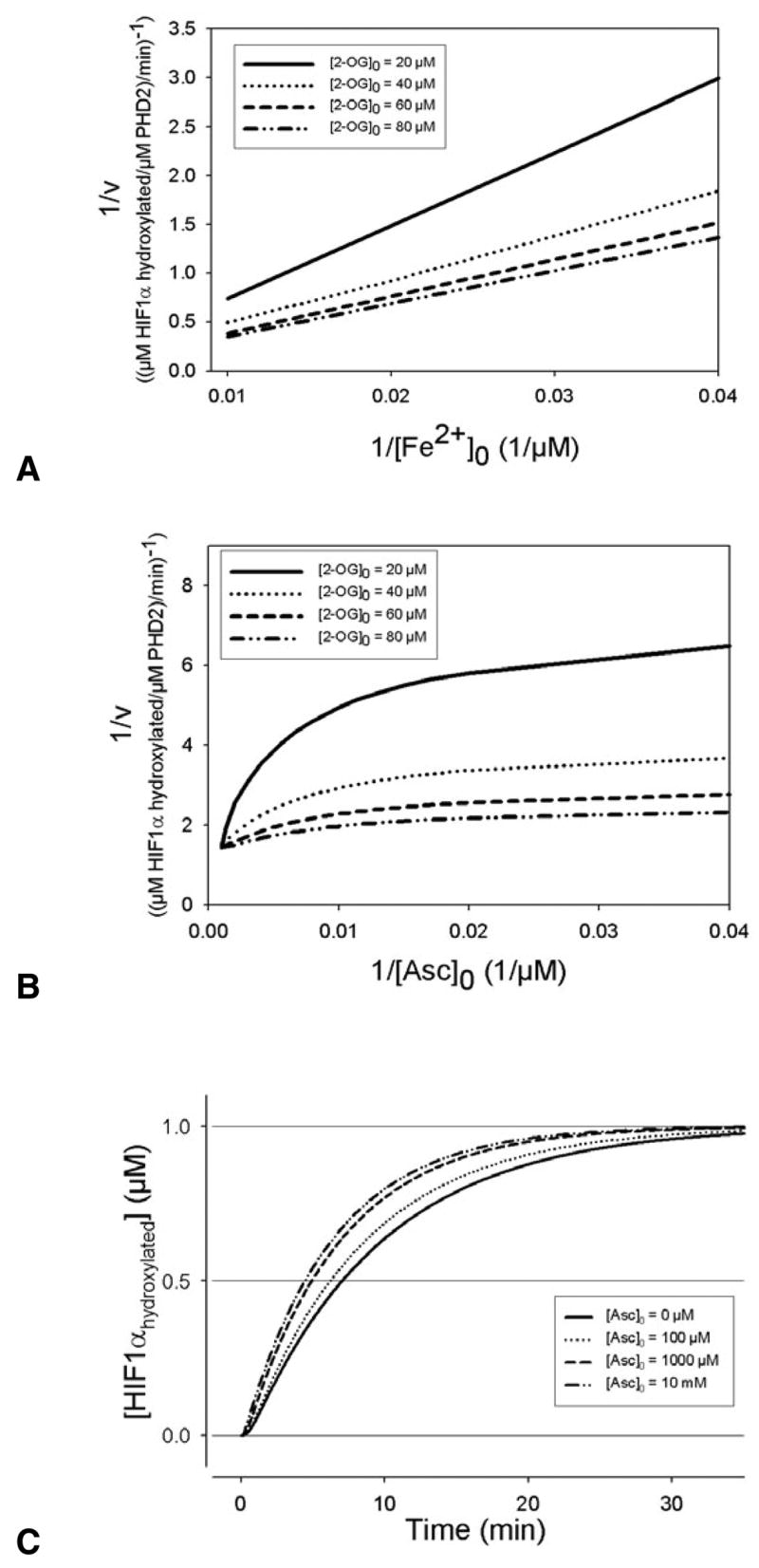
(A) Effect of Fe2+ on the rate of prolyl hydroxylase (PHD2) reaction at different concentrations of 2-oxoglutarate (2-OG). Iron binds uncompetitively to PHD2, as indicated by lines that will intersect in the double reciprocal plot. (B) The double reciprocal plot of ascorbate concentrations at different levels of 2-OG show parallel lines above 1/[Asc]0=0.01 μM, while at higher ascorbate concentrations the lines begin to converge. This indicates ascorbate predominantly reacting with Fe3+, and not binding to the hydroxylases, at low ascorbate concentrations; at higher ascorbate levels, there is significant ascorbate reacting in iron reduction and the overall hydroxylation reaction. (C) The effect of ascorbate on the hydroxylation reaction. When there is no ascorbate, the reaction occurs though it takes more than 30 minutes for HIF1α to be fully degraded.
The model was compared with independent experimental data from several sources to validate the oxygen dependency and the time course of HIF1α hydroxylation by PHD2 (Fig. 3). One form of validation was the amount of VHL captured at different O2 levels. Tuckerman et al. measured the activity of endogenous HIF-PHDs (from MDA-MB-435 cell extracts) by a pVHL capture assay (Tuckerman et al., 2004). Oxygen concentrations in the simulations, all ≤200 μM, are below the Km values of PHD2 for O2 of 250 μM (Table 2). This is reflected in Fig. 3A, where the model is compared with published data (Tuckerman et al., 2004). The 21% oxygen values show a near-linear increase in the fraction of the maximal pVHL capture with time; it is reasonable to conclude (Tuckerman et al., 2004), that all of the experiments were conducted below the threshold for oxygen saturation.
Fig. 3.
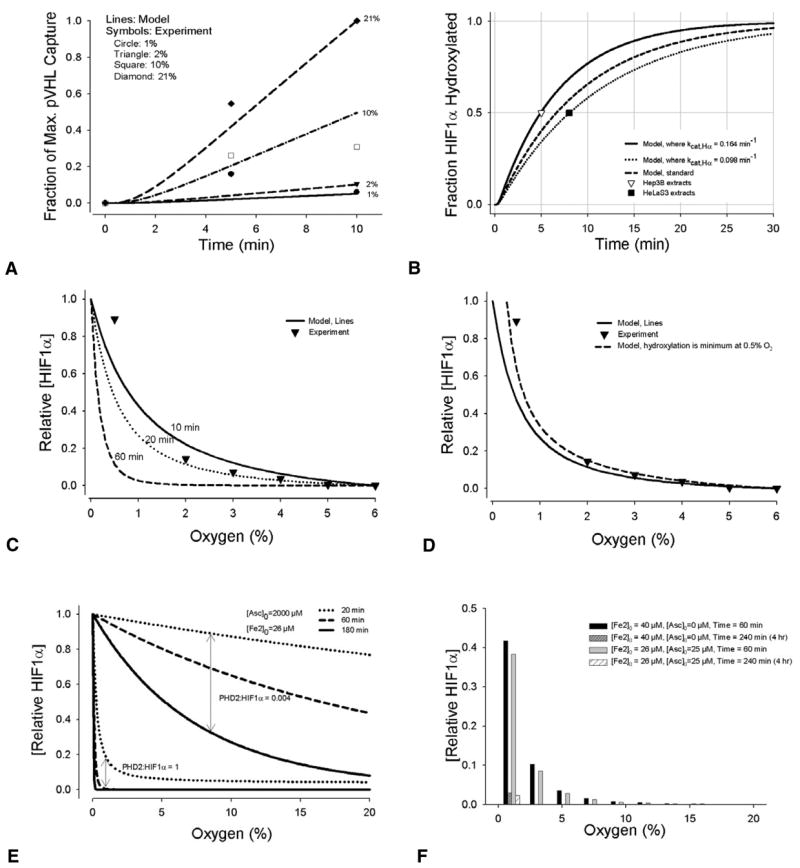
Model comparisons with experiments. (A) HIF1α hydroxylation by PHDs is related to cellular O2 levels (1, 2, 10 and 21%) in the model (lines). Results are compared with independent experimental data (Tuckerman et al., 2004), showing HIF1α modification by PHD2 measured by relative VHL capture at 0, 5 and 10 minutes (symbols). (B) Model predictions of the minimum percentage of HIF1α hydroxylated by PHD2 in normoxia (21% oxygen), lines. This is compared with experiments that measured HIF1α half-life in cells that were initially hypoxic, and at time zero in 20–21% oxygen; experimental values: t1/2=5–8 minutes (Berra et al., 2001; Jewell et al., 2001); t1/2<5 minutes (Huang et al., 1998). (C) Comparison of relative HIF1α accumulation predicted by the model at different oxygen levels with in vitro data (Jiang et al., 1996). Data points in Fig. 3C and 3D are the mean of two experiments. [O2]=0–59 μM in the model corresponds to ~0–6% in the experiment. Initial model conditions were default values (Table 1). 10, 20, and 60 minutes correspond to the duration of the hydroxylation reaction. Both the experimental data and model results were normalized to the value obtained at 6% O2 ([O2]=59 μM). (D) Comparison with the same experiment, using the time of 20 minutes in the model. A line is provided showing what the model would predict if the lowest hydroxylation rate was set at 0.5% O2 rather than 0% O2. Intranuclear hydroxylation during anoxia is one possible mechanism by which HIF1α nuclear levels decrease below 0.5%, as shown by Jiang et al (Jiang et al., 1996). The model currently does not account for additional changes during anoxia. The delay accounting for the time it takes unhydroxylated HIF1α in the cytoplasm to move into the nucleus is assumed constant across all O2 levels. (E) Model results for HIF1α expression with ascorbate supplementation. The effects of PHD2:HIF1α concentration ratios on HIF1α expression are shown. (F) Model results showing HIF1α expression with iron or ascorbate supplementation after 1 hour and 4 hours of normoxia.
Table 2.
Range of initial concentration values where there is a non-linear oxygen response curve and hypoxic levels of oxygen yield a different HIF1α hydroxylation rate than normoxic levels
| [PHD2]0 (μM) | [Fe2+]0 (μM) | [2-OG]0 (μM) | [O2] when switch occurs (μM) | |
|---|---|---|---|---|
| Steep oxygen response curve | >0.03 | in excess, 50 | in excess, 1000 | 40 |
| in excess, 1 | >0.1 | in excess, 1000 | 30–40 | |
| in excess, 1 | in excess, 50 | >10 | 30–40 |
For all initial conditions, [Asc]0=1000 μM; [HIF1α]0=1 μM. Values were determined by sensitivity analysis. The conditions correspond to Fig. 4A, but where the initial concentration of one hydroxylation cofactor (PHD2, Fe2+ or 2-OG) was gradually decreased to determine at what concentration it becomes a limiting determinant of the switch response.
Model calculations for HIF1α half-life in normoxia fall within the range of experimental data (Fig. 3B). Experimental half-life of HIF1α was estimated as 5–8 minutes from reoxygenated (1–2% O2 to 20% O2) CCL39 and NHE-1 cell lysates (Berra et al., 2001). [Using oxygen solubility in water at 37°C = 1.30 μmol/l/mmHg (Tuckerman et al., 2004), exposure to 1% oxygen corresponds to an O2 level of 9.9 μM; 21% to an O2 level of 207 μM.] This complements independent finding in lysates from Hep3B cells exposed to 21% oxygen after hypoxia (1% oxygen) where the half-life of HIF1α was found to be <5 minutes (Huang et al., 1998); and from HeLaS3 cells reoxygenated at 20% from hypoxia (0.5% oxygen), where the half-life of HIF1α was 8 minutes (Jewell et al., 2001).
A third form of validation was the comparison of the predicted relative HIF1α accumulation of the model at different oxygen levels, with data from HeLa cell nuclear extracts (Jiang et al., 1996) (Fig. 3C). Studies have shown that HIF1 expression is maximal at low oxygen concentrations in vivo [e.g. at 0 or 1% O2, in normal ferret lung ventilated for 4 hours (Yu et al., 1998)], and in vitro assays indicated the most pronounced changes in HIF1 expression occur at O2 levels considered physiologically relevant (0–5%) (Jiang et al., 1996). To compare the computational model with these experiments, a constant [HIF1α]0 was assumed, and [HIF1α] that was not hydroxylated was taken as a relative measure of HIF1α nuclear accumulation.
Fourth, the effects of ascorbate and iron in the model are qualitatively comparable to HIF1α expression observed in human prostate adenocarcinoma (PC3) cells (Knowles et al., 2003). Fig. 3E shows the model results of supplementing the system with 2000 μM ascorbate. Relative HIF1α values are a fraction of the maximum HIF1α expression during hypoxia, without supplementation. For different cell types, including PC3 cells, the PHD2:HIF1α concentration ratio has not been quantified. Fig. 3E provides an example of how the ratio affects the accumulation of HIF1α, during the first 3 hours of exposure to different O2 levels. A PHD2:HIF1α ratio of 0.004 is an estimate from measurements in breast carcinoma cells (Tuckerman et al., 2004). In normoxia, ascorbate and iron supplementation have similar effects on suppressing HIF1α expression (Fig. 3F). Comparable experimental results in PC3 cells show decreased expression of HIF1α over time with ascorbate supplemented at 2000 μM, and no appreciable expression of HIF1α in normoxia after 4 hours of supplementation with 25 μM ascorbate or >40 μM FeCl2 (40 μM FeCl2 added to medium containing ~26 μM Fe2+), see figure 2C,D in Knowles et al. (Knowles et al., 2003).
Sensitivity analysis
Sensitivity analysis was performed to confirm estimates for the unknown kinetic rate constants. The parameter of interest was varied over a minimum range of 1000-fold, while the remaining parameters were held constant. Calculated HIF1α half-life values were compared with experimental data, and this was used to narrow the range of reasonable parameter values. Details of the analysis are in provided in the Materials and Methods. Estimated values for five kinetic binding rates were determined: kcat,Hα=0.098–0.164 minute−1; koff,Fe2=36 minute−1; koff,DG=10.8 minute−1; koff,O2=10.8 minute−1; koff,AS=3.6 minute−1; koff,Hα=0.7 minute−1. Binding of Fe2+, 2-OG and O2 to PHD2 are the more reversible steps in the hydroxylation reaction, with significant off-rates relative to on-rate binding. The final step in the hydroxylation, binding to HIF1α is largely irreversible, as indicated by a low koff,Hα value and a significant kcat,Hα.
A recent study reported the apparent Km for Fe2+ of 0.03 μM (Hirsila et al., 2005). Using this estimate, as opposed to the Km of 2 μM, estimated from binding of factor-inhibiting HIF (FIH) with Fe2+ in the hydroxylation of HIF1α (Koivunen et al., 2004), the model predicts a higher specific activity for PHD2 than found in vitro (Hirsila et al., 2005) (see supplementary material Fig. S1). This discrepancy may reflect different concentrations of PHD2 and HIF1α relative to other compounds in the hydroxylation reaction; the model used initial conditions where quantitative concentrations of PHD2 were reported (Tuckerman et al., 2004).
Sensitivity analysis for all kinetic parameters found from experiments, was also performed using the protocol described for the estimated parameters (see Materials and Methods). Over a wide range of feasible Km,Fe2, Km,DG, Km,O2, Km,Asc and Km,Hα values (supplementary material Figs S2, S3), the changes in oxygen sensitivity were consistent, and the model features described below can be considered robust over these values.
Oxygen sensing
In many in vitro cell extract experiments monitoring HIF1α reactions, there is an excess concentration of initial 2-OG, iron, ascorbate and PHD2. When any of these compounds was limiting, the sensitivity to oxygen in the model was uniform at all O2 levels from 0–200 μM. Fig. 4 shows how initial reactant concentrations affect HIF1α hydroxylation at different O2 concentrations. When [PHD2]0 is in excess, the response to decreasing O2 results in a steep change in hydroxylation upon reaching hypoxia – whereas 21 and 10% O2 levels result in similar amounts of HIF1αhydroxylated; at hypoxic levels of 1%, the amount hydroxylated at 20 minutes is half of that at normoxia (Fig. 4A). However, when [PHD2]0 is low, the response to a 20% drop in O2 levels is far less sensitive (Fig. 4D). Sensitivity to oxygen is measured by the slope of the [HIF1αhydroxylated] vs [O2] curve. A constant slope represents a uniform sensitivity across O2 levels. Iron has a similar effect on HIF1α at different O2 concentrations. When iron is available in excess, the response curve is steep. At initial iron concentrations of 0.05 μM, or one-thousandth of the default value, the amount of HIF1α hydroxylated is linearly related to the O2 level (Fig. 4C). Changes in the concentration of ascorbate did not have such a significant effect on hydroxylation (Fig. 4F). The steepness of the response decreased when two or more compounds were limiting factors in the HIF1α hydroxylation (Fig. 5). Table 2 shows the theoretical range of initial conditions where steep, switch-like changes in hydroxylation occur.
Fig. 4.
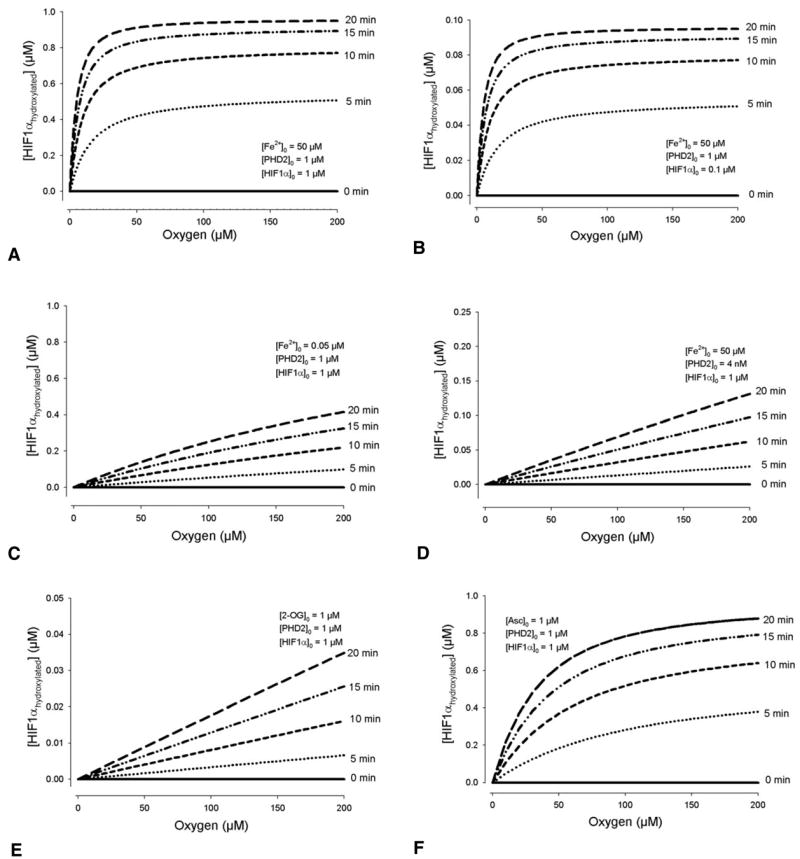
Model conditions where initial concentrations determine whether there is a switch-like response to O2 levels in the amount of HIF1α hydroxylated, or a gradual one. (A) When all enzymes are in excess, a steep drop in hydroxylation occurs as [O2] falls below 30 μM. For a range of initial unhydroxylated HIF1α between 0 and 1 μM, a steep, switch-like response is present. (B) An example with [HIF1α]0=0.1 μM. In comparison, when iron (C), PHD2 (D) or 2-oxoglutarate (E) is limiting, the hydroxylation shows no apparent switch-like behavior. (F) The effect of ascorbate is mixed. In A–E, [Asc]0=1000 μM, in excess. At low levels of the compound (below its Km for HIF1α, which is 180 μM), the oxygen response curve shows a more gradual, but non-linear reduction in hydroxylation. [Asc]0=1 μM is shown as an example.
Fig. 5.
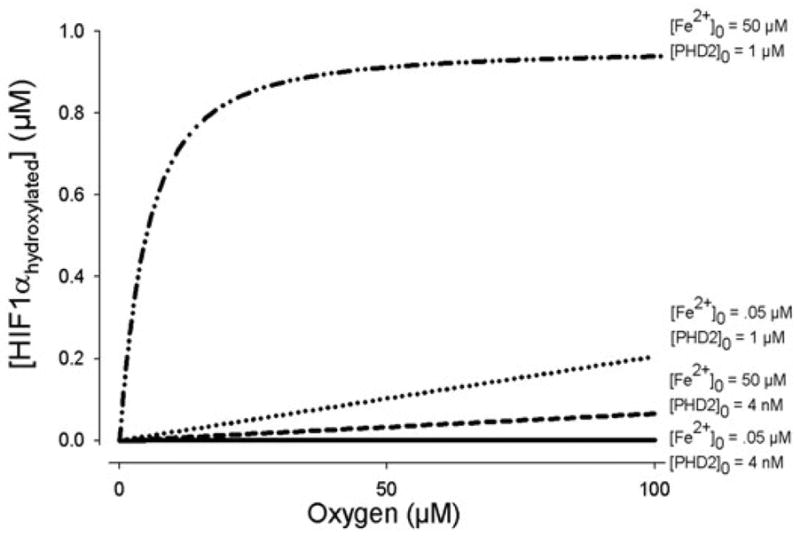
Comparisons of O2 response curves. When both PHD2 and Fe2+ are limiting reactants ([Fe2+]0=0.05, [PHD2]0=4 nM), the slope of the HIF1 hydroxylation curve (at 20 minutes) is significantly less than when either one of the compounds separately limit the reaction. These lines are compared with the oxygen response when all compounds are in excess. The effects of the changing sensitivity to oxygen are significant in hypoxia (below ~30 μM). For each line, [HIF1α]0=0.1 μM.
Chronic hypoxia: HIF1α and PHD2 synthesis
The assumed maximal HIF1α half-life range of 5–8 minutes in normoxia is consistent with at least three experiments (Berra et al., 2001; Huang et al., 1998; Jewell et al., 2001). However, the half-life of HIF1α upon reoxygenation depends on conditions such as duration of hypoxic exposure. For example, exposure to low oxygen levels beyond 6 hours, appears to decrease HIF1α half-life (G. Semenza, Johns Hopkins University, Baltimore, MD, personal communication). For short-term hypoxic exposure, the model assumes a maximum half-life of 5–8 minutes. The cited HIF1α half-life studies exposed cells to hypoxia for 1 hour (Jewell et al., 2001), 4 hours (Berra et al., 2001), and 4–6 hours (Huang et al., 1998) before reoxygenation. Beyond 4–6 hours of hypoxia, synthesis of HIF1α and PHD2 proteins occur. Fig. 6 shows the variability of HIF1α hydroxylation under conditions of chronic hypoxia, where a synthesis production term was added to the mass balance equations for HIF1α and PHD2 in the model. The simulated curves represent both the synthesis of HIF1α and its hydroxylation by increasing amounts of PHD2. In vivo, systems can adapt to chronic conditions, decreasing HIF1α expression within days of hypoxic exposure. A balance of HIF1α and PHD2 synthesis is a possible contributing mechanism.
Fig. 6.
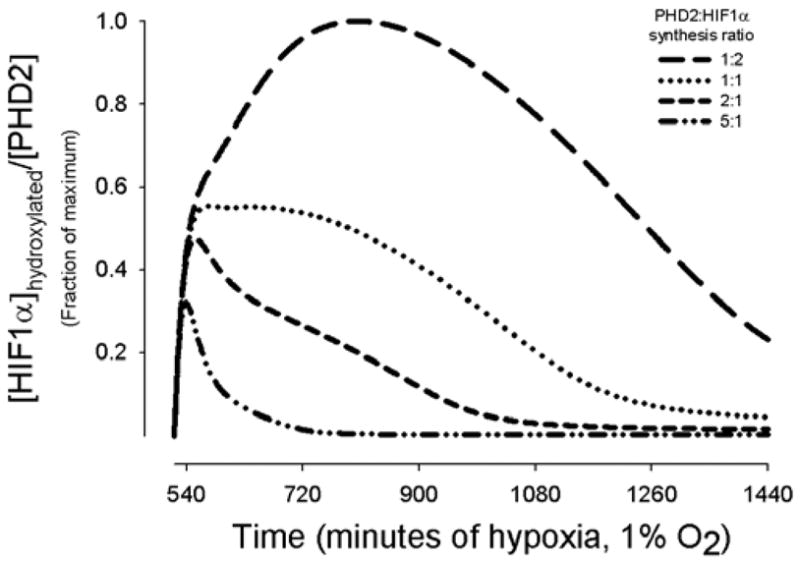
Effect of chronic hypoxia of 9 to 24 hours on HIF1α hydroxylation. Model results show the amount of hydroxylated [HIF1α] per [PHD2] for four different ratios of PHD2 synthesis rate to HIF1α synthesis rate relative to the maximum [HIF1α]hydroxylated/[PHD2]. HIF1α synthesis is a function of [O2] and duration of hypoxia, whereas PHD2 synthesis is a function of [HIF1α] and duration of hypoxia. HIF1α accumulation begins at 4 hours, and measurable PHD2 synthesis follows at 8 hours.
Therapeutic strategies to enhance HIF1α hydroxylation
Increased HIF1α nuclear expression has been associated with poor prognosis in several cancers (Nomura et al., 2004; Zagzag et al., 2000), and decreased susceptibility to radiotherapy (Vordermark and Brown, 2003; Vordermark et al., 2004). By enhancing HIF1 hydroxylation, the proteasome degradation rate of HIF1 can be increased, leading to a drop in nuclear accumulation of the dimer and associated hypoxia-dependent transcriptional activation. Targeting cofactors in the PHD reactions is one viable approach to increasing the hydroxylation rate. As our computational model predicted different oxygen sensitivity based on intracellular concentrations of key cofactors in the HIF1 reactions, we hypothesized that the microenvironment would also dictate the effectiveness of therapeutic strategies, and computationally, we would be able to predict the relative efficacy of each strategy at the molecular level.
Using the model, we tested two proposed approaches to increasing HIF1α hydroxylation: (1) increasing intracellular iron concentration (McCarty, 2003; Siddiq et al., 2005; Wartenberg et al., 2003) while simultaneously increasing intracellular ascorbate (Jones et al., 2006; Knowles et al., 2006; Knowles et al., 2003) and (2) increasing the expression of the main cytosolic prolyl hydroxylase, PHD2, directly (Fig. 7). Results show that increasing ascorbate is a proportionately more effective way to increase hydroxylation (Fig. 7A,B), when all other compounds are not limiting factors. At low levels of iron, doubling the amount of ascorbate from the standard in vitro level of 1000 μM, increases hydroxylation by as much as 60% at 50 μM O2; when iron is above 2 μM, the effect of additional ascorbate diminishes to a constant 3% increase in hydroxylated HIF1α (Fig. 7C,D).
Fig. 7.
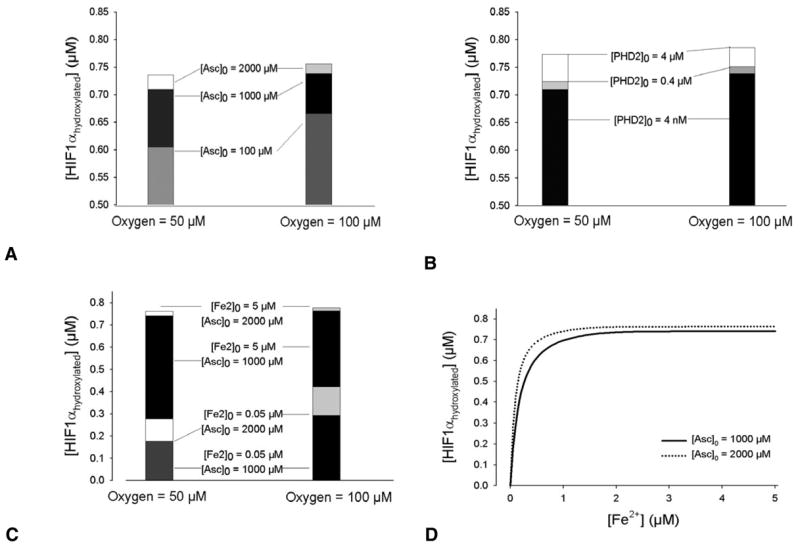
Testing potential anti-angiogenic strategies targeting HIF1α hydroxylation during normoxia and hypoxia. The effect on hydroxylation by addition of ascorbate (A), PHD2 (B), and iron and ascorbate (C) is shown for [O2]=50 and 100 μM. Initial concentrations of the compounds not shown are default values (Table 1). (D) Effect of doubling ascorbate concentration on HIF1α hydroxylation as a function of iron, at [O2]=50 μM. For [Fe2+]>5 μM, the increase in hydroxylated HIF1α when [Asc]0 is increased from 1000 to 2000 μM, remains 0.02 μM. For each reaction, t=10 minutes. Model predictions are based on in vitro values. Physiological in vivo concentrations are also variable, although in general lower. Ascorbate concentrations are estimated as 25–50 μM (Knowles et al., 2003); tissue Fe2+ levels may be as low as 10−12 μM (Bullen et al., 1978), whereas intracellular iron complexes are ~3–200 μM (Arredondo et al., 1997; Cooper et al., 1996; Hirsila et al., 2005), the fraction that is freely available for binding to PHD2 depends on cell type; absolute in vivo PHD2 concentrations are yet unknown – in cell extracts, they are in the nanomolar range.
Discussion
A rationale for developing the HIF model was to test several hypotheses on the signaling pathway, as it relates to hypoxic response, and to then use the results to evaluate therapeutic approaches targeting the onset of angiogenesis. An existing hypothesis of a switch-like change in HIF1α expression in response to a decrease in O2 levels to a critical level was tested (Kohn et al., 2004). The model demonstrated that based on the molecular environment and characteristics of the cell type, the response to hypoxia varies considerably.
There are several ways that cells are hypothesized to sense and respond to oxygen. The sensor of O2 levels has been independently attributed to iron (Postovit et al., 2005), HIF1α and HIF1α PHDs experimentally, but how their responses differ and when, has yet to be understood. The model shows two classes of HIF1 oxygen responses: a steep drop and a gradual drop in HIF1α hydroxylation in response to decreasing O2 levels. When all hydroxylation reactants are in excess, a steep drop in hydroxylation occurs during hypoxia. When 2-OG, Fe2+ or PHD2 are limiting, model results show the gradual response – a near-linear relationship between HIF1α hydroxylation and O2 level, i.e. a constant sensitivity to O2; this reflects the saturation kinetics used to represent the binding of HIF1α to these compounds (Fig. 4C–E). HIF1α hydroxylation is reduced when two or more required compounds are limiting (Fig. 5); what is notable is not only a significant reduction in the net amount of HIF1α hydroxylated, but a significant decrease in the relative sensitivity to O2 levels, defined by the steepness of the slope of the [HIF1αhydroxylated] vs [O2] curve.
Characterizing the role of ascorbate in the hydroxylation is complicated by its reaction with oxidized iron. At low levels of ascorbate (i.e. 1 μM, one-thousandth of the default initial value), HIF1α hydroxylation follows the curve shown in Fig. 4F. The decline in HIF1α hydroxylation at low O2 levels is less steep than the case where all compounds are in excess, however, it is not approaching linearity. This reflects the dual role of ascorbate: reactivating PHD2, by reducing the accumulating Fe3+; and, binding independently to the saturable enzyme complex formed by the binding of PHD2 with Fe2+, 2-OG and O2 (Majamaa et al., 1986). The decarboxylation of 2-OG without subsequent hydroxylation (termed uncoupled decarboxylation), which is catalyzed by PHDs and requires ascorbate, is not yet considered in the model. From the above observations, HIF1-activated cellular responses can largely be divided into two categories depending on the molecular environment: a steep, switch-like response to O2 levels, and a gradual one. The model shows that independently Fe2+ and PHD2 can act as the determinant of which response occurs. The high sensitivity of HIF1α hydroxylation to these compounds, notably at low (<5 μM) levels of oxygen, suggests that both iron and PHD2 play the role of hypoxic sensor.
Cellular in vivo concentrations of these compounds are difficult, if not yet possible, to measure. The concentration of HIF1α in vivo is thought to be in the sub-nanomolar range (Tuckerman et al., 2004). Based on the relative concentrations of other reactants in the HIF1 system, the response to O2 could be anticipated by the model. Although the quantitative values are largely unknown in vivo, it is known that iron and PHD2 concentrations are variable and affected by conditions such as hypoxia and anemia (Berra et al., 2003; Wardrop and Richardson, 1999). From the model, it would be predicted that this variability confers an advantage. With the HIF system being highly sensitive to the effects of multiple O2-dependent compounds, the response to O2 levels could be finely tuned, as well as robust.
Furthermore, the computational prediction that increasing ascorbate has significantly more effect at iron levels below 2 μM compared with higher iron levels, might later be extrapolated to characterizing tumor responsiveness to therapy based on in vivo microenvironments.
Using oxygen solubility in water, 37°C STP, 1.30 μmol/l/mmHg, [O2] of 21% corresponds to an O2 concentration of ~200 μM for in vitro experiments using cell lysates (Tuckerman et al., 2004). This is below the Km of oxygen reacting with PHD2, which was reported as 250 μM (Hirsila et al., 2003). Typical in vivo tissue concentrations are significantly lower, in the range of 6–25 μM, corresponding to 5–20 mmHg tissue pO2. The high Km indicates that the HIF1α reaction response remains highly sensitive to tissue O2 levels all the way from 0% oxygen to normoxia, under certain conditions. The model supports research on HIF1α nuclear expression changes being greatest at oxygen levels below 5% (Jiang et al., 1996), while showing how the most pronounced response by HIF1α to acute hypoxia can be achieved only with sufficient Fe2+, 2-OG and PHD2 (Fig. 4A,B). Varying the relative concentrations of the compounds involved in HIF1 hydroxylation, Fe2+ and PHD2 in particular, alters this sensitivity.
Other forms of variability and specificity could add to the flexibility of the HIF1 system in responding to oxygen. Although, as mentioned, PHD2 is the main PHD enzyme present in the cell cytoplasm during normoxia, the concentration of each PHD isoform varies by cellular microenvironment (Appelhoff et al., 2004; Berra et al., 2003). Hypoxia and estrogen change the relative concentrations of the PHD isoforms, and their relative contribution to the hydroxylation of HIF1α (Appelhoff et al., 2004).
While discussing other factors that influence HIF1α hydroxylation, it is appropriate to mention the limitations of the presented model. Five independent kinetic rates used in the model are unknown, and their values were estimated computationally. Their estimated values appear consistent with experimental results, and the key features of the hydroxylation reaction (steep and gradual responses to hypoxia) are robust over a range of all kinetic parameters. Additional experiments would be useful to validate the binding on- and off-rates. Kinetic rate constants taken from in vitro experiments may depend on slight differences in pH or other experimental variables (e.g. relative concentration of proteins) that have a limited degree of controllability within the cell. The enzyme used experimentally for the binding reactions was a minimal HIF1α peptide (residues 556–574) (Hirsila et al., 2003). Although increasing peptide length does not seem to affect the binding affinities in vitro to pVHL (Hon et al., 2002) or PHD2 (Hirsila et al., 2003) with HIF1α, the possibility that it may in vivo, cannot be ruled out. Additionally, estimates for binding kinetic rates are assumed sequential – this may or may not be what is actually measured by experiments, e.g. reactions may include PHD2-Fe2+ or PHD2-Fe2+-2-OG binding to HIF1α, etc., not just the completely modified PHD2 enzyme.
From the model, we predict several key characteristics about the mechanisms involved in the HIF1 pathway and apply the results to evaluate proposed therapies. Doing so, we provide a molecular representation of hypoxic response that merits further exploration experimentally and computationally. Future modeling studies include representing the independent hydroxylation of HIF1α on its Asn803 residue by factor-inhibiting HIF. To further approximate in vivo conditions, the model will need to represent the effect of pH, such as the acidic conditions found in tumors or in muscle during exercise, and its influence on VHL nucleolus sequestration. HIF2α and the different isoforms of PHD will become a part of the model, initially through modification of the binding rate constants and incorporation of spatial concentration heterogeneity. Incorporating the different sensitivity of the HIF1α degradation binding domains to iron and oxygen may yield a better mechanistic understanding of how the cell copes independently with a drop in iron or oxygen by altering PHD binding (Lee et al., 2005). Recently characterized compounds that influence the hydroxylation of HIF1α and its nuclear accumulation could be added, including OS-9, a protein that binds to HIF1α and the PHDs (Baek et al., 2005), and SUMO-1 protein, which covalently binds to HIF1α and affects its stability and transcriptional regulation (Bae et al., 2004). Although its role has yet to be fully elucidated, p53 binds to HIF1α in anoxia (~0–0.2% O2) and can promote its degradation (Fels and Koumenis, 2005); representing this binding in the model may account for the in vitro observation that a maximum in HIF1α nuclear expression occurs at 0.5% O2 (Jiang et al., 1996). Research into this maximum would consider the relative concentrations of PHD isoforms, and their binding site specificity as a function of O2 level (Chan et al., 2005), as well as the effect of reactive oxygen species (Schroedl et al., 2002).
It is the authors’ anticipation that the computational predictions will stimulate new experiments. An immediate proposal for an in vitro assay would be to quantitatively compare the effects of iron, ascorbate and PHD2 enzyme levels on the hydroxylation rate across a spectrum of O2 concentrations in a range of cell types. This would give an indication of whether indeed the response to O2 is of two natures, and whether concentrations of HIF1α co-factors determine if there is a steep drop in hydroxylation or a gradual decrease. Fig. 7C,D provide valuable predictions for therapeutic studies. Experiments to assess the relationship of ascorbate supplementation (e.g. 25, 1000e.g. 25, 2000 μM) in relationship to iron availability at different O2 levels (e.g. 0, 2, 5, 10, 20%) in cancer (e.g. MDA-MB-435) and endothelial cells (e.g. HUVEC, HBEC) would follow-up on the predictions of the model, and in vitro experiments (Jones et al., 2006; Knowles et al., 2006) to verify in which microenvironments the proposed therapies work effectively, and in what cells these microenvironments are found would be useful. Imaging techniques could be used to assess the variability present in tumor microenvironments – providing a basis for intra-tumor concentrations of compounds in the HIF1 pathway and allowing in vivo application of the computational model.
Different cell types may respond very differently to intracellular hypoxia than the model predicts, and the above experiments could help determine this. Deviations from predictions may reflect unaccounted mechanisms of the HIF1α pathway, and specifically, different reactions in the hydroxylation and degradation of HIF1α. There remains a question of whether the action of ascorbate is as described in the model (reducing Fe3+ to Fe2+ and binding the PHD2–2-OG–Fe2+–O2 intermediate complex), and whether this varies by cell type and in vivo conditions. The possibility of ascorbate working as a pro-oxidant at low concentrations and an antioxidant at high concentrations intracellularly involves alternate reactions affecting Fe2+, H2O2, O2 and HIF1α hydroxylation in the model, which could be assessed using techniques to determine ascorbate and H2O2 relationships (Kramarenko et al., 2006), in combination with VHL capture assays and HIF1α quantitative assessment.
The presented computational model adds a novel perspective to understanding the molecular details of how cells sense oxygen. It demonstrates how iron and PHD2 can determine whether the response to an acute hypoxic exposure is a pronounced or gradual change in the amount of HIF1αhydroxylated. This knowledge is applied to predict the response to proposed therapies targeting the HIF1 pathway. The model provides evidence as to how a changing microenvironment can significantly alter cell susceptibility to drugs that target cofactors in the HIF1α hydroxylation reactions. Even before experimental techniques allow measurements sensitive enough to detect intracellular molecular concentrations changes, a HIF1 pathway computational model can be used to gain an understanding of the molecular mechanisms underlying pre-angiogenic hypoxic response.
Materials and Methods
Formulation of computational model
From a comprehensive analysis of experimental data, we represent the hydroxylation of HIF1α by PHDs and the ubiquitylation of hydroxylated HIF1α by VHL (Fig. 1). In normoxia, PHD2 is the dominant PHD isoform that hydroxylates HIF1α and determines HIF1α concentrations in a range of cell types (Berra et al., 2003). We modeled the hydroxylation of HIF1α by PHD2 in the cell cytoplasm. The compounds involved in binding to PHD2 in preparation for the hydroxylation of HIF1α include iron, 2-oxoglutarate (2-OG), oxygen and ascorbate. The modified PHD2 then binds and hydroxylates HIF1α. Hydroxylated HIF1α is recognized and ubiquitylated by VBC, the complex that includes VHL bound to Elongins B and C, Cul2 and Rbx1. Equations 1–4 describe the overall scheme of HIF1α degradation. This includes HIF1α hydroxylation (Eqn 1), independent reactions of iron and ascorbate (Asc) (Eqns 2 and 3), and the binding of HIF1α to VHL (Eqn 4). Table 3 lists the compounds included in the model.
Table 3.
Model parameters and their abbreviations
| Variable | Abbreviation |
|---|---|
| Concentration of A | [A] |
| A·B | Binding of A and B |
| Ascorbate (Asc) | AS |
| Iron: Fe2+, Fe3+ | Fe2, Fe3 |
| Prolyl Hydroxylases (PHD) | PD2 |
| Hypoxia inducible factor (HIF1α) hydroxylated | Hαh |
| HIF1α unhydroxylated | Hα |
| von Hippel Lindau (VHL) | VL |
| HIF1α degradation products | HαD |
| 2-oxoglutarate (2-OG) | DG |
| Oxygen | O2 |
| HIF1β | Hβ |
| Elongin B | EB |
| Elongin C | EC |
| Cullins 2 | Cul2 |
| Hydrogen peroxide | H2O2 |
| dehydro-ascorbate | dAS |
Overall biochemical reaction of HIF1α hydroxylation:
| (1) |
Iron oxidation by reaction with hydrogen peroxide:
| (2) |
Iron reduction by ascorbate (Al-Ayash and Wilson, 1979; Williams and Yandell, 1982):
| (3) |
Reaction of VHL complex (VHL·Elongin B·Elongin C·Cul2·Rbx1) with HIF1α. HIF1α is polyubiquitylated and subsequently degraded:
| (4) |
The hydroxylation reactions follow enzyme-substrate binding kinetics. Governing equations are determined from mass balances surrounding the substrate and the intermediate enzyme-substrate complexes. Equation 5 shows an example of the kinetic reaction (Eqn 5A) and accompanying kinetic model (Eqn 5B) for initial steps in PHD2 hydroxylation of HIF1α (Eqn 5B). The full kinetic model can be found in Eqns 9–26. A combination of enzyme-substrate saturation assumptions was used for the binding of iron, ascorbate, oxygen and 2-oxoglutarate to PHD2, PHD2 hydroxylation of HIF, and VHL-mediated ubiquitylation. Model inputs are initial compound concentrations, including cellular O2 levels (Table 1). Output is HIF1α levels in the cell cytoplasm.
Table 1.
Parameters for the degradation of HIF1α in normoxia
| Constant | Value | Temperature (°C) | Reference |
|---|---|---|---|
| [H2O2]0 | 0.20 μM* | 37 | Gonzalez-Flecha and Demple, 1997 |
| [O2]0 | 200 μM | 37 | Tuckerman et al., 2004 |
| [Fe3+]0 | 0 μM | 37 | – |
| [Fe2+]0 | 50 μM | 37 | Tuckerman et al., 2004 |
| [2-OG]0 | 1000 μM | 37 | Tuckerman et al., 2004 |
| [Asc]0 | 1000 μM | 37 | Tuckerman et al., 2004 |
| [HIF1α]0 | 1 μM | 37 | Tuckerman et al., 2004 |
| [PHD2]0 | 1 μM; 4 nm (where stated for in vitro comparisons) | 37 | Tuckerman et al., 2004 |
| kon,Fe2 | 18 μM−1 min−1, estimate | 37 | – |
| koff,Fe2 | 36 min−1, estimate | 37 | – |
| kcat,Fe2 | 0, assumed | 37 | – |
| Km,Fe2 | 2 μM (0.03 μM) | 37 | Hirsila et al., 2005; Koivunen et al., 2004 |
| kon,DG | 1.8×10−1 μM−1 min−1, estimate | 37 | – |
| koff,DG | 10.8 min−1 estimate | 37 | – |
| kcat,DG | 0, assumed | 37 | – |
| Km,DG | 60 μM | 37 | Hirsila et al., 2003 |
| kon,O2 | 4.3×10−2 μM−1 min−1, estimate | 37 | – |
| koff,O2 | 10.8 min−1, estimate | 37 | – |
| kcat,O2 | 0, assumed | 37 | – |
| Km,O2 | 250 μM | 37 | Hirsila et al., 2003 |
| kon,AS | 1.8×10−2 μM−1 min−1, estimate | 37 | – |
| koff,AS | 3.6 min−1, estimate | 37 | – |
| kcat,AS | 0, assumed | 37 | – |
| Km,AS | 180 μM | 37 | Hirsila et al., 2003 |
| kon,Hα | 1.1×10−1 μM−1 min−1, estimate calculated | 37 | – |
| koff,Hα | 0.7 min−1, estimate | 37 | – |
| kcat,Hα | 0.098–0.164 min−1, estimate 0.08–10 min−1 min/max from experiments† | 37; 30 | Kersteen et al., 2004; Tuckerman et al., 2004 |
| Km,Hα | 7 μM | 37 | Hirsila et al., 2003 |
| kFe3 | 1.1×10−2 μM−1 min−1, 1.6×10−2 μM−1 min−1 | 30 | Beltran et al., 1998; Lovstad, 2003; Millero and Sotolongo, 1989‡ |
| kASFe | 6×10−3 μM−1 min−1 | 37 | Buettner and Jurkiewicz, 1996 |
| kon,VL | 42 μM−1 min−1 | 25 | Hon et al., 2002 |
| koff,VL | 1.3 min−1 | 25 | Hon et al., 2002 |
Values are experimentally determined or estimated, or estimated from model calculations as noted.
Assumed, as an approximation. Range of 0.13–0.25 μM given in the cited reference. Within this range, the effects on varying H2O2 initial concentration on the model response are relatively negligible.
Maximum is estimated from Kersteen et al. (Kersteen et al., 2004), where kcatHα=13±1 min−1 for (Pro-Pro-Gly) binding to PHD2; 9.2±1.2 min−1 for dansyl-GFPG-OE to PHD2. The model default value, 1 min−1, is closer to the minimum, estimated from Tuckerman et al. (Tuckerman et al., 2004).
For the Fenton reaction at pH 3–4, kFe3 is 76.52 M−1 s−1 (Beltran et al., 1998); this value is approximately threefold higher at pH 8 (Millero and Sotolongo, 1989). For pH 6.7 (the pH assumed for all other model kinetic parameters), kFe3 was estimated from Beltran et al. (Beltran et al., 1998) as 6.7/8*3*76.52 M−1s−1=1.1×10−2 μM−1 min−1, which corresponds with that reported by Lovstad (Lovstad, 2003).
PHD2r is the PHD2 enzyme modified following its reaction to iron.
| (5A) |
| (5B) |
PHD hydroxylation of HIF1α
While HIF1 was discovered in 1993, and its degradation kinetics have only been characterized in the past few years, the reactions of PHDs and lysyl hydroxylases have been analyzed for their roles in collagen hydroxylation since the mid-1960s (Adams and Frank, 1980; Gunsalus et al., 1975). During hydroxylation, sequential binding of collagen prolyl hydroxylase has been observed in the order of iron, 2-oxoglutarate, oxygen and peptide (Myllyla et al., 1977). The hydroxylation reaction of PHDs with HIF1 is thought to occur in the same sequence (M. Hirsila, Characterization of the novel human prolyl 4-hydroxlyases and asparaginyl hydroxylase that modify the hypoxia-inducible factor. PhD Thesis University of Oulu, Finland, 2004; http://herkules.oulu.fi/isbn9514275756/index.html?lang=en). Reflecting this in the model, we represent the binding of PHD2 with the substrates iron (Eqns 10–12), 2-oxoglutarate (Eqns 13 and 14), and oxygen (Eqns 15 and 16) sequentially. According to experiments (Hirsila et al., 2003), the reactions of the hydroxlyase binding to its cofactors follow saturation curves consistent with Michaelis-Menten kinetics. This is reflected in our model. Each reaction of a compound with PHD2 in the computational model is defined by three kinetic parameters: kon, koff and kcat (e.g. see Eqn 12). In pseudo-steady-state conditions, where the fluxes of intermediate compounds (enzyme-substrate) are constant, the Michaelis-Menten constant, Km, relates these three parameters:
| (6) |
If no intermediate products are formed, then the catalytic rate constant for production formation kcat are set to zero, and Km can be estimated by the dissociation constant, KD:
| (7) |
In the human type I collagen prolyl hydroxylase reactions with iron, 2-oxoglutarate, oxygen and the peptide substrate (Pro-Pro-Gly)10, the reported Km values are close to the dissociation constants, and the assumption of their equality (Eqn 7) appears valid (Hieta et al., 2003; Myllyla et al., 1977). The HIF-PHDs and the collagen PHDs share the same hydroxylation reaction sequence and binding cosubstrates. At first glance, it seems reasonable to make the same equilibrium binding assumptions for HIF-PHDs as for the collagen PHDs. However, the overall PHD2 hydroxylation of HIF1α is not at equilibrium and probably unidirectional (Chan et al., 2005; Willam et al., 2004). For the model presented here, the binding was assumed bi-directional between PHD2, iron, 2-oxoglutarate, oxygen (Epstein et al., 2001) and ascorbate, with negligible intermediates formed (Eqn 9) (Goda et al., 2003). For the intermediate complexes of PHD2 and its cofactors, this means the catalytic production terms involving kcat,Fe2, kcat,DG, kcat,O2 and kcat,AS were set to zero during model runs.
The final step of the PHD2 reaction sequence, the hydroxylation of HIF1α by the modified PHD2 enzyme complex, was first modeled using the assumption of near irreversibility, and Briggs-Haldane, where kcat≫koff. Assuming pseudo-steady-state conditions, characteristics of Briggs-Haldane kinetics hold (Briggs and Haldane, 1925; Cornish-Bowden, 2004):
| (8) |
Effects of larger relative koff rates were also tested, and the current model includes a final irreversible hydroxylation step that considers all three constants, kon, koff and kcat, to be significant (Eqns 19–21).
Ascorbate and iron reactions
Ascorbate is assumed to be a co-reactant with compounds in the hydroxylation reaction, as has been shown with collagen PHDs (Majamaa et al., 1986). Hydroxylation proceeds without ascorbate when there is sufficient iron in the form of Fe2+ present (Myllyla et al., 1984; Tuderman et al., 1977). To account for the possibility of the reaction proceeding without ascorbate, the ascorbate-independent binding of HIF1α to PHD2 · Fe2+ · 2-OG · O2 and subsequent hydroxylation is part of the model (third term in Eqn 20). However, eventually without any ascorbate, Fe2+ can be oxidized to its Fe3+ (or Fe4+) form, leaving insufficient iron to bind to PHD2 and halting the hydroxylation reaction. The role of ascorbate in the model is to bind to O2 and reduce Fe3+ back to Fe2+. We represent the Fe2+ reaction with H2O2 (Eqns 2, 10 and 22) and Fe3+ reduction by ascorbate (Eqns 3, 17 and 22) in the model.
HIF1α ubiquitylation and degradation
Following hydroxylation, VHL ubiquitylation of HIF1α is likely also not at equilibrium. However VHL binding is notably reversible (kcat,HαV=0, in Eqns 23–26). The deubiquitylating enzyme, VDU2, interacts with pVHL, mediates the backward reaction and stabilizes HIF1α (Li et al., 2005). We make the kinetic assumption that proteosome degradation of HIF1α after ubiquitylation is approximately first order in ubiquitylated HIF1α. This simplification seems a valid approximation from reoxygenation experiments (Huang et al., 1998), although it leaves to future studies the exploration of how decay rate varies with the ratio of free to bound or modified HIF1α.
Model parameters
The model rate constants are given in Table 1. Ten of the constants are derived from experimental data (Buettner and Jurkiewicz, 1993; Hirsila et al., 2003; Hirsila et al., 2005; Hon et al., 2002; Kersteen et al., 2004; Lovstad, 2003; Tuckerman et al., 2004). Km was available from experimental data while individual kon and koff were calculated from the model for all but the VHL binding step, where on and off rates were known experimentally. Values for the unknown parameters were estimated as described above and shown in supplementary figures. Default values for kinetic constants and initial conditions are shown in Table 1 with corresponding references. For experiments, O2 levels are given in percentages, mmHg, or micromolar quantities. The last is reserved for cell culture experiments, where O2 concentration is calculated in solution. Where appropriate for comparison, we converted model results to the measured experimental units. For cell culture experiments characterizing conditions in cell lysates, values given as percent oxygen or mmHg were equated to micromolar quantities based on an oxygen solubility in water (Tuckerman et al., 2004).
Numerical solution
The system of nonlinear differential equations presented in Eqns 9–26 was solved using Mathworks Matlab software. The ode23s solver, based on a modified Rosenbrock formula, was used to find a solution for the series of seventeen differential equations. For the time integration, the solver used adjustable time steps with default absolute error tolerance in the solution of 10−6 μM.
Governing equations
Overall reaction sequence, incorporating the assumptions of pseudo-steady-state and bidirectionality in the binding of prolyl hydroxylase with iron, deoxyglutarate, oxygen and ascorbate; assumptions of pseudo-steady-state and unidirectionality in the hydroxylation of HIF1α; and reversibility in the ubiquitylation of hydroxylated HIF1α by the VHL complex where Hα is unhydroxylated HIF1α and Hαh is hydroxylated HIF1α (the following equations use the notation defined in Table 3):
| (9) |
kcat terms for the binding of prolyl hydroxylase 2 (PD2) with its cofactors (Fe2, DG, O2, AS) are included in the kinetic model for completeness. kcat represents the catalytic rate constants for production of intermediate complexes with PD2. These rates are set to zero using the assumptions described in the Materials and Methods.
Production terms qo in Eqn 12 and q in Eqn 20 are nonzero only in chronic hypoxia (>6 hours, Fig. 6). Estimates of these functions are based on experiments (Appelhoff et al., 2004; Berra et al., 2003; D’Angelo et al., 2003).
Reactions of PD2 and Fe2 and Fe2 with H2O2
| (10) |
| (11) |
| (12) |
Reaction of PD2·Fe2 and DG
| (13) |
| (14) |
Reaction of PD2·Fe2·DG and O2
| (15) |
| (16) |
Reaction of PD2·Fe2·DG·O2 and AS
| (17) |
| (18) |
Reaction of PD2·Fe2·DG·O2·AS (PD2mod) and Hα
Note for reactions 19 and 20, these equations were modified to allow reaction with an intermediate uncoupled to AS
| (19) |
| (20) |
| (21) |
Reaction of Fe3 with AS and O2
| (22) |
Related reactions:
Addition of VHL binding
| (23) |
The following reactions represent degradation kinetics involving ubiquitylation and proteasome degradation of HIF1α. The current model is limited to the VHL complex binding to hydroxylated HIF1α, where degradation is assumed a first order reaction.
| (24) |
| (25) |
| (26) |
Parameter estimation
Ten parameters (five of them independent), were unknown experimentally: kon,Fe2, koff,Fe2, kon,DG, koff,DG, kon,O2, koff,O2,, kon,AS, koff,AS, kon,Hα and koff,Hα. The following protocol was used to estimate these parameters. Initial rough estimates for kon and koff, for the reactions of iron, 2-OG and ascorbate with PHD2, were found from binding of these reactants with other substrates (collagen prolyl hydroxylase, lysyl hydroxylase). Fe2+ oxidization and reduction rate constants were estimated respectively from values for the Fenton reaction and from (Buettner and Jurkiewicz, 1996). Sensitivity analysis confirmed estimates for kinetic constants. With the assumption kcat=0 for intermediate steps, kon rates were determined by koff/Km. In the case of kon,Hα and koff,Hα, where kcat,Hα was a significant value estimated from experiments, the relationship Km=(kcat+koff)/kon was used.
Sensitivity analysis
For all estimated parameters, the kinetic parameter of interest was varied over a minimum range of 1000-fold, while the remaining parameters were held constant. Calculated HIF1α half-lives were compared with experimental data (Berra et al., 2001; Huang et al., 1998; Jewell et al., 2001), to narrow the range of reasonable kinetic parameter values (supplementary material Fig. S4).
The estimated range for the catalytic constant kcat,Hα (Eqn A12) was relatively small: 0.098–0.164 minute−1; this corresponds well with a 0.1 minute−1 approximation from experiments (Tuckerman et al., 2004). Testing ±100-fold differences in kcat showed the model’s high sensitivity to this parameter (supplementary material Fig. S4i). The remaining kinetic parameters estimated from the model were koffs for the binding of iron (Eqns 10–12), 2-OG (Eqns 13,14), oxygen (Eqns 15,16) and ascorbate (Eqns 17,18) with PHD2. These were determined using the time of 5 minutes for half [HIF1α]0 = 1 μM to be hydroxylated (this is based on experiments and the estimate that ubiquitylation of hydroxylated HIF1α takes ≤3 minutes; see Model validation section). With this assumption, the estimated minimum kFe,off is 36 minute−1; if the 5–8 minute HIF1α half-life range is considered, the range of kFe,off is 0.019–36 minute−1 (supplementary material Fig. S4ii). Kinetic off-rate ranges were also determined for the PHD2 enzyme complex binding to 2-OG (supplementary material Fig. S4iii), 2-OG (supplementary material Fig. S4iv), and ascorbate (supplementary material Fig. S4v). To test the assumption of irreversibility in the binding of modified PHD2 with HIF1α, a range of koff values were explored (supplementary material Fig. S4vi). The graph shows the feasibility of a nearly irreversible reaction, with koff,Hα close to zero giving a half-life within the experimental range expected. However the sensitivity of the HIF1α half-life to koff,Hα was low, and another comparison was needed to help limit its value.
A second means to confirm estimates for kinetic constants was a comparison of PHD2 specific activity with that from Tuckerman et al. (Tuckerman et al., 2004), which estimated a lower limit of 20 mol HIF1α hydroxylated/mol PHD2/minute, as a constant rate over the first 6 minutes in hypoxic MDA-MB-435 cell extracts. Using initial concentrations consistent with those reported in the experiment, [HIF1α]0=1 μM; [Ascorbate]0=1 mM; [α-ketoglutarate]0=1 mM (model, [2-oxoglutarate]0=1 mM); [FeCl2]0=50 μM (model, [Fe2+]0=50 μM); [PHD2]0=4 nM; [O2]0=209 μM (ambient, 21%), and varying the kinetic parameters of interest, the model was compared with experiments to determine minimum estimates for kcat,Hα, koff,Fe2, koff,DG, koff,O2, koff,AS and koff,Hα using the best fit determined by linear regression of the computational curves and least squares analysis comparisons to the data (supplementary material Fig. S5). The model’s lower estimates for the kinetic rates corresponded well to the specific activity value from experiments. An exception was seen consistently at two minutes, where lower estimates for the model’s kinetic constants underestimated the reported specific activity. Using the range of kcat,Hα corresponding to the HIF1α half-life of 5–8 min, the higher kcat,Hα values increased the predicted specific activity for the same koffs.
After determining estimated kinetic constants, we investigated the sensitivity of the hydroxylation reaction to initial concentrations of reactants (supplementary material Fig. S6). Fig. S1 in supplementary material shows model estimates for PHD2 specific activity and HIF1α half-life using the newly reported Km,Fe2 value of 0.03 μM, and the corresponding best fit kon and koff rates for the other binding reactions. Alternate choices for the set of kon and koff rates, using the maximum HIF1α half-life of 8 minutes, resulted in higher estimates for PHD2 specific activity.
Sensitivity analysis was also performed for all parameters found from experiments: Km,Fe2, Km,DG, Km,O2, Km,AS, Km,Hα, kFe3, kASFe, kon,VL and koff,VL using the protocol described for estimated parameters (supplementary material Fig. S2 show examples of Km,Fe2 and Km,DG). Changes in kFe3 and kASFe, for the reactions of Fe2+ with H2O2 and ascorbate, had negligible effects on hydroxylation at default initial conditions; as determined by the model, constants for ubiquitylation (kon,VL and koff,VL) have no effect on hydroxylation. Sensitivity analysis shows the switch-like, steep drop in HIF1α hydroxylation at low O2 levels is a consistent feature (supplementary material Fig. S3). The range of kinetic values used for analysis was 0.2Km–4Km, where Km is the experimental value (Table 1). Comparison of PHD2 specific activity to experiments determined this was an appropriate range (supplementary material Fig. S2ii,iv). An exception was Km,Fe2, where the range was 0.025Km,Fe2–4Km,Fe2 (supplementary material Fig. S2ii). Changes in Km,Fe2 had minimal effects on the hydroxylation curve (supplementary material Fig. S3i). For Km,DG, Km,O2, Km,AS, and Km,Hα, increasing Km values increase the time for hydroxylation, thereby decreasing the steepness of the HIF1α response (supplementary material Fig. S3ii). Increasing Km values also make the system more sensitive to changes in oxygen at higher O2 levels.
Numerical methods
A script was written in Mathworks Matlab to run the model repeatedly and perform sensitivity analysis over the described parameter range.
Note on the definition of switch-like
The term ‘switch’ in its mathematical use is a gate, defining two states of a system (e.g. ‘on’ or ‘off’). For biological purposes, switch takes a broader definition – a distinct change in conditions that triggers a physiological state change (e.g. HIF1 or a threshold concentration of HIF1α protein as the angiogenic switch). Here we define ‘switch-like’ as a threshold change in HIF1α protein concentration in response to a specified decrease in O2 levels; this definition is to distinguish the change from a gradual increase in protein levels. Switch-like properties can be determined by calculating the slope of the fraction of [HIF1αhydroxylated] vs [O2] curve. A constant slope indicates a uniform response to increasing hypoxia across all O2 levels. A gradual response in this article (Fig. 4C,D,E and Fig. 5), refers to a constant slope of
A switch-like response refers to a slope that changes abruptly from a near constant zero at normoxia to greater than 0.016 at low O2 levels (Fig. 4A,B). Experimentally, the absolute threshold distinguishing this difference in response would vary by cell type and experimental set-up; regardless, a distinct difference in response would be measurable by assessing changes in HIF1α expression with changing O2 levels.
Supplementary Material
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/119/16/3467/DC1
Acknowledgments
This work was supported by NIH Grants HL79653 and HL18292. The authors thank G. Semenza, R. Pili, D. Qian, Z. Bhujwalla and J. Myllyharju for helpful discussions.
References
- Adams E, Frank L. Metabolism of proline and the hydroxyprolines. Annu Rev Biochem. 1980;49:1005–1061. doi: 10.1146/annurev.bi.49.070180.005041. [DOI] [PubMed] [Google Scholar]
- Al-Ayash AI, Wilson MT. The mechanism of reduction of single-site redox proteins by ascorbic acid. Biochem J. 1979;177:641–648. doi: 10.1042/bj1770641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ, Gleadle JM. Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia-inducible factor. J Biol Chem. 2004;279:38458–38465. doi: 10.1074/jbc.M406026200. [DOI] [PubMed] [Google Scholar]
- Arredondo M, Orellana A, Garate MA, Nunez MT. Intracellular iron regulates iron absorption and IRP activity in intestinal epithelial (Caco-2) cells. Am J Physiol. 1997;273:G275–G280. doi: 10.1152/ajpgi.1997.273.2.G275. [DOI] [PubMed] [Google Scholar]
- Bae SH, Jeong JW, Park JA, Kim SH, Bae MK, Choi SJ, Kim KW. Sumoylation increases HIF-1alpha stability and its transcriptional activity. Biochem Biophys Res Commun. 2004;324:394–400. doi: 10.1016/j.bbrc.2004.09.068. [DOI] [PubMed] [Google Scholar]
- Baek JH, Mahon PC, Oh J, Kelly B, Krishnamachary B, Pearson M, Chan DA, Giaccia AJ, Semenza GL. OS-9 interacts with hypoxia-inducible factor 1alpha and prolyl hydroxylases to promote oxygen-dependent degradation of HIF-1alpha. Mol Cell. 2005;17:503–512. doi: 10.1016/j.molcel.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Beltran FJ, Gonzalez M, Rivas FJ, Alvarez P. Fenton reagent advanced oxidation of polynuclear aromatic hydrocarbons in water. Water Air Soil Pollut. 1998;105:685–700. [Google Scholar]
- Berra E, Roux D, Richard DE, Pouyssegur J. Hypoxia-inducible factor-1 alpha (HIF-1 alpha) escapes O(2)-driven proteasomal degradation irrespective of its subcellular localization: nucleus or cytoplasm. EMBO Rep. 2001;2:615–620. doi: 10.1093/embo-reports/kve130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berra E, Benizri E, Ginouves A, Volmat V, Roux D, Pouyssegur J. HIF prolyl-hydroxylase 2 is the key oxygen sensor setting low steady-state levels of HIF-1alpha in normoxia. EMBO J. 2003;22:4082–4090. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blouw B, Song H, Tihan T, Bosze J, Ferrara N, Gerber HP, Johnson RS, Bergers G. The hypoxic response of tumors is dependent on their microenvironment. Cancer Cell. 2003;4:133–146. doi: 10.1016/s1535-6108(03)00194-6. [DOI] [PubMed] [Google Scholar]
- Bos R, van Diest PJ, de Jong JS, van der Groep P, van der Valk P, van der Wall E. Hypoxia-inducible factor-1alpha is associated with angiogenesis, and expression of bFGF, PDGF-BB, and EGFR in invasive breast cancer. Histopathology. 2005;46:31–36. doi: 10.1111/j.1365-2559.2005.02045.x. [DOI] [PubMed] [Google Scholar]
- Briggs GE, Haldane JBS. A note on the kinetics of enzyme action. Biochem J. 1925;19:339. doi: 10.1042/bj0190338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buettner GR, Jurkiewicz BA. Ascorbate free radical as a marker of oxidative stress: an EPR study. Free Radic Biol Med. 1993;14:49–55. doi: 10.1016/0891-5849(93)90508-r. [DOI] [PubMed] [Google Scholar]
- Buettner GR, Jurkiewicz BA. Catalytic metals, ascorbate and free radicals: combinations to avoid. Radiat Res. 1996;145:532–541. [PubMed] [Google Scholar]
- Bullen JJ, Rogers HJ, Griffiths E. Role of iron in bacterial infection. Curr Top Microbiol Immunol. 1978;80:1–35. doi: 10.1007/978-3-642-66956-9_1. [DOI] [PubMed] [Google Scholar]
- Chan DA, Sutphin PD, Yen SE, Giaccia AJ. Coordinate regulation of the oxygen-dependent degradation domains of Hypoxia-inducible factor 1{alpha} Mol Cell Biol. 2005;25:6415–6426. doi: 10.1128/MCB.25.15.6415-6426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper CE, Lynagh GR, Hoyes KP, Hider RC, Cammack R, Porter JB. The relationship of intracellular iron chelation to the inhibition and regeneration of human ribonucleotide reductase. J Biol Chem. 1996;271:20291–20299. doi: 10.1074/jbc.271.34.20291. [DOI] [PubMed] [Google Scholar]
- Cornish-Bowden A. Fundamentals of Enzyme Kinetics. London: Portland Press; 2004. [Google Scholar]
- D’Angelo G, Duplan E, Boyer N, Vigne P, Frelin C. Hypoxia up-regulates prolyl hydroxylase activity: a feedback mechanism that limits HIF-1 responses during reoxygenation. J Biol Chem. 2003;278:38183–38187. doi: 10.1074/jbc.M302244200. [DOI] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, et al. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- Esposito BP, Epsztejn S, Breuer W, Cabantchik ZI. A review of fluorescence methods for assessing labile iron in cells and biological fluids. Anal Biochem. 2002;304:1–18. doi: 10.1006/abio.2002.5611. [DOI] [PubMed] [Google Scholar]
- Fels DR, Koumenis C. HIF-1alpha and p53: the ODD couple? Trends Biochem Sci. 2005;30:426–429. doi: 10.1016/j.tibs.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Goda N, Dozier SJ, Johnson RS. HIF-1 in cell cycle regulation, apoptosis, and tumor progression. Antioxid Redox Signal. 2003;5:467–473. doi: 10.1089/152308603768295212. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Flecha B, Demple B. Homeostatic regulation of intracellular hydrogen peroxide concentration in aerobically growing Escherichia coli. J Bacteriol. 1997;179:382–388. doi: 10.1128/jb.179.2.382-388.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groulx I, Lee S. Oxygen-dependent ubiquitination and degradation of hypoxia-inducible factor requires nuclear-cytoplasmic trafficking of the von Hippel-Lindau tumor suppressor protein. Mol Cell Biol. 2002;22:5319–5336. doi: 10.1128/MCB.22.15.5319-5336.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus IC, Pederson TC, Sligar SG. Oxygenase-catalyzed biological hydroxylations. Annu Rev Biochem. 1975;44:377–407. doi: 10.1146/annurev.bi.44.070175.002113. [DOI] [PubMed] [Google Scholar]
- Hewitson KS, Schofield CJ. The HIF pathway as a therapeutic target. Drug Discov Today. 2004;9:704–711. doi: 10.1016/S1359-6446(04)03202-7. [DOI] [PubMed] [Google Scholar]
- Hieta R, Kukkola L, Permi P, Pirila P, Kivirikko KI, Kilpelainen I, Myllyharju J. The peptide-substrate-binding domain of human collagen prolyl 4-hydroxylases. Backbone assignments, secondary structure, and binding of proline-rich peptides. J Biol Chem. 2003;278:34966–34974. doi: 10.1074/jbc.M303624200. [DOI] [PubMed] [Google Scholar]
- Hirsila M, Koivunen P, Gunzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- Hirsila M, Koivunen P, Xu L, Seeley T, Kivirikko KI, Myllyharju J. Effect of desferrioxamine and metals on the hydroxylases in the oxygen sensing pathway. FASEB J. 2005;19:1308–1310. doi: 10.1096/fj.04-3399fje. [DOI] [PubMed] [Google Scholar]
- Hon WC, Wilson MI, Harlos K, Claridge TD, Schofield CJ, Pugh CW, Maxwell PH, Ratcliffe PJ, Stuart DI, Jones EY. Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL. Nature. 2002;417:975–978. doi: 10.1038/nature00767. [DOI] [PubMed] [Google Scholar]
- Huang LE, Gu J, Schau M, Bunn HF. Regulation of hypoxia-inducible factor 1alpha is mediated by an O2-dependent degradation domain via the ubiquitin-proteasome pathway. Proc Natl Acad Sci USA. 1998;95:7987–7992. doi: 10.1073/pnas.95.14.7987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivan M, Kondo K, Yang H, Kim W, Valiando J, Ohh M, Salic A, Asara JM, Lane WS, Kaelin WG., Jr HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science. 2001;292:464–468. doi: 10.1126/science.1059817. [DOI] [PubMed] [Google Scholar]
- Jewell UR, Kvietikova I, Scheid A, Bauer C, Wenger RH, Gassmann M. Induction of HIF-1alpha in response to hypoxia is instantaneous. FASEB J. 2001;15:1312–1314. [PubMed] [Google Scholar]
- Jiang BH, Semenza GL, Bauer C, Marti HH. Hypoxia-inducible factor 1 levels vary exponentially over a physiologically relevant range of O2 tension. Am J Physiol. 1996;271:C1172–C1180. doi: 10.1152/ajpcell.1996.271.4.C1172. [DOI] [PubMed] [Google Scholar]
- Jones DT, Trowbridge IS, Harris AL. Effects of transferrin receptor blockade on cancer cell proliferation and hypoxia-inducible factor function and their differential regulation by ascorbate. Cancer Res. 2006;66:2749–2756. doi: 10.1158/0008-5472.CAN-05-3857. [DOI] [PubMed] [Google Scholar]
- Josko J, Mazurek M. Transcription factors having impact on vascular endothelial growth factor (VEGF) gene expression in angiogenesis. Med Sci Monit. 2004;10:RA89–RA98. [PubMed] [Google Scholar]
- Kamura T, Koepp DM, Conrad MN, Skowyra D, Moreland RJ, Iliopoulos O, Lane WS, Kaelin WG, Jr, Elledge SJ, Conaway RC, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science. 1999;284:657–661. doi: 10.1126/science.284.5414.657. [DOI] [PubMed] [Google Scholar]
- Kamura T, Sato S, Iwai K, Czyzyk-Krzeska M, Conaway RC, Conaway JW. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci USA. 2000;97:10430–10435. doi: 10.1073/pnas.190332597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kersteen EA, Higgin JJ, Raines RT. Production of human prolyl 4-hydroxylase in Escherichia coli. Protein Expr Purif. 2004;38:279–291. doi: 10.1016/j.pep.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Knowles HJ, Raval RR, Harris AL, Ratcliffe PJ. Effect of ascorbate on the activity of hypoxia-inducible factor in cancer cells. Cancer Res. 2003;63:1764–1768. [PubMed] [Google Scholar]
- Knowles HJ, Mole DR, Ratcliffe PJ, Harris AL. Normoxic stabilization of hypoxia-inducible factor-1alpha by modulation of the labile iron pool in differentiating U937 macrophages: effect of natural resistance-associated macrophage protein 1. Cancer Res. 2006;66:2600–2607. doi: 10.1158/0008-5472.CAN-05-2351. [DOI] [PubMed] [Google Scholar]
- Kohn KW, Riss J, Aprelikova O, Weinstein JN, Pommier Y, Barrett JC. Properties of switch-like bioregulatory networks studied by simulation of the hypoxia response control system. Mol Biol Cell. 2004;15:3042–3052. doi: 10.1091/mbc.E03-12-0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koivunen P, Hirsila M, Gunzler V, Kivirikko KI, Myllyharju J. Catalytic properties of the asparaginyl hydroxylase (FIH) in the oxygen sensing pathway are distinct from those of its prolyl 4-hydroxylases. J Biol Chem. 2004;279:9899–9904. doi: 10.1074/jbc.M312254200. [DOI] [PubMed] [Google Scholar]
- Koshiji M, Huang LE. Dynamic balancing of the dual nature of HIF-1alpha for cell survival. Cell Cycle. 2004;3:853–854. doi: 10.4161/cc.3.7.990. [DOI] [PubMed] [Google Scholar]
- Kramarenko GG, Wilke WW, Dayal D, Buettner GR, Schafer FQ. Ascorbate enhances the toxicity of the photodynamic action of Verteporfin in HL-60 cells. Free Radic Biol Med. 2006;40:1615–1627. doi: 10.1016/j.freeradbiomed.2005.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KH, Choi E, Chun YS, Kim MS, Park JW. Differential responses of two degradation domains of HIF-1alpha to hypoxia and iron deficiency. Biochimie. 2005;88:163–169. doi: 10.1016/j.biochi.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Li Z, Wang D, Messing EM, Wu G. VHL protein-interacting deubiquitinating enzyme 2 deubiquitinates and stabilizes HIF-1alpha. EMBO Rep. 2005;6:373–378. doi: 10.1038/sj.embor.7400377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Los M, Jansen GH, Kaelin WG, Lips CJ, Blijham GH, Voest EE. Expression pattern of the von Hippel-Lindau protein in human tissues. Lab Invest. 1996;75:231–238. [PubMed] [Google Scholar]
- Lovstad RA. A kinetic study on iron stimulation of the xanthine oxidase dependent oxidation of ascorbate. Biometals. 2003;16:435–439. doi: 10.1023/a:1022523920998. [DOI] [PubMed] [Google Scholar]
- Majamaa K, Gunzler V, Hanauske-Abel HM, Myllyla R, Kivirikko KI. Partial identity of the 2-oxoglutarate and ascorbate binding sites of prolyl 4-hydroxylase. J Biol Chem. 1986;261:7819–7823. [PubMed] [Google Scholar]
- Marti HH. Erythropoietin and the hypoxic brain. J Exp Biol. 2004;207:3233–3242. doi: 10.1242/jeb.01049. [DOI] [PubMed] [Google Scholar]
- McCarty MF. Turning an ‘Achilles’ Heel’ into an asset–activation of HIF-1alpha during angiostatic therapy will increase tumor sensitivity to iron-catalyzed oxidative damage. Med Hypotheses. 2003;61:509–511. doi: 10.1016/s0306-9877(03)00229-9. [DOI] [PubMed] [Google Scholar]
- Milkiewicz M, Hudlicka O, Verhaeg J, Egginton S, Brown MD. Differential expression of Flk-1 and Flt-1 in rat skeletal muscle in response to chronic ischaemia: favourable effect of muscle activity. Clin Sci. 2003;105:473–482. doi: 10.1042/CS20030035. [DOI] [PubMed] [Google Scholar]
- Millero FJ, Sotolongo S. The oxidation of Fe(II) with H2O2 in seawater. Geochim Cosmochim Acta. 1989;53:1867–1873. [Google Scholar]
- Myllyla R, Tuderman L, Kivirikko KI. Mechanism of the prolyl hydroxylase reaction. 2 Kinetic analysis of the reaction sequence. Eur J Biochem. 1977;80:349–357. doi: 10.1111/j.1432-1033.1977.tb11889.x. [DOI] [PubMed] [Google Scholar]
- Myllyla R, Majamaa K, Gunzler V, Hanauske-Abel HM, Kivirikko KI. Ascorbate is consumed stoichiometrically in the uncoupled reactions catalyzed by prolyl 4-hydroxylase and lysyl hydroxylase. J Biol Chem. 1984;259:5403–5405. [PubMed] [Google Scholar]
- Nomura M, Nomura N, Newcomb EW, Lukyanov Y, Tamasdan C, Zagzag D. Geldanamycin induces mitotic catastrophe and subsequent apoptosis in human glioma cells. J Cell Physiol. 2004;201:374–384. doi: 10.1002/jcp.20090. [DOI] [PubMed] [Google Scholar]
- Postovit LM, Sullivan R, Adams MA, Graham CH. Nitric oxide signalling and cellular adaptations to changes in oxygenation. Toxicology. 2005;208:235–248. doi: 10.1016/j.tox.2004.11.022. [DOI] [PubMed] [Google Scholar]
- Powell FL. Functional genomics and the comparative physiology of hypoxia. Annu Rev Physiol. 2003;65:203–230. doi: 10.1146/annurev.physiol.65.092101.142711. [DOI] [PubMed] [Google Scholar]
- Schroedl C, McClintock DS, Budinger GR, Chandel NS. Hypoxic but not anoxic stabilization of HIF-1alpha requires mitochondrial reactive oxygen species. Am J Physiol Lung Cell Mol Physiol. 2002;283:L922–L931. doi: 10.1152/ajplung.00014.2002. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hydroxylation of HIF-1: oxygen sensing at the molecular level. Physiology Bethesda. 2004;19:176–182. doi: 10.1152/physiol.00001.2004. [DOI] [PubMed] [Google Scholar]
- Siddiq A, Ayoub IA, Chavez JC, Aminova L, Shah S, Lamanna JC, Patton SM, Connor JR, Cherny RA, Volitakis I, et al. HIF prolyl 4-hydroxylase inhibition: a target for neuroprotection in the central nervous system. J Biol Chem. 2005;280:41732–41743. doi: 10.1074/jbc.M504963200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckerman JR, Zhao Y, Hewitson KS, Tian YM, Pugh CW, Ratcliffe PJ, Mole DR. Determination and comparison of specific activity of the HIF-prolyl hydroxylases. FEBS Lett. 2004;576:145–150. doi: 10.1016/j.febslet.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Tuderman L, Myllyla R, Kivirikko KI. Mechanism of the prolyl hydroxylase reaction. 1 Role of co-substrates. Eur J Biochem. 1977;80:341–348. doi: 10.1111/j.1432-1033.1977.tb11888.x. [DOI] [PubMed] [Google Scholar]
- Vordermark D, Brown JM. Endogenous markers of tumor hypoxia predictors of clinical radiation resistance? Strahlenther Onkol. 2003;179:801–811. doi: 10.1007/s00066-003-1150-9. [DOI] [PubMed] [Google Scholar]
- Vordermark D, Katzer A, Baier K, Kraft P, Flentje M. Cell type-specific association of hypoxia-inducible factor-1 alpha (HIF-1 alpha) protein accumulation and radiobiologic tumor hypoxia. Int J Radiat Oncol Biol Phys. 2004;58:1242–1250. doi: 10.1016/j.ijrobp.2003.11.030. [DOI] [PubMed] [Google Scholar]
- Wang GL, Jiang BH, Rue EA, Semenza GL. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci USA. 1995;92:5510–5514. doi: 10.1073/pnas.92.12.5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardrop SL, Richardson DR. The effect of intracellular iron concentration and nitrogen monoxide on Nramp2 expression and non-transferrin-bound iron uptake. Eur J Biochem. 1999;263:41–49. doi: 10.1046/j.1432-1327.1999.00447.x. [DOI] [PubMed] [Google Scholar]
- Wartenberg M, Wolf S, Budde P, Grunheck F, Acker H, Hescheler J, Wartenberg G, Sauer H. The antimalaria agent artemisinin exerts antiangiogenic effects in mouse embryonic stem cell-derived embryoid bodies. Lab Invest. 2003;83:1647–1655. doi: 10.1097/01.lab.0000098424.38003.ff. [DOI] [PubMed] [Google Scholar]
- Willam C, Nicholls LG, Ratcliffe PJ, Pugh CW, Maxwell PH. The prolyl hydroxylase enzymes that act as oxygen sensors regulating destruction of hypoxia-inducible factor alpha. Adv Enzyme Regul. 2004;44:75–92. doi: 10.1016/j.advenzreg.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Williams N, Yandell J. Outer-sphere electron transfer of ascorbate anions. Aust J Chem. 1982;35:1133–1144. [Google Scholar]
- Yu AY, Frid MG, Shimoda LA, Wiener CM, Stenmark K, Semenza GL. Temporal, spatial, and oxygen-regulated expression of hypoxia-inducible factor-1 in the lung. Am J Physiol. 1998;275:L818–L826. doi: 10.1152/ajplung.1998.275.4.L818. [DOI] [PubMed] [Google Scholar]
- Zagzag D, Zhong H, Scalzitti JM, Laughner E, Simons JW, Semenza GL. Expression of hypoxia-inducible factor 1alpha in brain tumors: association with angiogenesis, invasion, and progression. Cancer. 2000;88:2606–2618. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material available online at http://jcs.biologists.org/cgi/content/full/119/16/3467/DC1


