Abstract
Rsp5p of Saccharomyces cerevisiae is a member of the C2-WW-HECT family of ubiquitin ligases and it interacts with targets via its WW domains. Spt23p and Mga2p are Rsp5p substrates and Rsp5p activates the OLE1 inducing functions of these membrane-localized transcription factors by ubiquitination. Although it is known that Rsp5p binds Mga2p and Spt23p via an imperfect WW domain-binding site (LPKY) that is located within the carboxy-terminal domain of the proteins, it remains unclear which WW domains mediate binding. We show that Rsp5p mutants harboring mutations in single WW domains are Spt23p/Mga2p binding and ubiquitination proficient. This is also the case for WW domains 1/2 and WW domains 1/3 mutants. However, disrupting WW domains 2 and 3 abrogates a physical and functional interaction with substrates in vitro and in cells. We also show that abrogation of WW domains 2 and 3 eliminates the activity of an Rsp5p dominant-negative mutant and an rsp5 WW domain 2/3 mutant is unable to rescue the proliferative defects of rsp5Δ cells. Interestingly, while rsp5Δ cells are able to grow on oleic acid containing YPD media, they as well as those transformed with the WW domain 2/3 mutant are unable to proliferate on oleic acid containing synthetic drop-out media. We conclude from these studies that WW domains 2 and 3 of Rsp5p play overlapping roles in binding to the LPKY site on Spt23p and Mga2p. Also, we propose that WW domains 2 and 3 perform yet to be defined essential function(s) outside of the OLE1 pathway when cells are grown in nutrient restrictive media.
Keywords: Rsp5p, WW domains, P/LPXY motif, ligase, ubiquitination
Introduction
Ubiquitination is a highly conserved post-translational protein modification process that results in the covalent attachment of a 76 amino acid poly-peptide called ubiquitin (Ub) to a Lysine of the targeted substrate (Hochstrasser, 1996; Hershko & Ciechanover, 1998). Ub conjugation is a multi-step process and requires the coordinated activity of at least three enzymes. These are an Ub-activating enzyme (E1), an Ub-conjugating enzyme (E2) and a specificity factor or complex termed an Ub ligase (E3). E3 enzymes have been divided into specific subclasses, including single subunit RING finger proteins, multi-subunit RING finger proteins, HECT domain containing proteins and more recently, U-box containing E3’s (Hochstrasser, 1996; Ardley & Robinson, 2005). The majority of E3 ligases promote the transfer of ubiquitin directly from the E2 enzyme to the substrate. In the case of HECT domain containing ligases, these proteins capture Ub via their carboxy-terminal Cys residue from the E2 prior to placement on the target. Many HECT domain-containing proteins share a modular structure consisting of an amino-terminal localized calcium-phosphate-lipid-binding domain (C2) and centrally localized protein interaction motifs termed WW domains (Harvey, & Kumar; 1999; Rotin, Staub, & Haguenauer-Tsapis, 2000; Ingham, Gish, & Pawson, 2004). These C2-WW-HECT proteins (also called the Nedd4 family) are found in budding and fission yeast and multiple paralogues are present in vertebrates.
Rsp5p of Saccharomyces cerevisiae is an essential protein and one of the most intensely studied of the Nedd4 family (www.yeastgenome.org). The gene has been isolated from numerous genetic screens and has been postulated to play an important role in a plethora of biochemical and cellular events. Surprisingly, we still have very little knowledge of direct Rsp5p targets and how their modification affects specific biochemical processes. Spt23p and Mga2p have emerged as direct and physiologically relevant targets of Rsp5p and the ligase is required for activation of their function (Hoppe et al., 2000; Shcherbik, Zoladek, Nickels, & Haines, 2003; Shcherbik, Kee, Lyon, Huibregtse, & Haines, 2004,). These homologous proteins are endoplasmic reticulum (ER) localized transcription factors that play overlapping roles in up-regulating the expression of OLE1, an essential gene required for the synthesis of oleic acid (Zhang, Skalsky, & Garfinkel, 1999; Hoppe et al, 2000). Rsp5p controls the expression and activation of these proteins in at least two ways. In the case of Spt23p, Rsp5p is required for proteasome-dependent processing, resulting in the generation of the 90-kD form of the protein (referred to as Spt23p90 in this study) and release of the processed protein from the ER (Hoppe et al, 2000; Rape et al, 2001). For Mga2p, Rsp5p is dispensable for processing and generation of 90-kD form of the protein (referred to as Mga2p90), but is required for mobilization of the processed product from the ER (Shcherbik, Zoladek, Nickels, & Haines, 2003; Shcherbik, & Haines, 2007). Consistent with a required role for Rsp5p in Spt23p and Mga2p function, the growth defects of rsp5Δ cells can be rescued, at least in part, on nutrient rich media by the addition of oleic acid (Hoppe et al, 2000).
Rsp5p interacts with substrates via its WW domains (Harvey, & Kumar, 1999; Rotin, Staub, Haguenauer-Tsapis, 2000; Ingham, Gish, & Pawson, 2004). WW domains are protein interaction modules that are approximately forty amino acids in length (Kay, Williamson, & Sudol, 2000; Macias, Wiesner, & Sudol, 2002; Dupre, Urban-Grimal, & Haguenauer-Tsapis, 2004) and its name refers to two strictly conserved tryptophan residues that are spaced 20–22 amino acids apart. In addition to these amino acids, WW domains contain two highly conserved tyrosine/phenylalanine residues that are located in the middle of the domain as well as a number of semi-conserved amino acids, some of which differ between the various groups (Kay, Williamson, & Sudol, 2000; Macias, Wiesner, & Sudol, 2002; Dupre, Urban-Grimal, & Haguenauer-Tsapis, 2004). Nevertheless, the amino acids that comprise this domain adopt a triple stranded anti-parallel □-sheet that forms a hydrophobic pocket for substrate binding. WW domains have been classified into at least four groups based on their binding specificity (Kato, Ito, Kawai, Nagata, & Tanokura, 2002; Kato et al, 2004). Group I binds to poly-proline motifs that contain a tyrosine residue separated by any amino acid (e.g. PPXY); groups II and III associates with longer poly-proline motifs; and group IV interacts with phosphorylated serine and threonine that are flanked by a proline residue.
Rsp5p harbors three group I WW domains and they play unique roles in Rsp5p function. Although disruption of any of the three impairs plasma membrane receptor ubiquitination-mediated endocytosis, mutation of WW domain 1 or 3, severely impairs transport of fluid phase markers to the vacuole (Dunn, & Hicke, 2001; Gajewska et al, 2001). In addition, mutations in WW domains 2 or 3 alter sorting of cargo to multivesicular bodies (Gajewska et al, 2001; Morvan, Froissard, Haguenauer-Tsapis, & Urban-Grimal, 2004) and mRNA nuclear export (Rodriguez, Gwizdek, Haguenauer-Tsapis & Dargemont, 2003). In terms of mediating interaction with substrates, WW domain 2 performs a dominant role in binding to the carboxy-terminal domain of Rbp1, the large subunit of RNA polymerase II (Wang, Yang, & Huibregtse, 1999). Considering that each individual WW domain does not perform an essential function (Wang, Yang, & Huibregtse, 1999; Dunn, & Hicke, 2001; Gajewska et al, 2001), it remains unclear if there is redundancy between individual WW domains in interacting with specific targets or if the ligase regulates distinct targets that play overlapping roles in essential pathways.
We have previously identified a single imperfect WW domain binding motif (i.e. LPKY) within Spt23p and Mga2p that is required for Rsp5p binding and ubiquitination in vitro and in vivo (Shcherbik, Kee, Lyon, Huibregtse, & Haines, 2004). Interestingly, a recent published protein interaction map for 12 of the WW domains present in Saccharomyces cerevisiae has shown that many yeast proteins harboring LPXY or a similar LPXF motif are able to interact with the various WW domains of Rsp5p (Hesselberth et al, 2006). Spt23p and Mga2p were not identified in this screen, presumable due to the insensitive nature of protein microarrays. Nevertheless, the goal of this study was to define which WW domains of Rsp5p mediate a physical and functional interaction via the LPKY motif of Spt23p and Mga2p in vitro and in cells.
Materials and Methods
Yeast strains and viability assays
InvSc1 (wild type) and rsp5Δ (Rsp5p deletion) strains have been described previously (Shcherbik, Zoladek, Nickels, & Haines. 2003). For viability assays, InvSc1 cells transformed with RSP5 expressing constructs were grown-up in synthetic dropout (SD) media containing glucose for 24 hrs. These cultures were streaked on SD media plates containing glucose or galactose, with or without 1 mM oleic acid, and incubated at 30°C. rsp5Δ strains were streaked on YPD plates or SD media plates, with or without 1 mM oleic acid, and incubated at 25°C.
Antibodies
Anti-FLAG M5 (Sigma Aldrich), anti-FLAG affinity gel M2 (Sigma Aldrich), anti-Myc 9E10 (Calbiochem), anti-HA 12CA5 (Roche Molecular Biochemicals) and anti-Ub P4D1 (Santa Cruz) were purchased from the indicated sources.
Plasmids
pYEplac181-MYCMGA2 was provided by S. Jentsch (Max Planck Institute of Biochemistry, Martinsried, Germany) while pGEX-6p-1-RSP5 was provided by J. Huibregtse (Institute for Cellular and Molecular Biology, Section of Molecular Genetics and Microbiology, University of Texas at Austin). pESC-FLAGSPT23, pESC-FLAGspt23pΔlpky , pYes-HARSP5 , and pYEplac181-MYCmga2Δlpky have been described previously (Shcherbik, Zoladek, Nickels, & Haines. 2003; Shcherbik, Kee, Lyon, Huibregtse, & Haines. 2004). pYes- HArsp5Δc was generated by introducing a stop codon upstream of the catalytic cystine using site directed mutagenesis (Shcherbik, Zoladek, Nickels, & Haines. 2003). pESC-FLAGspt23pΔlpky and pYEplac181-MYCmga2Δlpky were generated by deleting the LPKY encoding sequences by PCR based site directed mutagenesis using pESC-FLAGSPT23 and pYEplac181-MYCMGA2 as templates. Recombinant Rsp5p WW mutants were generated by PCR based site directed mutagenesis using pGEX-6p-1-Rsp5 as the template. The specific amino acid substitutions were replacement of the conserved tryptophan and proline residues with phenylalanine and alanine respectively as has been previously described (Gajewska et al, 2001). WW domain mutants in pYes-HARSP5 and pYes-HArsp5Δc were generated by replacing the WT sequences with the mutated sequences generated in pGEX-6p-1-Rsp5, using PstI and NsiI.
Protein purification
E. coli BL21 DE3 RIL were transformed with pGEX-6p-1-Rsp5 (WT or mutant constructs) or pGEX-6p-1-Ubc1. Transformed cells were grown for 12–16 hrs, diluted 1:5 and grown for 4 additional hrs, all at 37°C. The cultures were induced with 100 □m IPTG for 4 hrs at 37°C. The cells were collected, resuspended in PBS containing 1% Triton X-100 and lysed by treatment with lysozyme (1μg/μl). The lysate was incubated with glutathione beads (Amersham) for 2-4 hrs. Bound proteins were washed with PBS and quantified by resolving on an 8% SDS-PAGE gel using BSA as a standard. Proteins bound to the beads were either used for in vitro binding assays or liberated with PreScission Protease (Amersham) for ubiquitination assays. Elution of the GST tagged proteins was done by incubating the glutathione bead bound proteins with PreScission Protease in a buffer containing 50 mM Tris-HCl, pH 7.0, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, for 2 to 4 hrs at 4°C.
In vitro binding assay
35S-labeled FLAGSpt23p and FLAGMga2p were generated in a coupled in vitro transcription translation rabbit reticulocyte system (TNT, Promega). Equal molar amounts of GST-Rsp5p preparations were incubated with 3μl of the in vitro translated proteins in PBS containing 0.1% Triton X-100 for 2 hrs at 4°C. The GST-Rsp5p-bound proteins were pelleted by centrifugation, washed extensively with PBS, and boiled in SDS-PAGE loading buffer. The proteins were resolved by SDS-PAGE and the amount of radiolabeled protein was determined by fluorography. Gels were rehydrated and Coomassie Blue stained to verify equal pull down of Rsp5p.
In vitro ubiquitination assays
35S-labeled FLAGSpt23p and FLAGMga2p were generated in a coupled in vitro transcription translation wheat germ extract system (TNT, Promega). Recombinant E2 (Ubc1p) and E3 (Rsp5p) were prepared as described above and liberated from the glutathione beads using PreScission Protease (Amersham). The various Rsp5p variants were quantified and equal molar amounts were used in the in vitro ubiquitination reactions. 35S-labeled proteins were incubated with E1 (Boston Biochemicals), Ubc1p (E2) and Rsp5p (E3) in a buffer containing 10 mM Tris pH 7.9, 50 mM NaCl, 5 mM ATP (Sigma), 5 mM MgCl2, 0.1 mM DTT, and 50 μg/ml ubiquitin (Boston Biochemicals). Reactions were incubated at room temperature for 1 hr and terminated by addition of SDS-PAGE loading buffer and boiling the samples. Proteins were resolved by SDS-PAGE gel and detected by fluorography. For the substrate free ubiquitination assays, Rsp5p was incubated with 175 μg/ml ubiquitin (Boston Biochemicals) in a buffer containing E1, E2, 25 mM Tris pH 7.9, 5 mM ATP (Sigma), 5 mM MgCl2, 0.125 mM DTT.
Cell lysis and immunoprecipitation
Yeast cells harboring the various expression constructs were grown on SD media 24 hours, shifted to SD media containing galactose as the carbon source for 36 hours, pelleted and lysed in RIPA buffer containing 150 mM NaCl, 1% NP-40, 0.5% deoxycholic acid, 0.1% SDS, and 50 mM Tris (pH 8.0) supplemented with protease inhibitors aprotinin, pepstatin A, leupeptin, and phenylmethylsulfonyl Fluoride. Acid washed glass beads (Sigma) were added to the cell suspension, and the samples were vortexed for 20 min at 4 °C. The samples were then centrifuged and the supernatant was used for further analysis. The proteins were quantified by Bradford assay (BioRad). Lysates were pre-incubated with 10 μl of protein G-Sepharose (Amersham Biosciences) for 2 hrs and incubated with either anti-FLAG affinity gel M2 overnight or anti-Myc antibody 9E10 for 2 hrs followed by protein G-Sepharose incubation overnight. Protein complexes were pelleted, washed extensively and eluted by boiling in SDS-PAGE loading buffer. Proteins were resolved by SDS-PAGE, transferred to nitrocellulose membranes and western blotting was performed with the indicated antibodies.
Results
WW domain 2 or 3 of Rsp5p mediates a physical and functional interaction with Spt23p and Mga2p in vitro
It remains unclear which WW domains of Rsp5p are required for mediating a physical and functional interaction with Spt23p and Mga2p in vitro and in cells. Therefore, an in vitro binding assay was developed using full-length recombinant Rsp5p and in vitro translated Spt23p and Mga2p. Included as controls were Spt23p and Mga2p mutants that are missing the LPKY Rsp5p binding site (Shcherbik, Kee, Lyon, Huibregtse, & Haines, 2004) and an Rsp5p mutant that lacks WW domain binding activity. Disruption of WW domain ligand binding function was performed by site directed mutagenesis and substitution of conserved proline and alanine residues in all three WW domains to phenylalanine and alanine, respectively, (Gajewska et al, 2001). As shown in Fig 1A, a robust interaction was detected between Rsp5p and Spt23p or Mga2p. We were unable to detect association between the substrates and an Rsp5p mutant harboring mutations in all three WW domains or WT (Wild-Type) Rsp5p and the LPKY Sp23p or Mga2p deletion mutants. These results suggest that Rsp5p interacts with Spt23p and Mga2p via the single LPKY WW domain binding site.
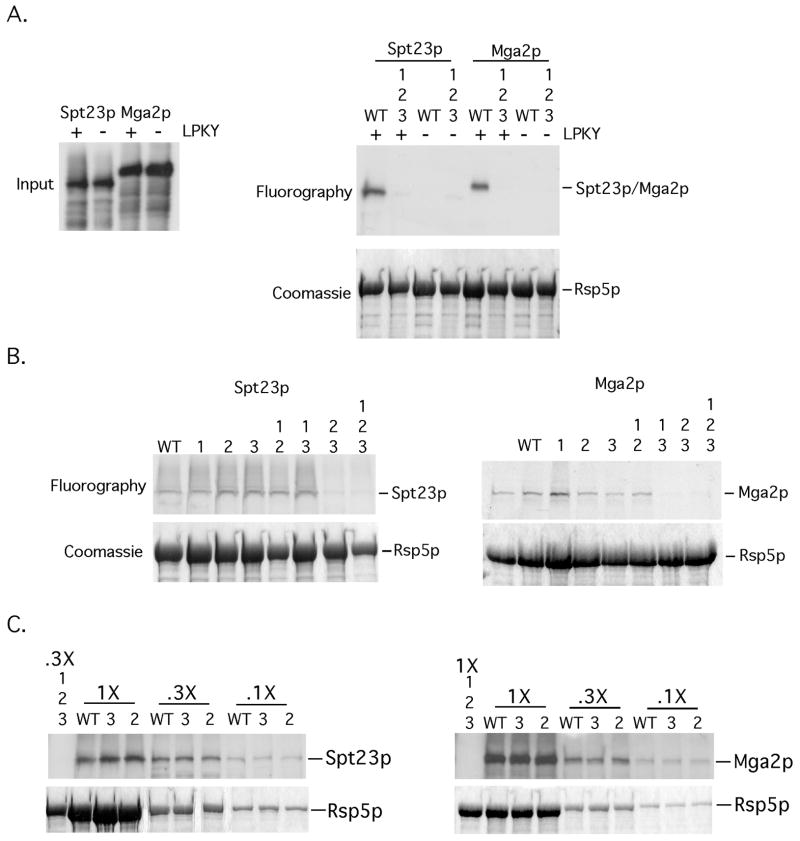
FIG 1. Mutations in WW domains 2 and 3 of Rsp5p abrogate an interaction with Spt23p and Mga2p in vitro.
(A) (left panel) Inputs of in vitro translated 35S-labeled Spt23p, Spt23pΔLPKY, Mga2p and Mga2ΔLPKY used for the in vitro binding experiments are depicted. (right panel) Equivalent amounts of bead-bound recombinant WT GST-Rsp5p or GST-Rsp5p harboring mutations in all three WW domains were incubated with equal amounts of in vitro translated material. Protein complexes were allowed to form, followed by centrifugation and washing of bead-bound material. Pelleted material was resuspended in loading buffer, resolved by SDS-PAGE and the amount of radiolabeled protein was determined by fluorography. The gel was rehydrated and stained with Coomassie Blue to show equal amounts of Rsp5p in the pelleted material. (B) The above-described binding assays were performed with the designated WW domain mutants and 35S-labeled Spt23p or Mga2p. (C) Binding assays were also performed with varying amounts of WT Rsp5p or Rsp5p mutants (WW2 and WW3) and 35S-labeled Spt23p or Mga2p.
To define which WW domain(s) of Rsp5p is required for interaction with the LPKY motif, we generated Rsp5p mutants harboring substitutions in individual or multiple WW domains and performed in vitro interaction assays. As shown in Fig 1B, disruption of individual WW domains did not disrupt an Rsp5p-Spt23p or Rsp5p-Mga2p binding. Also, Rsp5p mutants harboring mutations in WW domain 1 and 2 or 1 and 3 are fully competent for substrate binding (Fig 1B). In contrast, an Rsp5p mutant harboring mutations in WW domains 2 and 3 was unable to interact with either Spt23p or Mga2p in vitro (Fig 1B). In addition, we were unable to detect any differences between WT, the WW domain 2 or WW domain 3 mutants in Spt23p and Mga2p binding using varying amounts of Rsp5p proteins (Fig 1C). These results suggest that WW domain 2 and 3 play overlapping roles in binding to the LPKY motif of Spt23p and Mga2p in vitro.
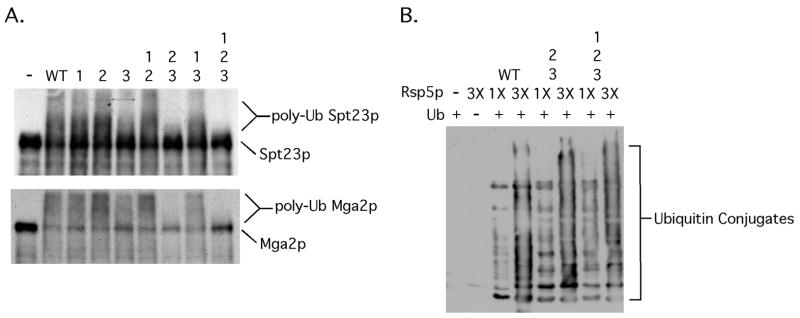
In vitro Spt23p and Mga2p ubiquitination assays were next carried out with the various WW domain Rsp5p mutants. As shown in Fig 2A, an Rsp5p mutant containing substitutions in WW domains 2 and 3 was unable to efficiently promote Spt23p or Mga2p ubiquitination. Rsp5p proteins harboring individual WW mutations as well as those harboring double mutations that include WW domain 1 were Spt23p and Mga2p ubiquitination proficient (Fig 2A). The observed slight reduction in Spt23p ubiquitination with the WW domain 3 mutant is not significant and within the range of experimental variation of 3 independent experiments and likely reflects a slight difference in Rsp5p inputs. Fig 2B shows that both the WW domain 2/3 and 1/2/3 mutants are fully functional in a substrate-free ubiquitination assay, suggesting that their inability to ubiquitinate Spt23p and Mga2p is not due to a generalized defect in ubiquitin ligase activity.
FIG 2. Mutations in WW domains 2 and 3 of Rsp5p prevent Rsp5p-dependent Spt23p and Mga2p ubiquitination in vitro.
(A) Ubiqitination assays were performed as described in the methods section with in vitro translated 35S-labeled Spt23p or Mga2p and equal amounts of the indicated WW domain mutant recombinant Rsp5p. (B) Enzymatic activity of Rsp5p (WT and Spt23p/Mga2p ubiquitination defective mutants over a range of inputs) was assessed using a substrate free ubiquitination assay. After completion of reactions, products were resolved by SDS-PAGE, transferred to nitrocellulose membrane and western blotting was performed with an anti-Ub antibody.
WW domain 2 and 3 are required for a physical and functional interaction with Spt23p and Mga2p in cells
Based on the above-presented findings, we next tested how disruption of WW domain 1 and WW domains 2 and 3 of Rsp5p affect an interaction between the ligase and Spt23p and Mga2p in cells. Because it is difficult to detect interactions between substrates and catalytically active ligase complexes, we utilized for these studies ligase deficient dominant-negative Rsp5p mutants lacking the carboxy-terminal residues (termed Rsp5pΔC’s). As shown in Fig 3, we were able to co-immunoprecipitate Rsp5p with antibodies directed against epitope tagged Spt23p and Mga2p using extract derived from cells expressing the WT WW domain or WW domain 1 mutant Rsp5pΔC. In contrast, we were not able to co-immunoprecipitate Rsp5p using cell extracts derived from yeast expressing the WW domain 2/3 mutant Rsp5pΔC or the triple WW domain mutant Rsp5pΔC (Fig 3). Fig 3 also shows that deletion of the LPKY motif of Spt23p and Mga2p eliminates Rsp5p binding, indicating that a single WW domain interaction motif within these proteins mediates an interaction with Rsp5p in cells. The observed slight reduction in binding between Mga2p and the Rsp5p WW domain 1 mutant (Fig 3B) is not significant and within the range of experimental variation of 3 independent experiments. It also deserves to be noted that there was more Spt23p90 (see inputs in for Fig 1A; running immediately below the cross reactive band marked by an *) in cells expressing the WW domains 2/3 and 1/2/3 mutants while the amount of Mga2p90 (see inputs of Fig 3B) is similar. These results are consistent with the idea that Rsp5p is required for Spt23p but not Mga2p processing (Hoppe et al, 2000; Shcherbik, Zoladek, Nickels, & Haines, 2003, Shcherbik, Kee, Lyon, Huibregtse, & Haines. 2004) and at least in the case of Spt23p, WW domain 2 or 3 are required for this interaction.
FIG 3. Mutations in WW domains 2 and 3 of Rsp5pΔC abrogate an interaction with Spt23p and Mga2p in vivo.
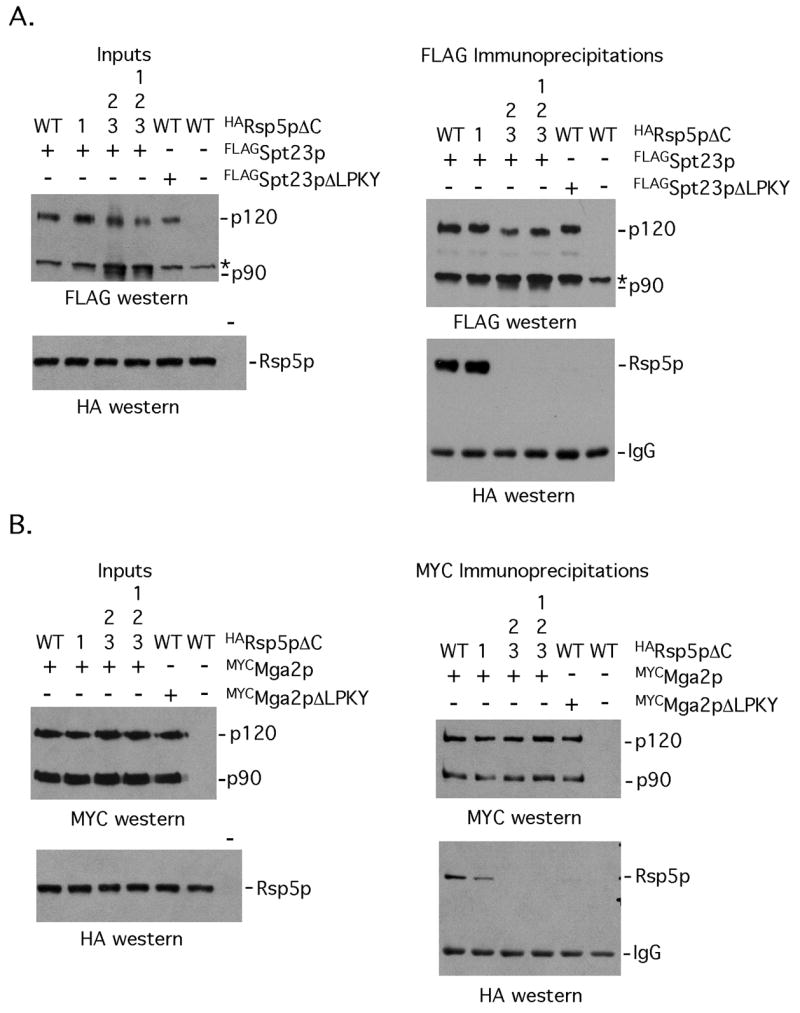
(A) Lysates from cells co-expressing FLAGSpt23p or FLAGSpt23pΔLPKY and HARsp5pΔC (WT for its WW domains or harboring the indicated WW domain mutations) were subjected to immunoprecipitation with anti-FLAG antibodies. The immunoprecipitated proteins were separated by SDS-PAGE and western blotting was carried out with anti-FLAG and anti-HA western blotting. The amount of epitope tagged Spt23p and Rsp5p proteins present in the extracts are shown in the left panel. (B) Interaction studies were performed as described in (A), except that cells expressing MYCMga2p were used for the studies. * denotes an anti-FLAG cross-reactive band.
In addition to the above presented cell-based interaction studies, we also tested if disruption of WW domains 2 and 3 eliminates this dominant negative activity of the catalytically defective Rsp5p mutant. As shown in Fig 4 increased amounts of ubiquitinated Spt23p and Mga2p are present in cells expressing the WW domain 2/3 mutant Rsp5pΔC or WW1/2/3 mutant Rsp5pΔC when compared to Rsp5pΔC. Also, disruption of WW domain 1 did not have any affects on Rsp5pΔC-mediated inhibition of Spt23p or Mga2p ubiquitination (Fig 4). Expression of Rsp5pΔC in cells inhibits both Spt23p and Mga2p ubiquitination by endogenous Rsp5p. We conclude from these studies that WW domains 2 and 3 mediate an interaction between Rsp5p and the membrane-bound transcription factors in vivo.
FIG 4. Mutations in WW domains 2 and 3 of Rsp5pΔC relieve its dominant-negative activity of inhibiting Spt23p/Mga2p ubiquitination.
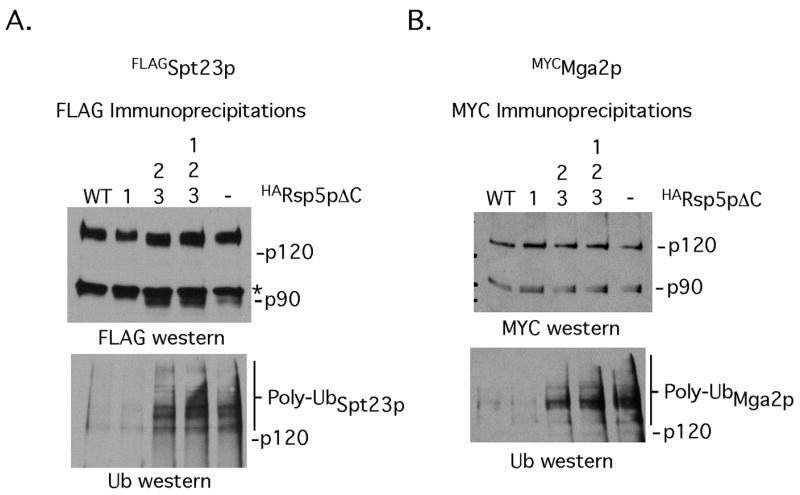
Lysates from cells harboring the indicated Spt23p, Mga2p and Rsp5pΔC expression constructs were subjected to immunoprecipitation with anti-FLAG (A) or anti-Myc (B) antibodies. The immunoprecipitated proteins were resuspended in SDS-PAGE loading buffer, resolved by SDS-PAGE, transferred to nitrocellulose membranes and probed with antibodies recognizing Spt23p or Mga2p (top panels) or Ub modified forms of the membrane-bound transcription factors.
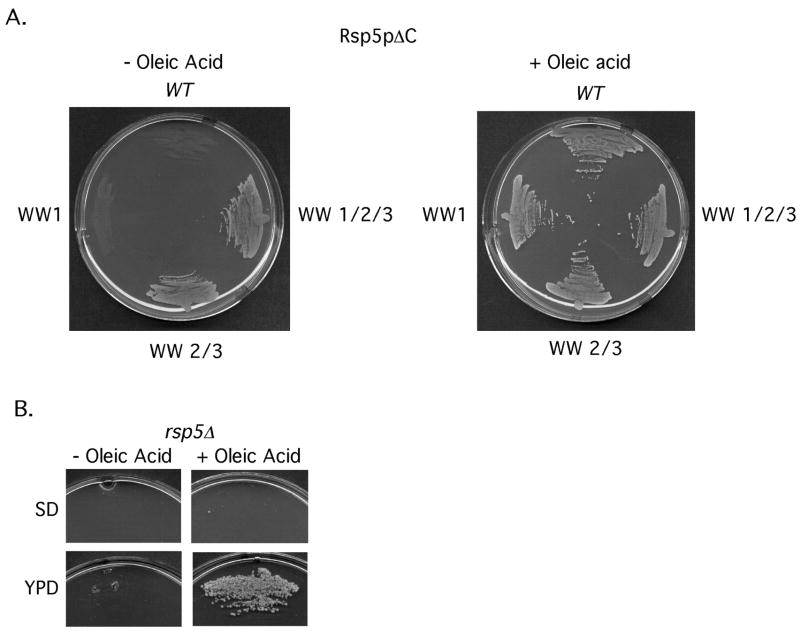
Abrogation of WW domains 2 and 3 eliminates the dominant-negative activity of Rsp5pΔC for oleic acid dependent cell growth and WW domains 2 and 3 of Rsp5p are required for yeast cell viability
Cells lacking both SPT23 and MGA2 require oleic acid for growth. Also, oleic acid supplementation of media rescues the proliferation defects of cells expressing Rsp5p dominant negative mutants (Hoppe et al, 2000; Shcherbik, Kumar, & Haines 2002), including the Rsp5pΔC mutant (Shcherbik, Zoladek, Nickels, & Haines, 2003). To determine if WW domains 2 and 3 are required for the dominant-negative activity of Rsp5pΔC, cells harboring galactose-inducible Rsp5pΔC mutants were placed on galactose media containing or lacking oleic acid. As shown in Fig 5A cells expressing Rsp5pΔC or WW domain 1 mutant Rsp5pΔC require oleic acid for growth. This was not the case for the WW domain 2/3 or WW 1/2/3 Rsp5pΔC mutants; cells expressing these proteins did not display any obvious growth defects (Fig 5A). We also tested how the various WW domain mutants (present within a single copy vector and under the transcription control of the native RSP5 promoter) rescue a RSP5 deletion rsp5Δ cells were grown -up in oleic acid media and transformed with the various WW domain mutant constructs. After transformation, cells were plated on SD media lacking or containing oleic acid. As shown in Table 1, all WW domain mutant constructs except the double WW domain 2/3 and triple WW 1/2/3 were able to confer growth of rsp5Δ cells on both media. Interestingly, we did not observe that oleic acid is able to rescue the proliferative defects of rsp5Δ cells transformed with these mutants as well as with the vector control. Thus, we wanted to determine if there is a difference between the ability of rsp5Δ cells to grown on oleic acid supplemented YPD media versus SD media. As shown in Fig 5B, rsp5Δ cells are able to grow on oleic acid supplemented YPD media but not on oleic acid containing SD media. We conclude from these experiments that oleic acid rescues the proliferation defect of rsp5Δ cells only when cultured in nutrient rich media and that WW domains 2 and 3 of Rsp5p play other essential functions (these could be overlapping or independent of one another) in addition to Spt23p and Mga2p activation when yeast are cultured in nutrient restrictive conditions or Spt23p and Mga2p might regulate other essential genes in addition to OLE1.
FIG 5. Mutations in WW domains 2 and 3 of Rsp5pΔC eliminate its growth inhibitory activity and WW domains 2 and 3 of Rsp5p are required for yeast cell viability.
(A) Cells expressing HARsp5pΔC (WT for its WW domains or harboring the indicated WW domain mutations) were streaked on SD media with or without 1mM oleic acid. (B) rsp5Δ cells were streaked on synthetic dropout media or YPD media −/+ 1 mM oleic acid.
Table 1.
| RSP5 | Growth on SD (− oleic acid) | Growth on SD (+ oleic) |
|---|---|---|
| V | - | - |
| WT | +++++ | +++++ |
| WW1 | +++++ | +++++ |
| WW2 | +++++ | +++++ |
| WW3 | +++++ | +++++ |
| WW1/2 | +++++ | +++++ |
| WW1/3 | +++++ | +++++ |
| WW2/3 | - | - |
| WW1/2/3 | - | - |
rsp5Δ cells were transformed with the indicated WT or WW domain mutant rsp5 constructs. V denotes the empty vector control. +++++ = more than 50 colonies per plate, - = less than 10 colonies.
Discussion
Rsp5p is required for the activation of the ER-bound transcription factors Mga2p and Spt23p. In the case of Spt23p, Rsp5p directed ubiquitination is required for proteasome dependent processing and release of processed Spt23p from the ER membrane (Hoppe et al, 2000; Rape et al, 2001). In the case of Mga2p, Rsp5p is dispensable for the processing signal, but is required for mobilization of Mga2p90 from the ER membrane (Shcherbik, Kee, Lyon, Huibregtse, & Haines, 2004). Studies published by our group previously have defined a single imperfect WW domain binding site, the LPKY motif, that is present within the carboxy-terminal domains of Mga2p and Spt23p that mediates an interaction with Rsp5p (Shcherbik, Kee, Lyon, Huibregtse, & Haines, 2004). In this report, we show that WW domains 2 and 3 of Rsp5p possess overlapping roles in mediating a physical and functional interaction with the LPKY motif of Mga2p and Spt23p both in vitro and in vivo. Also, we provide evidence that WW domain 1 of Rsp5p is incapable of binding to this motif. Interestingly, while WW domains 2 and 3 do show the greatest degree of similarity between the three WW domains (73% identical, 76% similar), it is very modest. WW domain 1 is 66% identical and 73% similar to WW domain 2 while it is 63% identical and 70% similar to WW domain 3. Thus, it is possible that binding specificity is governed by a small percentage of amino acids present that are contained with the WW domains and the atypical LPKY motif. Alternatively, specificity may be mediated by sequences that are outside of the traditionally designated WW domain region and/or WW domain recognition sequence. Further structural and mutagenesis based studies will be required to address these various possibilities. Notwithstanding, although all three WW domains have been classified as a group I WW domain in terms of their ability to recognize the PPXY motif (Sudol, Chen, Bougeret, Einbond, & Bork, 1995; Einbond, & Sudol, 1996; Wang, Yang, & Huibregtse, 1999; Chang, Cheang, Espanel, & Sudol. 2000), it is clear from these studies that WW domain 1 is clearly different from WW domains 2 and 3 in terms of its ability to recognize the LPKY motif-containing region of Mga2p and Spt23p.
We previously documented that overexpression of a Nedd4 (the mammalian ortholog of Rsp5p) mutant harboring mutations in WW domain 1 or an Rsp5p WW 1 domain deletion mutant suppressed the growth of yeast and the growth inhibitory activity of these mutants could be overcome by OLE1 overexpression or inclusion of oleic acid in the media (Shcherbik, Kumar, & Haines, 2002). In addition, these mutants were Spt23p binding proficient and inhibited Spt23p processing (Shcherbik, Kumar, & Haines, 2002). Based on these studies we proposed that WW domain 1 interacts with a co-factor in Rsp5p/Nedd4-mediated ubiquitination events. Curiously, we have found no evidence that the WW domain 1 Rsp5p mutant used here (harboring only two amino amino acid substitutions instead of a large scale deletion mutant) has dominant negative activity (data not shown). Moreover, considering that WT Rsp5p and the WW domain 1 Rsp5p mutant is fully competent in promoting Mga2p and Spt23p ubiquitination in vitro, it is highly probable that Rsp5p-dependent binding and ubiquitination of these proteins in vitro and in vivo is only dependent on the presence of WW domain 2 or 3. The dominant activity of the WW domain 1 Rsp5p deletion mutant could be due to conformation problems resulting in a binding proficient, yet ubiquitination deficient protein.
Our viability data is also consistent with the idea that WW domains 2 or 3 mediate binding to Mga2p and Spt23p since mutation of both of these domains eliminates the dominant negative activity of Rsp5pΔC. Also, transformation of rsp5Δ cells with the WW domain 2/3 mutant is unable to rescue the growth defect of these cells. Interestingly, rsp5Δ cells that have been transformed with plasmid encoding WW domain 2/3 mutant, the triple WW domain mutant, or the vector alone control are unable to proliferate on oleic acid containing SD media when cells are grown at 25°C. Moreover, while parental rsp5Δ cells are able to grow on oleic acid supplemented YPD media, they are unable to do so on oleic acid containing SD media. It deserved to be noted however that oleic acid does rescue the growth inhibitory activity of Rsp5pΔC on SD media. It is thus conceivable that Rsp5p has ligase independent functions. Alternatively and perhaps a more likely scenario is that overexpression of Rsp5pΔC does not completely abrogate the function of endogenous Rsp5p and the residual activity allows for growth on oleic acid containing media.
Previous studies have shown that a Rsp5p truncation mutant containing only WW domain 3 and the HECT domain is a able to rescue the growth defects of rsp5Δ cells (Hoppe et al. 2000) and that essential function(s) of Rsp5p can be supplied by the presence of domains 2 or 3 (Gajewska et al, 2001). The results presented here are consistent with both of these studies. However, other experiments have shown that all three WW domains are dispensable for yeast viability when cells are grown at non-stress temperatures (Dunn, & Hicke, 2001) while another study has demonstrated that WW domain 2 and 3 are required to complement a temperature sensitive rsp5 mutant when cells are grown to elevated temperatures. While we do not have an explanation for the results of the former study, it is likely that growth a normal temperatures requires WW domain 2 or 3 while proliferation at elevated temperatures requires the function of both WW domain 2 and 3 of Rsp5p.
The essential pathway(s) that requires the functions of Rsp5p’s WW domains 2 and 3 when cells are grown in SD media remain to be defined. Considering that SD media lacks lipids and a recent report implicating Mga2p and Spt23p in regulating a number of lipid metabolism genes, including those involved in ergosterol biosynthesis (Auld, Brown, Casolari, Komili, & Silver, 2006), it is plausible that Mga2p and/or Spt23p mediate this OLE1-independent activity of Rsp5p. However, we have not found that introduction of spt23 or mga2 mutants lacking the encoded transmembrane domain rescue the proliferative defects of rsp5Δ cells when they are plated in the presence or absence of oleic acid. In addition, supplementing SD media with oleic acid and ergosterol does not rescue the growth defect of rsp5Δ cells and there is no difference in the growth rate of rsp5Δ yeast when grown in YPD containing oleic acid versus YPD supplemented with both oleic acid and ergosterol. We therefore propose that WW domains 2 and 3 perform yet to be defined essential (this could be overlapping or distinct) function(s) outside of the OLE1 pathway that is not mediated by Spt23p or Mga2p when cells are grown in nutrient restrictive media and it will be of interest for future studies to identify other direct Rsp5p targets that function within these pathways.
Acknowledgments
This work is supported by NIH grant GM070769 to DSH. We also thank Natalia Shcherbik for her technical expertise and critically reviewing of the manuscript.
Abbreviations
- ER
Endoplasmic Reticulum
- ub
ubiquitin
- cys
cystine
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ardley HC, Robinson PA. E3 ubiquitin ligases. Essays in Biochemistry. 2005;41:15–30. doi: 10.1042/EB0410015. [DOI] [PubMed] [Google Scholar]
- Auld KL, Brown CR, Casolari JM, Komili S, Silver PA. Genomic association of the proteasome demonstrates overlapping gene regulatory activity with transcription factor substrates. Molecular Cell. 2006;21:861–871. doi: 10.1016/j.molcel.2006.02.020. [DOI] [PubMed] [Google Scholar]
- Chang A, Cheang S, Espanel X, Sudol M. Rsp5 WW domains interact directly with the carboxyl-terminal domain of RNA polymerase II. The Journal of Biological Chemistry. 2000;275:20562–20571. doi: 10.1074/jbc.M002479200. [DOI] [PubMed] [Google Scholar]
- Dunn R, Hicke L. Domains of the Rsp5 ubiquitin-protein ligase required for receptor-mediated and fluid-phase endocytosis. Molecular Biology of the Cell. 2001;12:421–435. doi: 10.1091/mbc.12.2.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre S, Urban-Grimal D, Haguenauer-Tsapis R. Ubiquitin and endocytic internalization in yeast and animal cells. Biochimica et Biophysica acta. 2004;1695:89–111. doi: 10.1016/j.bbamcr.2004.09.024. [DOI] [PubMed] [Google Scholar]
- Einbond A, Sudol M. Towards prediction of cognate complexes between the WW domain and proline-rich ligands. FEBS Letters. 1996;384:1–8. doi: 10.1016/0014-5793(96)00263-3. [DOI] [PubMed] [Google Scholar]
- Gajewska B, Kaminska J, Jesionowska A, Martin NC, Hopper AK, Zoladek T. WW domains of Rsp5p define different functions: determination of roles in fluid phase and uracil permease endocytosis in Saccharomyces cerevisiae. Genetics. 2001;157:91–101. doi: 10.1093/genetics/157.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey KF, Kumar S. Nedd4-like proteins: an emerging family of ubiquitin-protein ligases implicated in diverse cellular functions. Trends in Cell Biology. 1999;9:166–169. doi: 10.1016/s0962-8924(99)01541-x. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annual Review of Biochemistry. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Hesselberth JR, Miller JP, Golob A, Stajich JE, Michaud GA, Fields S. Comparative analysis of Saccharomyces cerevisiae WW domains and their interacting proteins. Genome Biology. 2006;7:R30. doi: 10.1186/gb-2006-7-4-r30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochstrasser M. Ubiquitin-dependent protein degradation. Annual Review of Genetics. 1996;30:405–39. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- Hoppe T, Matuschewski K, Rape M, Schlenker S, Ulrich HD, Jentsch S. Activation of a membrane-bound transcription factor by regulated ubiquitin/proteasome-dependent processing. Cell. 2000;102:577–586. doi: 10.1016/s0092-8674(00)00080-5. [DOI] [PubMed] [Google Scholar]
- Ingham RJ, Gish G, Pawson T. The Nedd4 family of E3 ubiquitin ligases: functional diversity within a common modular architecture. Oncogene. 2004;23:1972–1984. doi: 10.1038/sj.onc.1207436. [DOI] [PubMed] [Google Scholar]
- Kato Y, Ito M, Kawai K, Nagata K, Tanokura M. Determinants of ligand specificity in groups I and IV WW domains as studied by surface plasmon resonance and model building. The Journal of Biological Chemistry. 2002;277:10173–10177. doi: 10.1074/jbc.M110490200. [DOI] [PubMed] [Google Scholar]
- Kato Y, Nagata K, Takahashi M, Lian L, Herrero JJ, Sudol M, Tanokura M. Common mechanism of ligand recognition by group II/III WW domains: redefining their functional classification. The Journal of Biological Chemistry. 2004;279:31833–31841. doi: 10.1074/jbc.M404719200. [DOI] [PubMed] [Google Scholar]
- Kay BK, Williamson MP, Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. The FASEB Journal. 2000;14:231–241. [PubMed] [Google Scholar]
- Macias MJ, Wiesner S, Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Letters. 2002;513:30–37. doi: 10.1016/s0014-5793(01)03290-2. [DOI] [PubMed] [Google Scholar]
- Morvan J, Froissard M, Haguenauer-Tsapis R, Urban-Grimal D. The ubiquitin ligase Rsp5p is required for modification and sorting of membrane proteins into multivesicular bodies. Traffic. 2004;5:383–392. doi: 10.1111/j.1398-9219.2004.00183.x. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Mechanisms underlying ubiquitination. Annual Review of Biochemistry. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez MS, Glide C, Haguenauer-Tsapis R, Argument C. The HECT ubiquitin ligase Rsp5p is required for proper nuclear export of mina in Saccharomyces cerevisiae. Traffic. 2003;8:566–575. doi: 10.1034/j.1600-0854.2003.00115.x. [DOI] [PubMed] [Google Scholar]
- Rotin D, Stub O, Haguenauer-Tsapis R. Ubiquitination and endocytosis of plasma membrane proteins: role of Nedd4/Rsp5p family of ubiquitin-protein ligases. The Journal of Membrane Biology. 2000;176:1–17. doi: 10.1007/s00232001079. [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Kumar S, Haines DS. Substrate proteolysis is inhibited by dominant-negative Nedd4 and Rsp5 mutants harboring alterations in WW domain 1. Journal of Cell Science. 2002;115:1041–1048. doi: 10.1242/jcs.115.5.1041. [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Zoladek T, Nickels JT, Haines DS. Rsp5p is required for ER bound Mga2p120 polyubiquitination and release of the processed/tethered transactivator Mga2p90. Current Biology. 2003;13:1227–1233. doi: 10.1016/s0960-9822(03)00457-3. [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Kee Y, Lyon N, Huibregtse JM, Haines DS. A single PXY motif located within the carboxyl terminus of Spt23p and Mga2p mediates a physical and functional interaction with ubiquitin ligase Rsp5p. The Journal of Biological Chemistry. 2004;279:53892–53898. doi: 10.1074/jbc.M410325200. [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Haines DS. Cdc48p(Npl4p/Ufd1p) binds and segregates membrane-anchored/tethered complexes via a polyubiquitin signal present on the anchors. Molecular Cell. 2007;25:385–397. doi: 10.1016/j.molcel.2007.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudol M, Chen HI, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module--the WW domain. FEBS Letters. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- Wang G, Yang J, Huibregtse JM. Functional domains of the Rsp5 ubiquitin-protein ligase. Molecular and Cellular Biology. 1999;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Skalsky Y, Garfinkel DJ. MGA2 or SPT23 is required for transcription of the delta9 fatty acid desaturase gene, OLE1, and nuclear membrane integrity in Saccharomyces cerevisiae. Genetics. 1999;151:473–483. doi: 10.1093/genetics/151.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]