Abstract
Preclinical data support the potential of the death-signaling receptors for TRAIL as targets for cancer therapy. However, it is unclear whether these death-signaling receptors suppress the emergence and growth of malignant tumors in vivo. Herein we show that TNF-related apoptosis-inducing ligand receptor (TRAIL-R), the only proapoptotic death-signaling receptor for TRAIL in the mouse, suppresses inflammation and tumorigenesis. Loss of a single TRAIL-R allele on the lymphoma-prone Eμ-myc genetic background significantly reduced median lymphoma-free survival. TRAIL-R–deficient lymphomas developed with equal frequency irrespective of mono- or biallelic loss of TRAIL-R, had increased metastatic potential, and showed apoptotic defects relative to WT littermates. In addition, TRAIL-R–/– mice showed decreased long-term survival following a sublethal dose of ionizing radiation. Histological evaluation of moribund irradiated TRAIL-R–/– animals showed hallmarks of bronchopneumonia as well as tumor formation with increased NF-κB p65 expression. TRAIL-R also suppressed diethylnitrosamine-induced (DEN-induced) hepatocarcinogenesis, as an increased number of large tumors with apoptotic defects developed in the livers of DEN-treated TRAIL-R–/– mice. Thus TRAIL-R may function as an inflammation and tumor suppressor in multiple tissues in vivo.
Introduction
TNF-related apoptosis-inducing ligand (TRAIL) or agonist antibodies targeting TRAIL receptors are promising proapoptotic anti-cancer drugs currently undergoing Phase I/II clinical evaluation in patients with solid malignancies or non-Hodgkin lymphomas. Five distinct TRAIL receptors have been identified in humans (DR4, DR5, DCR1, DCR2, and the soluble osteoprotegrin), of which only DR4 and DR5 are capable of initiating cell death through an intracellular death domain following trimerization by TRAIL. The death domain facilitates intracellular interaction with specific adaptor proteins, such as Fas-associated death domain (FADD) (1, 2), containing specific sequences necessary for the binding of important proapoptotic effector proteins such as caspases-8 and -10 that subsequently undergo autoactivation through cleavage to enzymatically active forms (3–5). Active caspases-8 and -10 trigger proteolysis and activation of apoptotic executioner caspases-3, -6, and -7, which in turn act on key cellular substrates, leading to cell death. This pathway is commonly referred to as the immune-mediated apoptotic pathway or the extrinsic cell death pathway. However, cross-talk between the extrinsic and the mitochondrial (intrinsic) cell death pathways through caspase-8–dependent cleavage of Bid might be required for efficient triggering of TRAIL-mediated apoptosis in certain cell types (6).
In contrast to humans, the mouse genome contains a single TRAIL-binding proapoptotic death receptor (TRAIL receptor [TRAIL-R]), which mimics human DR4 and DR5 (7). The physiological role of TRAIL-R has remained unclear until the recent development of TRAIL-R–/– mice, demonstrating that TRAIL-R is not essential for normal embryonic development but is involved in the regulation of innate immunity and the DNA damage response (8, 9). Indeed, TRAIL-R expression has been shown to be regulated by the tumor suppressor p53 following genotoxic stress (10). TRAIL-R–/– cells ex vivo show increased NF-κB activation and elevated expression of IL-12, IFN-α, and IFN-γ following (some) toll-like receptor stimuli and are compromised with regard to apoptosis in, for example, the spleen and thymus following ionizing irradiation. Thus TRAIL-R deficiency may, in addition to providing protection from cell death, also regulate proinflammatory factors through an as yet uncharacterized molecular mechanism.
Although previous studies have suggested the death-signaling TRAIL receptors as useful targets in cancer therapy, no data have demonstrated an etiological role in cancer for TRAIL-R deficiency in vivo. TRAIL-R–/– mice do not spontaneously develop malignancy at a higher frequency than their WT littermates early in life, and the lack of TRAIL-R in a p53-null or APCmin genetic background does not lead to increased susceptibility for lymphomas or intestinal polyps, respectively (11). In contrast, mice lacking the TRAIL ligand suggest that TRAIL might protect against tumorigenesis, as these mice suffer from a greater than 25% increased risk of developing lymphoid malignancies after 500 days of age (12). Moreover, TRAIL has been shown to suppress experimental and spontaneous metastases through NK cell–mediated cytotoxicity and to protect against the development of methylcholanthrene-induced fibrosarcomas (13–15). However, it cannot be excluded that some of the phenotypes observed in TRAIL-null animals may be a result of ameliorated nonapoptotic signaling through the 2 TRAIL decoy receptor homologs present in mice (Dcr1 and Dcr2), unidentified TRAIL-interacting receptors, or receptor-independent functions of TRAIL (16).
Given the previous in vivo findings in TRAIL-null mice and the role of TRAIL-R downstream of p53, we reasoned TRAIL-R might still be an important modulator of tumorigenesis initiated by oncogenic events affecting the p53 pathway. TRAIL-R signaling may also be selected against during tumorigenesis, given that approximately 50% of all human tumor cell lines show resistance to TRAIL-mediated apoptosis. To test this hypothesis we interbred the TRAIL-R–null trait onto a Burkitt lymphoma/leukemia–prone (Eμ-myc mice) genetic background, in which the deregulated c-myc expression has been shown to trigger p53 through p19ARF (17, 18). We also subjected TRAIL-R–deficient mice to a single sublethal dose of γ-irradiation, which has been previously shown to trigger expression of TRAIL-R– and p53-dependent apoptosis in vivo (9, 18, 19), and treated 7-day-old pups with DNA-damaging hepatocarcinogen diethylnitrosamine (DEN), which triggers the development of hepatocellular carcinoma (HCC) in a similar manner to that of human HCC (20).
We show that TRAIL-R suppresses chronic inflammation, tumorigenesis and lymphomagenesis in vivo. Our observations have implications for cancer etiology as well as for the treatment of certain malignancies with TRAIL, as novel diagnostic tools may need to be developed to assess the likelihood of successful TRAIL treatment of human malignancies.
Results
Monoallelic loss of TRAIL-R promotes lymphomagenesis and metastasis.
Loss of p53 or p19ARF is a common event in c-myc–driven lymphomas, leading to a more aggressive disease and to resistance to treatment with cyclophosphamide in vivo (17, 18). We hypothesized that aberrations in the p53-dependent regulation of the immune-mediated extrinsic cell death pathway, through the deletion of TRAIL-R, could affect myc-driven lymphomagenesis by loss of a putative tumor suppressor activity. To test this hypothesis we crossed mice bearing the Eμ-myc transgene with mice lacking TRAIL-R (the unique proapoptotic TRAIL receptor in the mouse genome). Hemizygous deletion of TRAIL-R (TRAIL-R+/–; n = 30) in the Eμ-myc background led to an increased rate of lymphoma formation as compared with WT (n = 11) animals, whereas no obvious difference with regard to disease-free survival between TRAIL-R+/– and TRAIL-R–/– (n = 14) animals could be found (Figure 1A). Furthermore, the median disease-free survival was significantly different between the TRAIL-R+/– mice and WT littermates as well as between the TRAIL-R–/– mice and WT littermates (87 days for TRAIL-R+/–, 82 days for TRAIL-R–/–, and 119 days for WT mice) in the Eμ-myc background (log-rank test, P = 0.0023 and P = 0.0003, respectively), with an associated increased lymphoma risk of 2.6-fold (95% CI, 1.5–5.9) for TRAIL-R+/– and 3.4-fold (95% CI, 2.4–17.4) for TRAIL-R–/– mice for lymphoma development (Figure 1A).
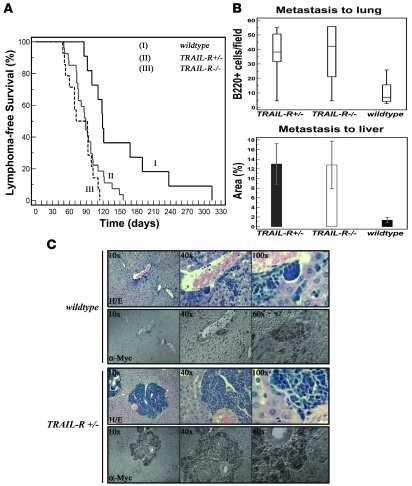
Figure 1. Monoallelic loss of TRAIL-R promotes c-myc–driven lymphomagenesis.
(A) Kaplan-Meier survival curves for the different genotypes (TRAIL-R–/–, n = 14; TRAIL-R+/–, n = 30; and WT, n = 11) on the Eμ-myc genetic background. Survival was markedly decreased in TRAIL-R+/– and TRAIL-R–/– mice (log rank test, P = 0.023 and P = 0.003, respectively) relative to WT littermates. Immunohistochemistry for c-myc+ (α-myc) (B and C) and B220+ showed increased metastasis of lymphoma cells (B and data not shown; n = 6/genotype) to liver (n = 5/genotype; average tumor emboli area percentage ± SEM; Student’s t test, P < 0.05) and lung (box-and-whisker plot shows relative median number of B220+ cells/field; Mann-Whitney U test, P < 0.05) in both TRAIL-R+/– and TRAIL-R–/– animals relative to WT animals on the Eμ-myc genetic background.
More aggressive disease was evident as well with increased metastasis, as detected by immunohistochemical analysis for c-Myc (Figure 1, B and C) and B220+ (B cell marker; data not shown) cells in nonlymphoid organs such as the livers and lungs of TRAIL-R–deficient animals. Quantitative analysis of immunohistochemically stained sections from the liver and lungs revealed an increased frequency of B220+ cells in the lungs of TRAIL-R+/– and TRAIL-R–/– mice (median B220+, 39 cells/field and 43 cells/field, respectively) relative to that of WT mice (median B220+, 7 cells/field) (Figure 1B). The average percent of liver surface occupied by metastatic lymphoma cells was assessed (Figure 1B) in animals that developed lymphoma. Analysis of H&E-stained sections revealed that TRAIL-R+/– and TRAIL-R–/– had a significantly (P < 0.05, Student’s t test) larger liver area occupied by invading lymphoma cells (13.0% and 12.8% of total analyzed liver area) relative to WT animals (1.4% of total analyzed liver area) on the Eμ-myc genetic background. Thus loss of one allele of TRAIL-R was sufficient to contribute to an increased metastatic potential of c-myc–driven lymphomas to lung and liver.
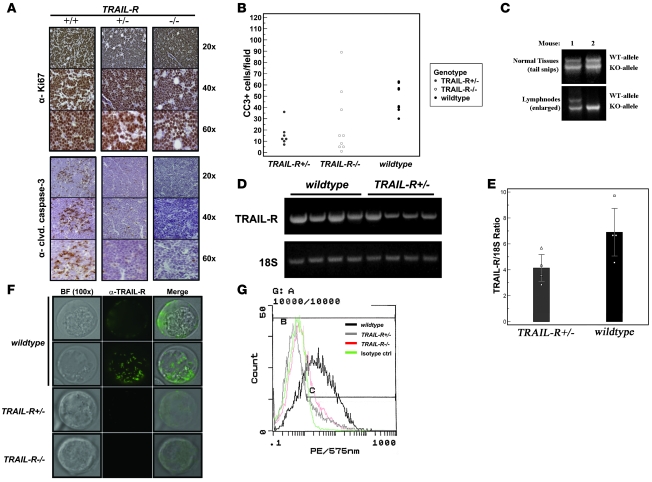
Next we investigated changes in lymphoma proliferation and cell death by performing immunohistochemistry for Ki-67 and cleaved caspase-3 (Figure 2A). Approximately 90%–95% of all cells present in the lymphomas stained positive for Ki-67, in concordance with the previous reports on the aggressiveness of the disease (21). However, we were unable to document any consistent changes in Ki-67 labeling that correlated with any particular genotype. In contrast, monoallelic loss of TRAIL-R led to a marked loss of cells labeling for cleaved (active) caspase-3 (Figure 2, A and B) in lymphomas, suggesting decreased levels of apoptosis may have contributed to the increased aggressiveness of the lymphomas arising in TRAIL-R–/– Eμ-myc animals.
Figure 2. TRAIL-R+/– lymphomas show apoptotic defects and reduced TRAIL-R mRNA expression.
(A) Immunohistochemistry shows abundant labeling of the proliferation marker Ki-67 (DAB, brown staining) in both WT (+/+) and TRAIL-R–deficient (+/– and –/–) lymphomas. Immunohistochemical staining (A) for cleaved caspase-3 and quantification thereof (B) shows fewer median number of cells expressing active caspase-3 (DAB, brown staining) in TRAIL-R+/– and TRAIL-R–/– lymphomas compared with that of WT lymphomas (Mann-Whitney U test, P < 0.05). (C) LOH was detected by PCR analysis of DNA isolated from Eμ-myc TRAIL-R+/– lymphomas in 20% (2 of 10) of the lymphomas (data not shown). (D) RT-PCR analysis on RNA isolated from Eμ-myc lymphomas of different genotypes shows decreased TRAIL-R mRNA expression in Eμ-myc TRAIL-R+/–lymphomas compared with RNA isolated from WT Eμ-myc lymphomas. (E) Relative quantitative RT-PCR analysis and densitometry shows that loss of 1 TRAIL-R allele reduced the mean (± SEM) TRAIL-R expression to approximately 60% that in WT lymphomas (n = 4 each genotype; Student’s t test, P < 0.05). (F) Immunofluorescence (FITC, green) on living Eμ-myc lymphoma cells of different TRAIL-R genotypes using the MD-5 antibody suggest expression on WT Eμ-myc lymphoma cells (α–TRAIL-R) with a heterogenous (speckled) membrane distribution as evident from the 2 different focal images. However, expression was barely detectable in TRAIL-R+/– Eμ-myc lymphoma cells and not present in TRAIL-R–/– Eμ-myc lymphoma cells. Representative images are shown from 2 independent lymphomas per genotype. Original magnification, ×100. (G) Flow cytometry analysis on Eμ-myc lymphoma cells suggests expression of TRAIL-R in WT cells but not TRAIL-R+/– and TRAIL-R–/– Eμ-myc lymphoma cells. Eμ-myc lymphoma cells from at least 2 different animals/genotype were analyzed.
Loss of heterozygosity (LOH) is a common event for many well-established tumor suppressor genes. Both INK4A/ARF and Trp53 are subject to LOH in the Eμ-myc model (18). A similar analysis on DNA isolated from enlarged lymph nodes from TRAIL-R+/– animals suggested that loss of the remaining TRAIL-R allele could only be detected in 20% (2/10) of the tumors analyzed (Figure 2C). Although this suggests that signaling through TRAIL-R could be modulated by additional mechanisms and that a TRAIL-resistant phenotype was selected for, LOH constitutes a rare mechanism that is unlikely to explain the lack of effect of TRAIL-R gene dosage. Using RT-PCR analysis to monitor changes in expression levels of TRAIL-R (Figure 2, D and E), we found that expression of TRAIL-R mRNA was intact in lymphomas lacking one allele of TRAIL-R. However, loss of one allele of TRAIL-R reduced expression of TRAIL-R mRNA by 60% of WT lymphomas (Figure 2E) and was associated with a near complete loss of surface expression of TRAIL-R as detected by flow cytometry and immunofluorescence (Figure 2, F and G). Potentially, reduced TRAIL-R expression (together with downstream mechanisms) may be sufficient to quench signaling through the TRAIL-R to the point at which it mimics complete loss of TRAIL-R on the Eμ-myc genetic background.
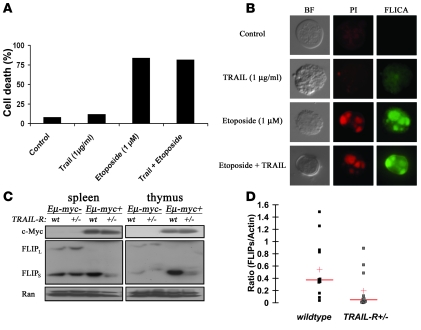
Sensitivity to TRAIL may be circumvented by several mechanisms, and our data on TRAIL-R+/– lymphomas on the Eμ-myc genetic background suggest that such mechanisms might occur in this model and that this may hamper the response to TRAIL. Therefore, we investigated TRAIL sensitivity in lymphomas in vitro. Indeed, challenging a WT Eμ-myc lymphoma cell line (p54) with recombinant murine TRAIL did not trigger death and only weakly triggered caspase activation as determined by FLICA analysis (Figure 3, A and B). No synergistic killing or caspase activation was observed in the presence of etoposide and TRAIL. We investigated biochemical changes in enlarged thymuses and spleens from WT and TRAIL-R+/– animals suffering from lymphoma. Cellular FLICE-inhibitory protein (c-FLIP) has been proposed to protect cells from TRAIL-induced cell death and has been shown to be directly repressed by c-Myc (22). Thus we reasoned that in this model c-FLIP expression might be selected for during lymphoma development. Changes in c-FLIP protein levels correlated with changes in the TRAIL-R genotype, where decreased expression of c-FLIPs was evident in enlarged thymuses and spleens from TRAIL-R+/– animals as compared with WT animals (Figure 3, C and D). This suggests that high levels of c-FLIPS could be selected for in order to circumvent TRAIL-mediated killing in lymphomas arising in WT animals.
Figure 3. WT Eμ-myc lymphomas are resistant to TRAIL in vitro and express high levels of FLIPs.
(A) Challenging isolated WT Eμ-myc lymphoma cells in vitro with TRAIL resulted in lack of cell death (as determined by tryphan blue staining) in comparison with treatment with etoposide. (B) In vitro cell death assays. Propidium iodide uptake (red staining) and fluorescent-labeled inhibitor of caspase activity (FLICA or FITC-VAD-FMK; green staining) suggest that TRAIL (1 μg/ml) only weakly triggers caspase activity and cell death relative to control in comparison with treatment with etoposide (1 μM) in living Eμ-myc lymphoma cells. (C) Total RNA isolated from WT western blot of splenic and thymic lymphomas shows increased expression of c-FLIPS in WT relative to TRAIL-R+/– lymphomas. (D) Densitometric analysis using NIH ImageJ of western blots from lymphomas (splenic and thymic) from WT (n = 10) and TRAIL-R+/– (n = 10) mice. The ratios of the FLIPs band in relation to the actin band (loading control) is shown. The red horizontal band represents median, and the red cross represents the mean. WT lymphomas have a higher FLIPs/actin ratio (P < 0.05, Student’s t test), suggesting a relatively higher FLIPs expression compared with TRAIL-R+/– lymphomas.
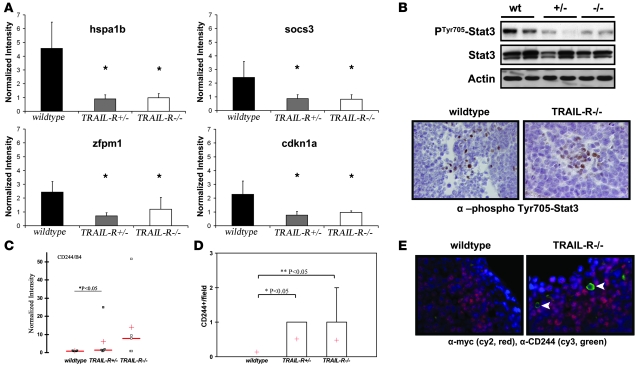
We hypothesized that changes in gene expression in Eμ-myc WT lymphomas relative to TRAIL-R–deficient lymphomas could reflect TRAIL-R–dependent downstream changes in gene expression or changes that may provide protective adaptations to intact TRAIL-R signaling. In order to address these questions, we performed expression profiling on a subset of lymphomas from WT, TRAIL-R+/– and TRAIL-R–/– animals. Fifty-nine genes were found to be differentially expressed and common among WT and TRAIL-R–/– animals (Supplemental Figure 1 and Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI29900DS1). Using the gene lists of differentially expressed genes for supervised expression-based clustering discriminated between WT and TRAIL-R–deficient lymphomas but did not cluster the different TRAIL-R–deficient genotypes (i.e., TRAIL-R+/– and TRAIL-R–/–) into distinct groups (Supplemental Figure 1). In addition, 4 genes were TRAIL-R dependently regulated with a false discovery rate of less than 26.7% and were annotated as signal transducers, oncogenes, or involved in cell death (according to the classification of the Gene Ontology Consortium) (Figure 4A). Interestingly, a number of genes were apoptosis and proliferation regulators (i.e., Hspa1b, Socs3, and p21), whereas others implicated a role in transcription (Zfpm1, also known as Fog-1). Hsp70 and Socs3 are downstream targets for the Jak/Stat pathway and have been implicated in the regulation of apoptosis (23, 24), whereas upregulation of the cyclin-dependent kinase inhibitor p21 is a common feature of activated Stat signaling and protection from apoptosis (25, 26). Thus we further hypothesized that TRAIL-R may either regulate Stat-signaling or that increased Stat-signaling may protect some WT lymphomas from immune-mediated cell death by TRAIL. Indeed, western blotting revealed increased levels of phosphorylated (Tyr705) Stat3 present in WT lymphomas as compared with those deficient in TRAIL-R (Figure 4B), and immunohistochemistry suggested that the lymphoma cells expressed phosphorylated Stat3 (Figure 4B). To the best of our knowledge, no report has previously described Stat3 phosphorylation as a result of TRAIL-R signaling, and treatment of the TRAIL-sensitive murine L929 cell line with recombinant TRAIL did not produce any increased levels of phosphorylated Stat3 (Supplemental Figure 2A). However, treating murine L929 cells and human colon cancer HCT116 cells with a combination of the pharmacologic Stat3 inhibitor JSI-124 (cucurbitacin I) at doses below or near the EC50 in combination with recombinant TRAIL triggered synergistic caspase-3 cleavage and cell death that was associated with a reduced expression of the Stat3-responsive gene product survivin in HCT116 cells (Supplemental Figure 2, B–E). This suggests that phosphorylated Stat3 may provide cell-intrinsic protection of cells from TRAIL-induced cell death.
Figure 4. Expression profiling of Eμ-myc lymphomas suggests that differentially expressed genes are regulated independent of TRAIL-R gene dosage as well as a role for Stat3 in WT lymphomas.
RNA samples from TRAIL-R+/– and TRAIL-R–/– Eμ-myc lymphomas (n = 5/genotype) were hybridized to MOE430A 2.0 Affymetrix arrays. Differentially expressed genes (upregulated >2-fold) were subjected to statistical analysis. A shows the top 4 genes that showed consistent changes over the different probe sets. Hspa1b, heat-shock protein 70; Socs3, suppressor of cytokine signaling 3; Zfpm1, friend of GATA1 (also known as Fog-1); Cdkn1a, cyclin-dependent kinase inhibitor 1a (p21). *P < 0.05, Student’s t test. (B) Immunoblotting suggests that Tyr705-phosphorylated Stat3 is elevated in WT Eμ-myc lymphomas compared with TRAIL-R–deficient lymphomas. Tyr705-phosphorylated Stat3 is expressed in a subset of cells within the lymphomas, as detected by immunohistochemistry. (C) CD244 is expressed at higher levels in Eμ-myc lymphomas of TRAIL-R+/– animals (Mann-Whitney U test, P < 0.05; n = 5/genotype). Red horizontal lines indicate medians; red crosses indicate means. (D and E) TRAIL-R–deficient lymphomas (n = 3/genotype; Mann-Whitney U test) show frequent infiltration of CD244+/c-myc cells (Cy3, green; Cy2, red) compared with WT lymphomas, suggesting expression of CD244 on nonlymphoma cells. Representative pictures are shown. Original magnification, ×60 (B); ×100 (E).
Because low levels of phosphorylated Stat3 have been associated with antitumor immunity (27), we hypothesized that blockage of Stat3 phosphorylation might correlate with increased infiltration of immune cells in TRAIL-R–deficient lymphomas. Indeed, analysis of the microarray data identified a subset of the TRAIL-R–deficient lymphomas expressing high levels of CD244/2B4 (Figure 4C), a surface molecule on NK cells, CD8+ T cells, eosinophils, and basophils (28). Indeed, c-myc–negative cells expressing CD244 were frequently identified in TRAIL-R–deficient lymphomas but not in WT lymphomas (Figure 4, D and E), suggesting increased presence of immune infiltrating cells in TRAIL-R–deficient Eμ-myc lymphomas that correlated with low levels of Tyr705-phosphorylated Stat3.
TRAIL-R–deficient animals are susceptible to sublethal γ-irradiation–induced pneumonitis.
Loss of TRAIL-R has been associated with an attenuated apoptotic response to irradiation, particularly in the spleen and thymus (9). The long-term relevance for the lack of apoptosis in these organs in the context of radiation therapy is unclear. However, ionizing radiation is an established carcinogen in a number of animal models as well as in humans (reviewed in ref. 29). In order to investigate a putative protective effect of TRAIL-R in radiation-induced carcinogenesis, mice were subjected to a sublethal dose of 4 Gy of whole-body irradiation at the age of 4–5 weeks (14 WT, 13 TRAIL-R+/–, and 23 TRAIL-R–/– mice) and monitored for up to 18 months.
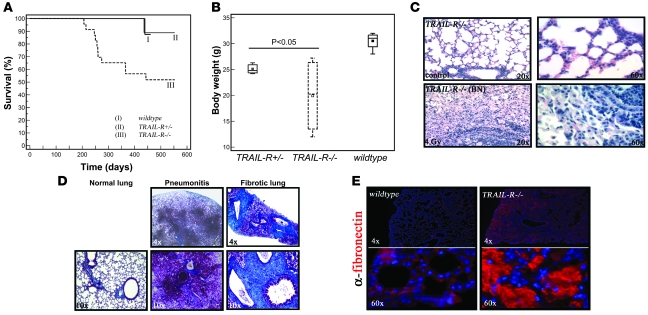
Lethality was only observed in the TRAIL-R–/– group of animals at 28–52 weeks following irradiation (Kaplan-Meier test, P = 0.010; Figure 5A), with lower body weight, labored breathing, and hunched body posture recorded on sex- and age-matched TRAIL-R–deficient animals at 28 weeks following ionizing irradiation as compared with WT controls (Figure 5B). This revealed systemic disease in irradiated TRAIL-R+/– and TRAIL-R–/– animals relative to WT littermates manifesting as lethality in the more severe cases. Histological examination of the lungs from control (nonirradiated WT), TRAIL-R–/–, and irradiated WT mice (39 weeks) (Figure 5C) showed no abnormalities, whereas TRAIL-R–/– mice that succumbed following sublethal irradiation showed emboli in respiratory bronchioles (Figure 5D) and loss of interstitial alveolar space accompanied by the deposition of fibronectin and collagen, as shown by immunohistochemistry and Masson’s trichrome staining (Figure 5, D and E). Lethality correlated with pulmonary CD3+ and B220– infiltrates, suggesting a T cell origin of the infiltrates (Figure 6A). The infiltrates were accompanied by a subset of cells expressing membrane/cytoplasmic TRAIL (Figure 6A). Substantial proliferation in alveolar/nonlymphoid cells was found as judged by increased labeling by the Ki-67 proliferation marker (Figure 6A), suggesting that the inflammation triggered hyperplasia. Evaluation classified the overall pulmonary condition as moderate to severe chronic bronchopneumonia.
Figure 5. TRAIL-R–/– animals show decreased survival following exposure to a single sublethal dose (4 Gy) of ionizing radiation.
(A) Survival following 4 Gy of whole-body irradiation was decreased in the TRAIL-R–/– group (n = 23) at 28–52 weeks following irradiation in comparison with the group of WT (n = 14) animals and TRAIL-R+/– (n = 13) animals (Kaplan-Meier log-rank analysis, P = 0.010). (B) However, decreased body weight was observed in both TRAIL-R+/– (n = 3) and TRAIL-R–/– (n = 4) animals relative to WT (n = 4) animals at 28 weeks following 4 Gy of ionizing irradiation (P < 0.05, Mann-Whitney U test). No weight difference was detected between genotypes in nonirradiated animals (data not shown). (C) A representative H&E staining of the lungs from lethargic TRAIL-R–/– animals irradiated with 4 Gy at 39 weeks prior to sacrifice. Extensive inflammatory emboli in the respiratory bronchioles and increased cellularity in the interstitial space was observed (C; ×20, lower left and right panels). Lungs were histochemically stained with Masson’s trichrome (D) in order to detect the presence of collagen (bright blue) and were analyzed by immunofluorescence for fibronectin (E; Cy3, red). Lungs from irradiated and lethargic TRAIL-R–/– animals showed severe pneumonitis (D) and extensive deposition of collagen and fibronectin (E).
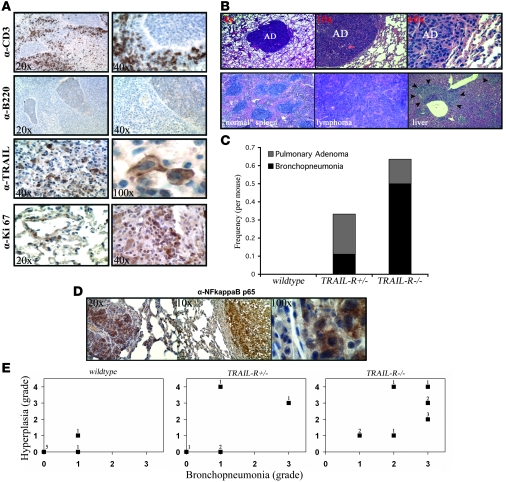
Figure 6. Loss of TRAIL-R leads to increased infiltration of CD3+ cells and tumorigenesis following sublethal irradiation.
(A) Immunohistochemistry on lungs from TRAIL-R–/– animals with infiltrates show increased numbers of CD3+ cells but little to no positive staining for B220. Infiltrates contained a number of TRAIL-positive cells expressing cytoplasmic and membrane-bound TRAIL and focal areas that stained positive for Ki-67. (B) Mice lacking 1 or 2 alleles of TRAIL-R show an increased incidence of pulmonary adenomas (top row). Microphotographs show a normal spleen and a splenic lymphoma with metastasis to the liver (bottom row). Lungs from 12 WT, 9 TRAIL-R+/–, and 18 TRAIL-R–/– mice were examined and stained immunohistochemically. (C) The frequency per mouse of pulmonary adenomas in relation to bronchopneumonia is shown. WT, n = 12; TRAIL-R+/–, n = 9; TRAIL-R–/–, n = 18. (D) Irradiated TRAIL-R–/– animals show correlation between bronchopneumonia and hyperplasia in their lungs. Hyperplastic adenomatous focal lesions (left panel) and pulmonary adenomas (middle and right panel) in the lungs of irradiated TRAIL-R–/– animals showed positive immunohistochemistry for NF-κB p65. Staining was observed in the cytoplasm and nucleus (right panel) of adenomatous cells. Representative photographs of investigated animals are shown. (E) Lungs of irradiated animals of the different TRAIL-R genotypes (WT, n = 7; TRAIL-R+/–, n = 5; TRAIL-R–/–, n = 10) were blindly classified according to inflammatory grade (bronchopneumonia grade) by the use of histological examination and immunohistochemistry for CD3 and fibronectin. Preneoplasia/neoplasia (hyperplastic) grading was based on combined macroscopic observations, histological findings (H&E staining), and immunohistochemistry for Ki-67. Grade 0, no or only scattered stained cells constituting less than 2% of the section; grade 1, heterogeneous staining with at least 20% of the section showing 2%–10% positive cells; grade 2, at least 20% of the section showing 11%–50% positive cells; grade 3, and at least 20% of the section showing more than 50% positive cells. For hyperplasia/neoplasia, presence of adenoma was classified as grade 4. The n-value represented by each data point is shown.
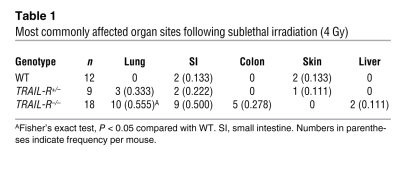
Overall, most organ sites affected (including any manifestation of inflammation, fibrosis, or neoplastic growth not present in nonirradiated animals) by sublethal irradiation were markedly different between TRAIL-R–/– and WT animals (Table 1). A tumor mass found unique to irradiated TRAIL-R–/– animals was a splenic lymphoma with extensive involvement of the liver and lungs observed in an irradiated TRAIL-R–/– animal. This lymphoma did not express surface markers of either B cells (B220) or T cells (CD3) (Figure 6B and data not shown). However, only lesions in the lung showed a statistically significant difference in rate per animal between irradiated TRAIL-R–/–(0.555 tumors per mouse) and WT (0) animals (Fisher’s exact test, P < 0.05). Interestingly, TRAIL-R–/–mice showed 0.136 tumors per mouse in the lungs, whereas the corresponding number for TRAIL-R+/– mice was 0.111 (see Table 1 and Figure 6, B and C). However, inflammation and/or fibrosis in the lung (i.e., bronchopneumonia) was more frequently observed in TRAIL-R–/– animals as compared with TRAIL-R+/– (0.500 compared with 0.111 tumors per mouse) mice. Hyperplastic adenomatous focal lesions and pulmonary adenomas (Figure 6D) in the lungs of irradiated TRAIL-R–/– animals frequently overexpressed NF-κB p65, a proinflammatory protein that is associated with tumor progression and metastasis (reviewed in ref. 30). Lungs of irradiated animals of the different TRAIL-R genotypes (WT, n = 7; TRAIL-R+/–, n = 5; TRAIL-R–/–, n = 10) were blindly classified according to inflammatory grade (bronchopneumonia grade) through the use of histological examination and immunohistochemistry for CD3 and fibronectin. The determination of preneoplasia/neoplasia (hyperplastic grade) was based on combined macroscopic observations, histological findings (H&E staining) and immunohistochemistry for Ki-67 (for more detail on classification, see Methods). TRAIL-R–/– animals showed correlation between the severity (grade) of bronchopneumonia and grade of hyperplasia in their lungs. (Figure 6E), suggesting that the presence of hyperplasia was intimately linked to chronic inflammation in the TRAIL-R–deficient genetic background. Thus the murine TRAIL-R plays an important role in regulating the long-term tissue response in the lung following a single sublethal dose of ionizing radiation.
Table 1 .
Most commonly affected organ sites following sublethal irradiation (4 Gy)
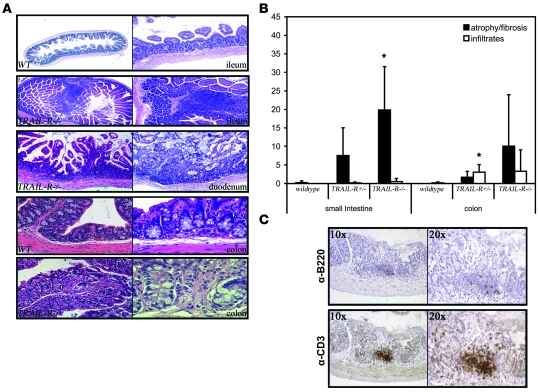
TRAIL-R–/– animals also showed chronic enterocolitis with some areas of ulceration, increased epithelial atrophy, erosion, and inflammatory infiltrates (Figure 7, A and B). These were typical radiation-induced lesions that, at sublethal doses, mostly heal 2–6 weeks after irradiation but may persist for some time. Indeed, we found some lesions in irradiated WT animals at similar time points following irradiation, but with significantly less severity (lower average percentage of total area) in the GI tract compared with irradiated TRAIL-R–/– animals (Figure 7B). Ulceration is, however, a highly abnormal feature after several months following irradiation, and its presence suggests that the TRAIL-R–/– animals suffer from chronic inflammation and impaired healing in the GI tract as well as in the lungs following tissue damage induced by ionizing irradiation. Infiltrates in the GI tract consisted of predominantly CD3+ cells and only a small number of B220+ cells (Figure 7C), suggesting increased infiltration of T cells.
Figure 7. TRAIL-R suppresses chronic colitis following sublethal irradiation.
(A) The small intestine (ileum) of WT and TRAIL-R–/– animals sublethally irradiated 32 weeks prior to sacrifice show increased lymphoid infiltration and luminal protrusions in the TRAIL-R–/– animals compared with WT mice. The proximal part of the small bowel (duodenum) showed epithelial atrophy and erosion in an irradiated TRAIL-R–/– animal at 32 weeks following irradiation. Normal colon of a WT animal at 32 weeks following 4 Gy of irradiation is compared with TRAIL-R–/– animals, which showed chronic enterocolitis following the same treatment. Original magnification, ×4 (first and second row, first column); ×10 (first and second row, second column); ×20 (third row); ×40 (fourth row) (fifth row, first column); ×60 (fifth row, second column). (B) The surface area covered with either lymphoid infiltrates or atrophic lesions was assessed from tissue sections of the small intestine and colon from WT (n = 4), TRAIL-R+/– (n = 3), and TRAIL-R–/– (n = 4) mice subjected to 4 Gy of whole-body irradiation. *P < 0.05, Student’s t test. (C) Representative immunohistochemistry for B220 and CD3 on a TRAIL-R–/– small intestine with atrophy shows abundant infiltration of CD3+ cells.
Loss of TRAIL-R leads to susceptibility to chemically induced hepatocarcinogenesis.
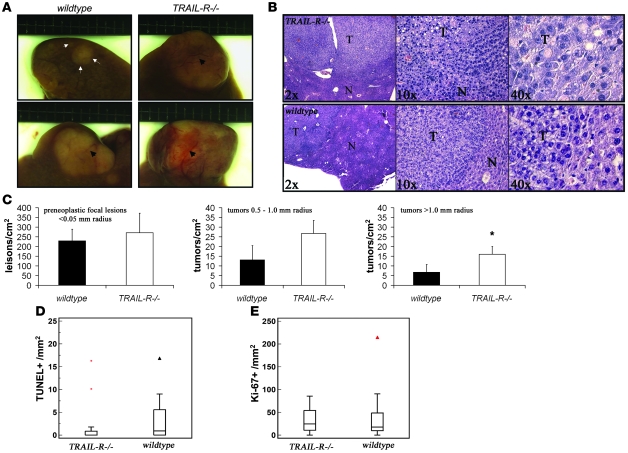
Ten male 7-day-old littermate WT and TRAIL-R–/– pups were challenged with the DNA-damaging hepatocarcinogen DEN. DEN has been shown to elicit a prominent activation of p53 in the mouse liver when administered i.p. (31). Given that TRAIL-R is a p53 target gene and is induced in the liver following DNA damage (ref. 10 and our unpublished observations), we reasoned that loss of TRAIL-R may contribute to increased initiation of hepatocarcinogenesis, as apoptosis and potentially removal of subsequent mutated cells in the organ may be perturbed. However, we observed only a slight increase in the incidence of DEN-induced HCC in TRAIL-R–/– mice compared with WT mice, where 80% (8/10) of the TRAIL-R–deficient animals developed HCC compared with 60% (6/10) of the WT animals at 10 months of age. TRAIL-R–/– mice developed a larger number of macroscopically visible liver nodules by gross observation (depicted in Figure 8A) than WT animals (11 and 8 per animal, respectively), indicating increased presence of neoplastic changes in the liver of TRAIL-R animals following DEN treatment. Detailed histological analysis of the livers from DEN-treated TRAIL-R–deficient and WT animals suggested that an increased number of lesions with a radius in excess of 1.0 mm was present in TRAIL-R–/– livers (Figure 8, B and C). However, no significant increase in the number of the DEN treatment–related preneoplastic lesions with a radius of less than 0.5 mm was observed in TRAIL-R–null animals (Figure 8C). Immunohistochemical analysis using the proliferation marker Ki-67 and staining for apoptotic cells using TUNEL suggested that the HCCs in the livers of TRAIL-R–/– mice had reduced numbers of TUNEL+ cells/mm2 compared with HCCs from WT animals (Figure 8D), whereas the number of Ki-67+ cells in the HCCs did not differ significantly (Figure 8E). This suggests that apoptosis is reduced in HCCs of TRAIL-R–/– animals compared with WT HCCs, whereas proliferation remains unaltered by the loss of the TRAIL-R.
Figure 8. Loss of TRAIL-R renders mice susceptible to DEN-induced hepatocarcinogenesis.
(A and B) TRAIL-R–/– mice (n = 10) injected with DEN (0.30 mmol/kg body weight) at 7 days of age showed an increased liver tumor load compared with WT littermates (n = 10) at 10 months after treatment. (A) Gross observations of small (top row, demarcated with arrows and arrowheads) and larger liver tumors in DEN-treated animals (bottom row, demarcated with arrowheads). (B) Microscopy of H&E-stained histological sections shows the location of a liver tumor (T) with surrounding normal (N) liver tissue. (C) Detailed histological analysis of livers (>0.5 cm2 liver area analyzed, excluding macroscopic lesions) from randomly selected littermates (n = 3/genotype) shows an increased number of lesions exceeding a radius of 1.0 mm in TRAIL-R–/– animals. *P < 0.05, Student’s t test. Means ± SD are shown. (D and E) Immunohistochemical analysis of proliferation (Ki-67 labeling; E) and cell death (TUNEL staining; D) in HCCs from WT (n = 17) and TRAIL-R–/– (n = 17) animals treated with DEN show reduced TUNEL labeling in TRAIL-R–/– HCCs (Mann-Whitney U test, P < 0.05) compared with WT HCCs, whereas similar levels of Ki-67–positive cells were seen in HCCs of both genotypes.
Discussion
The role of TRAIL-R in lymphomagenesis.
Given the importance of the p53 pathway in B cell (Burkitt) lymphoma (32), we chose to test the impact TRAIL-R deficiency on lymphoma development in spontaneous lymphoma-prone Eμ-myc mice. Eμ-myc mice lacking 1 or 2 alleles of TRAIL-R developed lymphomas more rapidly than WT Eμ-myc mice (without 1 allele, 82 days; without 2 alleles, 87 days; WT, 119 days). TRAIL-R–deficient lymphomas harbored apoptotic defects and showed increased metastases to the liver and lungs (Figure 1B and Figure 2, A and B). Interestingly, no statistically significant difference between the rate of lymphoma formation, the number of cleaved caspase-3–positive cells in the lymphomas, and metastasis was found between TRAIL-R+/– and TRAIL-R–/– animals in the Eμ-myc background (Figure 1 and Figure 2, A and B), suggesting that monoallelic loss of TRAIL-R is sufficient to promote myc-driven lymphomagenesis and that this correlates with loss of TRAIL-R–mediated cell death.
Apoptotic defects due to alterations within the extrinsic/intrinsic pathways have previously been investigated in the Eμ-myc model. Overexpression of Bcl-2 in concert with the Eμ-myc transgene triggers more rapid lymphomagenesis, with tumors showing hallmarks of primitive hematopoietic cells (33). Knockdown of Puma, a proapoptotic p53 target gene, in transplanted Eμ-myc–derived hematopoietic stem cells triggers lymphomas at a similar rate to that of p53 knockdown in recipient animals (34). In a study of the role of the extrinsic pathway, Eμ-L-myc mice, which are prone to develop both B and T cell lymphomas, carrying 2 defective alleles for the death receptor Fas/Apo-1 (lpr/lpr) showed enhanced B and T cell lymphomagenesis (35). Taken together, this suggests that both the extrinsic and intrinsic pathways have significant importance in repressing lymphomagenesis.
Data using 45 human B cell non-Hodgkin lymphoma cell lines suggest that monoallelic deletion of 650-kb region on chromosome 8p21.3, including TRAIL-R1 and TRAIL-R2, is a frequent event (36). No altered promoter methylation on the remaining alleles was found, whereas downregulation of DR4 and DR5 mRNA was found with concomitant loss of cell surface expression of TRAIL receptor proteins and resistance to apoptosis induced by FLAG-tagged TRAIL in vitro that was correlated with monoallelic deletion. No information on the c-myc status in those cell lines was provided. Our data may in part suggest that monoallelic loss might be evolutionarily conserved from mouse to human and sufficient for increased susceptibility to (B cell) lymphomagenesis. Indeed, only a small number (2/10) of the TRAIL-R+/– lymphomas undergo LOH (Figure 2C) and this does not appear sufficient to explain the lack of difference in median lymphoma-free survival between Eμ-myc TRAIL-R+/– and TRAIL-R–/– animals. Taken together with previous data on human B cell non-Hodgkin lymphomas (36), we suggest that gene dosage–dependent reduction in TRAIL-R mRNA expression may be sufficient to quench TRAIL-mediated signaling to promote lymphomagenesis.
TRAIL is currently undergoing clinical evaluation for the treatment of B cell non-Hodgkin lymphoma. Despite encouraging results in human cell lines, relatively little is known about the molecular mechanisms that sensitize cells to TRAIL-mediated cell death. Our previous data have suggested that direct downregulation of c-FLIPs by c-myc in human tumor cells contributes to TRAIL sensitivity (22). Interestingly, we found that c-FLIPs was increased in vivo in lymphomas from WT mice in comparison with those of TRAIL-R–deficient mice. One interpretation of these data could be that the relatively high c-FLIPs levels protect from cell death through the extrinsic pathway during lymphomagenesis in the presence of an intact TRAIL-R locus and might be a selection factor. However, it is possible that loss of 1 allele of TRAIL-R might alter the requirements for c-FLIPS expression due to the altered gene dosage and this may abrogate signaling through TRAIL-R by, for example, LOH or posttranscriptional/translational mechanisms. Indeed, the WT lymphoma cell line generated (p54), challenged in vitro with TRAIL, did not undergo apoptosis, and 50% of all human Burkitt lymphoma cell lines display resistance to TRAIL (37). This could potentially have implications for the use of TRAIL in the treatment of (B cell) lymphomas and requires novel diagnostic tools to be developed to assess TRAIL sensitivity.
Expression profiling of lymphomas from the different TRAIL-R genotypes suggested increased levels of a subset of established Stat3 target genes (socs3, hsp70, and p21) in WT lymphomas relative to TRAIL-R–deficient lymphomas (Figure 4A) (23–26). Indeed, the levels of phosphorylated Stat3 were found to be increased in WT lymphomas, suggesting that increased Stat3 activity may be associated with an intact TRAIL-R locus. Stat3 is frequently activated in malignant cells and has been shown both to suppress antitumor immunity by reducing the number of immune cells present in the tumor tissue and to reduce killing of tumor cells (27). On the other hand, Stat3 activity can be negatively regulated through the IFN-γ–STAT1 pathway, for example, through inflammatory cells invading malignant tissues. Increased infiltration of CD244+ cells, a surface molecule on, for example, NK cells, was found in TRAIL-R–deficient lymphomas (Figure 4, D and E), suggesting that the low levels of phosphorylated Stat3 correlated with increased tumor infiltration of immune cells. Increased Stat3 activity may potentially protect WT lymphomas from infiltration of NK cells, a potent source of membrane-bound TRAIL (38). Further experiments are required to clarify this, and we are currently transplanting lymphomas onto different TRAIL-R genetic backgrounds to investigate the involvement of TRAIL-R–deficient immunity in promoting lymphoma metastasis.
The role of TRAIL-R in the pulmonary irradiation response.
TRAIL-R–/– mice have decreased survival starting at 29 weeks following irradiation and display a unique tissue phenotype in the lung relative to WT littermates following irradiation (Figures 5 and 6) that includes chronic inflammation, fibrosis with fibronectin deposition, and accelerated tumorigenesis. TRAIL-R deficiency was also associated with increased chronic colitis and atrophy of the GI tract following sublethal irradiation (Figure 7), suggesting chronically impaired healing of the GI-tract as well as the lung in TRAIL-R–deficient mice following exposure to a sublethal dose of ionizing irradiation.
The in vivo role of TRAIL and its proapoptotic receptor in the lung is largely unclear. TRAIL is upregulated during allergic inflammation and can be detected in bronchoalveolar lavage fluids obtained from the lung (39). Some evidence suggests that TRAIL may under some circumstances function as an antiinflammatory cytokine, since ablation of the TRAIL gene in mice renders them susceptible to various conditions associated with inflammation, including arthritis, hepatitis, and type 1 diabetes, presumably through the regulation of T cell homeostasis (40–42).
Irradiated TRAIL-R–/– mice frequently show an abundance of CD3+ infiltrates (Figure 6A), suggesting that T cells are involved in radiation-induced pneumonitis in our model. It is thus possible that TRAIL-R–/–mice are sensitized to sporadic radiation-induced pneumonitis that is considered to be similar to hypersensitivity pneumonitis. Indeed, BAL fluids from such patients contain activated CD4+ T helper cells, and hypersensitive pneumonitis may rely preferentially on the expansion of CD8+ T cells, a process previously shown to be dependent on TRAIL-mediated cell death (43). Furthermore, it is worth noting that the finding of chronic inflammation in the sublethally irradiated TRAIL-R–deficient mice provides a new model system to study the late toxic effects of ionizing radiation. Importantly, while cytokines have been previously implicated and effects of TGF-β signaling may contribute to the late effects of radiation, we believe this is the first report to implicate TRAIL receptor signaling in this process.
The role of TRAIL-R in suppressing DEN-induced hepatocarcinogenesis.
In order to substantiate a role for TRAIL-R in suppressing the development of malignancies, we subjected TRAIL-R–deficient mice to an “organ-specific” chemical carcinogenesis protocol using DEN to induce HCC. TRAIL-R–null animals developed more liver tumors with a radius exceeding 1.0 mm compared with that of WT littermates (Figure 8C). Interestingly, no obvious difference in the number of smaller preneoplastic focal lesions that emerged following treatment with DEN was observed in TRAIL-R–/– animals compared with WT animals. This may suggest, given the connection between TRAIL-R and p53 and the DNA-damaging nature of DEN (10), that loss of TRAIL-R promotes neoplastic growth in the liver rather than the emergence of preneoplasia associated with DEN-induced DNA damage. TRAIL-R–deficient HCCs displayed reduced numbers of TUNEL+ cells but similar numbers of Ki-67–positive cells compared with WT HCCs (Figure 8, D and E), suggesting that loss of TRAIL-R was associated with blockade of apoptosis but also with unaltered proliferation in these malignancies.
Chronic inflammation is a frequent risk factor for the development of malignancies in the colon, lung, liver, and lymphoid organs (30). Several mechanisms have been suggested for the transformation of nonmalignant cells along the axis of carcinogenesis, where increased proliferation and altered epigenetic programming might contribute to initiation of preneoplasia, whereas cytokine triggering of the expression of chemotactic factors may contribute to metastasis. Indeed, the tumor suppressor p53, one of the transcriptional activators of TRAIL-R, has been implicated in suppressing inflammation through inhibition of NF-κB (44). Indeed, p53 mutations can be detected in patients suffering from idiopathic pulmonary fibrosis (45). Interestingly, we were only able to document lung tumors (a rare occurrence on the C57BL/6 background; ref. 46) in irradiated TRAIL-R+/– and TRAIL-R–/–, whereas no tumors were found in WT animals (Table 1). Indeed, bronchopneumonia was accompanied by increased deposition of extracellular matrix, proliferation, and preneoplasia/neoplasia in the lungs of TRAIL-R–/– animals (Figure 5, C–E, and Figure 6), 2 factors that are associated with an increased risk for the development of cancer. More animals and additional experiments are required, however, to substantiate that TRAIL-R protects from irradiation-induced carcinogenesis in the lung through the suppression of inflammation and fibrosis.
In conclusion, we believe this to be the first demonstration of tumor susceptibility upon TRAIL-R knockout in the mouse. We document that monoallelic loss of TRAIL-R is sufficient to significantly decrease the lymphoma-free survival and to increase metastasis in the Eμ-myc Burkitt lymphoma mouse model. TRAIL-R also suppresses bronchopneumonia/fibrosis in the lung and promotes survival following sublethal irradiation. A role for TRAIL-R in the etiology of HCC can also be envisioned, as DEN-induced hepatocarcinogenesis is suppressed by TRAIL-R. To the best of our knowledge, no previous study has shown that TRAIL-R can suppress tumorigenesis in vivo. While some in vivo data may support a role for TRAIL ligand in an etiological context, previous studies can not exclude a role for TRAIL signaling through any of the known TRAIL receptors present in the mouse. Further experiments will be required to determine what cell types are most dependent on the presence of TRAIL-R in suppressing inflammation and tumorigenesis. However, the increased levels of c-FLIP and phosphorylated Stat3 signaling in WT lymphomas and the infiltration of CD244+ cells in TRAIL-R–deficient lymphomas and infiltration of CD3+ cells and proliferation (hyperplasia) in the lungs of TRAIL-R–/– animals provide additional important mechanistic insights. Other mechanistic insights include the finding that hyperplastic adenomatous focal lesions and pulmonary adenomas from irradiated TRAIL-R–/– mice overexpress proinflammatory NF-κB p65 that has been associated with tumor progression and poor prognosis. Finally, it is important to note that the finding of chronic inflammation in the sublethally irradiated TRAIL-R–deficient mice provides a new model system to study the late toxic effects of ionizing radiation.
Methods
Mice.
Mice harboring targeted mutations of the TRAIL-R alleles (previously described in ref. 9) on the C57BL/6 background were crossed with WT C57BL/6 mice. Littermates from intercrosses between TRAIL-R+/– C57BL/6 mice were genotyped as previously described (9). Hemizygous C57BL/6J-Tg(IghMyc)22Bri/J (Eμ-myc mice; The Jackson Laboratory; ref. 47) were purchased and crossed with TRAIL-R–/– mice. Litters were genotyped and hemizygous Eμ-myc TRAIL-R+/– mice were crossed with TRAIL-R+/– mice. Transgenic F1 litters were monitored twice a week by palpation of the cervical lymph nodes, and enlarged lymph nodes of at least 5 mm in diameter were evaluated for malignant disease.
Cell lines.
The murine L929 fibrosarcoma cell line and the human colon carcinoma cell line HCT116 were obtained from the ATCC. The cell lines were cultured and passaged in MEM and McCoy’s 5A (GIBCO), respectively, supplemented with 10% FBS and 1% penicillin and streptomycin.
Ionizing irradiation treatment.
At 3–4 weeks of age, mice received a single 4-Gy dose of whole-body γ-irradiation from a 137Cs source (at 1.4 Gy/min). After irradiation, mice were monitored twice weekly and mice showing clinical signs of disease (e.g., hunched posture, labored breathing, immobility) or a tumor load exceeding 10% of body weight were euthanized according to approved University of Pennsylvania IACUC protocol. Mice not showing clinical signs of disease were euthanized at 18 months of age.
Chemical hepatocarcinogenesis.
WT and TRAIL-R–/– littermate mice on the C57BL/6 background were injected i.p. with 0.30 mg/kg body weight of DEN (Sigma-Aldrich) at 7 days of age. Animals were sacrificed 12 months following treatment, and livers were removed and separated into individual lobes. Externally visible tumors (>0.5 mm) were counted and investigated by stereomicroscopy. Large lobes were fixed in 4% paraformaldehyde overnight and embedded in paraffin. For detailed histological and immunohistochemical analysis, the livers of 3 littermate animals of each genotype were cut in 4-μm sections and analyzed as described in Histology and immunohistochemistry and Quantitation of histological and immunohistochemical findings.
Microarray analysis.
Total RNA was extracted from frozen tissues of 5 Eμ-myc lymphomas per TRAIL-R genotype (i.e., WT, TRAIL-R+/–, and TRAIL-R–/–) using the RNeasy mini kit (Qiagen). Complementary DNA was synthesized from total RNA using a dT primer tagged with a T7 promoter. Complementary RNA was synthesized by transcription in vitro and labeled with biotinylated nucleotides (Enzo Biochem). All hybridizations were performed using the MOE430A 2.0 GeneChip (Affymetrix). Gene lists, filtered by a minimum of 2-fold change (1-way ANOVA, P < 0.05), were generated with the GeneSpring GX 7.3.1 software (Agilent), which resulted in the creation of gene structures specific to WT and TRAIL-R–deficient mice (total of 59 genes, 17 upregulated and 42 downregulated). Also, more stringent exclusion criteria were applied by the use of significance analysis of microarrays (SAM) software version 2.0 (with an approximate 26.7% false discovery rate) and filtering against gene lists for signal transduction pathways, apoptosis-related genes, and oncogenes obtained from the Gene Ontology Consortium in order to detect 6 differentially expressed genes.
Culturing of lymphoma cells.
Upon the detection of palpable tumors or other signs of declining health, mice were sacrificed and enlarged lymphoid organs (thymus, spleen, and mesenteric and peripheral lymph nodes) were removed and single-cell suspensions were prepared. Lymphoma cells were cultured in RPMI 1640 with 10% FBS, MEM, and 1% streptomycin/penicillin as described previously (48). After 16 passages, cells were resistant to limiting dilution and were then subjected to treatment with 1 μg/ml of TRAIL (BioMol) and 1 μM of etoposide (Sigma-Aldrich) as a cell line.
Histology and immunohistochemistry.
Following necropsy, the skin, mammary glands, superficial lymph nodes, mesenteric lymph nodes, testes/ovaries, colon, cecum, ileum, jejunum, duodenum, liver, pancreas, spleen, kidneys, heart, lungs, esophagus, thymus, bone marrow (femur), and brain were sampled and fixed in 4% paraformaldehyde overnight at 4°C. Tissues were embedded in paraffin and cut into 4-μm sections and stained with H&E or Masson’s trichrome. Sections were blinded as to their genotype and evaluated. Immunohistochemistry was performed by rehydrating slides and subjecting them to antigen retrieval through boiling in 1 mM citric acid buffer (pH 6.0). Endogenous peroxidases were blocked, and in the case of use of the Vecta Elite ABC peroxidase kit (Vector Laboratories), slides were submerged in 3% H2O2. Slides were blocked using appropriate blocking reagents and subjected to primary antibodies overnight at 4°C. Primary antibodies used were rabbit anti-CD3 (1:1000; Novocastra), mouse anti-B220 (1:150; AbCam), rabbit anti-fibronectin (1:100; Novocastra), rabbit anti-cleaved caspase-3 (1:100, Cell Signaling; 1:200, BD Transduction Laboratories), rabbit anti–Ki-67 (1:1000; Novocastra), rabbit anti-TRAIL (1:100; AbCam), mouse α-CD244 (eBioscience), rabbit anti–NF-κB p65 (1:400; Santa Cruz Biotechnology Inc.), mouse anti-survivin (1:200; Cell Signaling), hamster anti-murine DR5 (1:100; BioLegend), and rabbit anti–c-Myc (1:200; Santa Cruz Biotechnology Inc.). Primary antibodies were detected using either appropriate biotinylated secondary antibodies raised in goat (1:400; Pierce Biotechnology), Cy3-conjugated antibodies raised in donkey (1:400; Jackson ImmunoResearch Labs) or avidin-FITC (1:50; Vector Laboratories). In addition, apoptosis was detected by TUNEL staining using the ApoptTag Plus peroxidase kit (Chemicon International). Representative depiction of histology and immunohistochemistry was made using the IP lab software.
Flow cytometry.
Freshly isolated viable lymphoma cells of different TRAIL-R genotypes were assessed for surface expression of TRAIL-R by flow cytometry using hamster anti-mouse TRAIL-R/DR5-PE (MD5-1; BioLegend) with isotype-specific hamster IgG (BioLegend) as a negative control antibody according to the manufacturer’s instructions. Surface fluorescence was measured using an Epics Elite flow cytometer (Beckman Coulter). HCT116 cells were treated with JSI-124 (Calbiochem), recombinant TRAIL (BioMol), or a combination of both compounds. Following this, cells were collected, fixed, stained with propidium iodide, and analyzed by flow cytometry as described above.
Quantitation of histological and immunohistochemical findings.
All slide sections were coded, analyzed, and quantitated blindly by counting 10 randomly selected ×400 fields of 3 nonserial sections of the same specimen. The percentage area of the GI tract occupied by atrophy/inflammatory infiltrates, the area of the liver occupied by preneoplasia/neoplasia, and the number of TUNEL+ cells/mm2 in HCCs was calculated using ImageJ (NIH ImageJ 1.62 software). Appropriate statistics were applied to the generated data (see Statistics). For classification of bronchopneumonia and hyperplasia/neoplasia in irradiated mice, lungs were graded based on macroscopic findings following necropsy, histology (H&E staining and Masson’s trichrome), and immunohistochemistry for CD3, fibronectin, and Ki-67. Necropsy records and slides were blinded to genotype. The following grading system was used: grade 0, no or only scattered stained cells constituting less than 2% of the section; grade 1, heterogenous staining with at least 20% of the section showing 2%–10% positive cells; grade 2, at least 20% of the section showing 11%–50% positive cells; grade 3, at least 20% of the section showing more than 50% positive cells. For the classification of hyperplasia/neoplasia, grade 4 represented the presence of adenoma.
Western blotting.
Enlarged lymphoid organs were removed from WT Eμ-myc and Eμ-myc TRAIL-R+/– mice were homogenized and sonicated in RIPA buffer (1× PBS, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM PMSF, Complete protease inhibitor cocktail [Roche]). Protein concentrations were determined by the Bradford method (Bio-Rad) and proteins separated on sodium dodecylsulfate 12.5% polyacrylamide gels and transferred to polyvinylidene difluoride membranes. Actin, Ran, c-Myc, c-FLIP, and phosphorylated Stat3 expression was detected by immunoblotting with mouse α-actin (C-2; Santa Cruz Biotechnology Inc.), mouse α-Ran (BD — Transduction Laboratories), rabbit α-myc (N-262; Santa Cruz Biotechnology Inc.), rabbit monoclonal α-Tyr705 phosphorylated Stat3 (Cell Signaling), and rat α-FLIP (dave-2; Axxora) antibodies. Membranes were incubated with horseradish peroxidase–conjugated secondary antibodies (1:4,000) and detected by the ECL procedure (Amersham).
Statistics.
Differences in survival were recorded by Kaplan-Meier analysis and Student’s t test. Statistical comparisons of proportions in all 3 groups or pairs of groups were made using the Fisher’s exact test. Non-normally distributed data were analyzed for statistically significant differences using the Mann-Whitney U test. All statistical analyses were performed using the MedCalc software version 8.2.1.0.
Supplementary Material
Acknowledgments
The authors thank Eric J. Bernhard for assistance with mouse irradiation in the early phases of this work. This work was presented in part at the 97th Annual American Association for Cancer Research (AACR) meeting in Washington, DC, in April 2006; the 13th International p53 Workshop in New York, New York, USA, in May 2006; and the AACR special conference on Mouse Models of Cancer in Boston, Massachusetts, USA, in October 2006. This work was supported in part by NIH grants CA75138 and CA98101.
Footnotes
Nonstandard abbreviations used: c-FLIP, cellular FLICE-inhibitory protein; DEN, diethylnitrosamine; HCC, hepatocellular carcinoma; LOH, loss of heterozygosity; TRAIL, TNF-related apoptosis-inducing ligand; TRAIL-R, TRAIL receptor.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:111–123 (2008). doi:10.1172/JCI29900
See the related Commentary beginning on page 100.
References
- 1.Boldin M.P., et al. A novel protein that interacts with the death domain of Fas/APO1 contains a sequence motif related to the death domain. J. Biol. Chem. 1995;270:7795–7798. doi: 10.1074/jbc.270.14.7795. [DOI] [PubMed] [Google Scholar]
- 2.Chinnaiyan A.M., O’Rourke K., Tewari M., Dixit V.M. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 3.Bodmer J.L., et al. TRAIL receptor-2 signals apoptosis through FADD and caspase-8. Nat. Cell Biol. 2000;2:241–243. doi: 10.1038/35008667. [DOI] [PubMed] [Google Scholar]
- 4.Kischkel F.C., et al. Apo2L/TRAIL-dependent recruitment of endogenous FADD and caspase-8 to death receptors 4 and 5. Immunity. 2000;12:611–620. doi: 10.1016/s1074-7613(00)80212-5. [DOI] [PubMed] [Google Scholar]
- 5.Sprick M.R., et al. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity. 2000;12:599–609. doi: 10.1016/s1074-7613(00)80211-3. [DOI] [PubMed] [Google Scholar]
- 6.Yin X.M., et al. Bid-deficient mice are resistant to Fas-induced hepatocellular apoptosis. Nature. 1999;400:886–891. doi: 10.1038/23730. [DOI] [PubMed] [Google Scholar]
- 7.Wu G.S., Burns T.F., Zhan Y., Alnemri E.S., El-Deiry W.S. Molecular cloning and functional analysis of the mouse homologue of the KILLER/DR5 tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor. Cancer Res. 1999;59:2770–2775. [PubMed] [Google Scholar]
- 8.Diehl G.E., et al. TRAIL-R as a negative regulator of innate immune cell responses. Immunity. 2004;21:877–889. doi: 10.1016/j.immuni.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 9.Finnberg N., et al. DR5 knockout mice are compromised in radiation-induced apoptosis. Mol. Cell. Biol. 2005;25:2000–2013. doi: 10.1128/MCB.25.5.2000-2013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wu G.S., et al. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat. Genet. 1997;17:141–143. doi: 10.1038/ng1097-141. [DOI] [PubMed] [Google Scholar]
- 11.Yue H.H., Diehl G.E., Winoto A. Loss of TRAIL-R does not affect thymic or intestinal tumor development in p53 and adenomatous polyposis coli mutant mice. Cell Death Differ. 2005;12:94–97. doi: 10.1038/sj.cdd.4401523. [DOI] [PubMed] [Google Scholar]
- 12.Zerafa N., et al. Cutting edge: TRAIL deficiency accelerates hematological malignancies. J. Immunol. 2005;175:5586–5590. doi: 10.4049/jimmunol.175.9.5586. [DOI] [PubMed] [Google Scholar]
- 13.Cretney E., et al. Increased susceptibility to tumor initiation and metastasis in TNF-related apoptosis-inducing ligand-deficient mice. J. Immunol. 2002;168:1356–1361. doi: 10.4049/jimmunol.168.3.1356. [DOI] [PubMed] [Google Scholar]
- 14.Takeda K., et al. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat. Med. 2001;7:94–100. doi: 10.1038/83416. [DOI] [PubMed] [Google Scholar]
- 15.Takeda K., et al. Critical role for tumor necrosis factor-related apoptosis-inducing ligand in immune surveillance against tumor development. J. Exp. Med. 2002;195:161–169. doi: 10.1084/jem.20011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schneider P., et al. Identification of a new murine tumor necrosis factor receptor locus that contains two novel murine receptors for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J. Biol. Chem. 2003;278:5444–5454. doi: 10.1074/jbc.M210783200. [DOI] [PubMed] [Google Scholar]
- 17.Eischen C.M., Weber J.D., Roussel M.F., Sherr C.J., Cleveland J.L. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 1999;13:2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schmitt C.A., McCurrach M.E., de Stanchina E., Wallace-Brodeur R.R., Lowe S.W. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 1999;13:2670–2677. doi: 10.1101/gad.13.20.2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fei P., Bernhard E.J., El-Deiry W.S. Tissue-specific induction of p53 targets in vivo. Cancer Res. 2002;62:7316–7327. [PubMed] [Google Scholar]
- 20.Verna L., Whysner J., Williams G.M. N-nitrosodiethylamine mechanistic data and risk assessment: bioactivation, DNA-adduct formation, mutagenicity, and tumor initiation. Pharmacol. Ther. 1996;71:57–81. doi: 10.1016/0163-7258(96)00062-9. [DOI] [PubMed] [Google Scholar]
- 21.Ferry J.A. Burkitt’s lymphoma: clinicopathologic features and differential diagnosis. Oncologist. 2006;11:375–383. doi: 10.1634/theoncologist.11-4-375. [DOI] [PubMed] [Google Scholar]
- 22.Ricci M.S., et al. Direct repression of FLIP expression by c-myc is a major determinant of TRAIL sensitivity. Mol. Cell. Biol. 2004;24:8541–8555. doi: 10.1128/MCB.24.19.8541-8555.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Auernhammer C.J., Bousquet C., Melmed S. Autoregulation of pituitary corticotroph SOCS-3 expression: characterization of the murine SOCS-3 promoter. Proc. Natl. Acad. Sci. U. S. A. 1999;96:6964–6969. doi: 10.1073/pnas.96.12.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephanou A., Latchman D.S. Transcriptional regulation of the heat shock protein genes by STAT family transcription factors. Gene Expr. 1999;7:311–319. [PMC free article] [PubMed] [Google Scholar]
- 25.Bellido T., O’Brien C.A., Roberson P.K., Manolagas S.C. Transcriptional activation of the p21(WAF1, CIP1, SDI1) gene by interleukin-6 type cytokines. A prerequisite for their pro-differentiating and anti-apoptotic effects on human osteoblastic cells. J. Biol. Chem. 1998;273:21137–21144. doi: 10.1074/jbc.273.33.21137. [DOI] [PubMed] [Google Scholar]
- 26.Chin Y.E., et al. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21 WAF1/CIP1 mediated by STAT1. Science. 1996;272:719–722. doi: 10.1126/science.272.5262.719. [DOI] [PubMed] [Google Scholar]
- 27.Wang T., et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat. Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 28.Boles K.S., et al. Molecular characterization of a novel human natural killer cell receptor homologous to mouse 2B4. Tissue Antigens. 1999;54:27–34. doi: 10.1034/j.1399-0039.1999.540103.x. [DOI] [PubMed] [Google Scholar]
- 29.Barcellos-Hoff M.H., Park C., Wright E.G. Radiation and the microenvironment — tumorigenesis and therapy. Nat. Rev. Cancer. 2005;5:867–875. doi: 10.1038/nrc1735. [DOI] [PubMed] [Google Scholar]
- 30.Karin M., Greten F.R. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nat. Rev. Immunol. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 31.Finnberg N., Stenius U., Hogberg J. Heterozygous p53-deficient (+/–) mice develop fewer p53-negative preneoplastic focal liver lesions in response to treatment with diethylnitrosamine than do wild-type (+/+) mice. Cancer Lett. 2004;207:149–155. doi: 10.1016/j.canlet.2003.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Gaidano G., et al. p53 mutations in human lymphoid malignancies: association with Burkitt lymphoma and chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. U. S. A. 1991;88:5413–5417. doi: 10.1073/pnas.88.12.5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Strasser A., Harris A.W., Bath M.L., Cory S. Novel primitive lymphoid tumours induced in transgenic mice by cooperation between myc and bcl-2. Nature. 1990;348:331–333. doi: 10.1038/348331a0. [DOI] [PubMed] [Google Scholar]
- 34.Hemann M.T., et al. Suppression of tumorigenesis by the p53 target PUMA. Proc. Natl. Acad. Sci. U. S. A. 2004;101:9333–9338. doi: 10.1073/pnas.0403286101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zornig M., Grzeschiczek A., Kowalski M.B., Hartmann K.U., Moroy T. Loss of Fas/Apo-1 receptor accelerates lymphomagenesis in E mu L-MYC transgenic mice but not in animals infected with MoMuLV. Oncogene. 1995;10:2397–2401. [PubMed] [Google Scholar]
- 36.Rubio-Moscardo F., et al. Characterization of 8p21.3 chromosomal deletions in B-cell lymphoma: TRAIL-R1 and TRAIL-R2 as candidate dosage-dependent tumor suppressor genes. Blood. 2005;106:3214–3222. doi: 10.1182/blood-2005-05-2013. [DOI] [PubMed] [Google Scholar]
- 37.Mouzakiti A., Packham G. Regulation of tumour necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in Burkitt’s lymphoma cell lines. Br. J. Haematol. 2003;122:61–69. doi: 10.1046/j.1365-2141.2003.04424.x. [DOI] [PubMed] [Google Scholar]
- 38.Kayagaki N., et al. Expression and function of TNF-related apoptosis-inducing ligand on murine activated NK cells. J. Immunol. 1999;163:1906–1913. [PubMed] [Google Scholar]
- 39.Robertson N.M., et al. TRAIL in the airways. Vitam. Horm. 2004;67:149–167. doi: 10.1016/S0083-6729(04)67009-9. [DOI] [PubMed] [Google Scholar]
- 40.Lamhamedi-Cherradi S.E., Zheng S., Tisch R.M., Chen Y.H. Critical roles of tumor necrosis factor-related apoptosis-inducing ligand in type 1 diabetes. Diabetes. 2003;52:2274–2278. doi: 10.2337/diabetes.52.9.2274. [DOI] [PubMed] [Google Scholar]
- 41.Lamhamedi-Cherradi S.E., Zheng S.J., Maguschak K.A., Peschon J., Chen Y.H. Defective thymocyte apoptosis and accelerated autoimmune diseases in TRAIL–/– mice. . Nat. Immunol. 2003;4:255–260. doi: 10.1038/ni894. [DOI] [PubMed] [Google Scholar]
- 42.Zheng S.J., Wang P., Tsabary G., Chen Y.H. Critical roles of TRAIL in hepatic cell death and hepatic inflammation. J. Clin. Invest. 2004;113:58–64. doi: 10.1172/JCI200419255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Janssen E.M., et al. CD4+ T-cell help controls CD8+ T-cell memory via TRAIL-mediated activation-induced cell death. Nature. 2005;434:88–93. doi: 10.1038/nature03337. [DOI] [PubMed] [Google Scholar]
- 44.Komarova E.A., et al. p53 is a suppressor of inflammatory response in mice. FASEB J. 2005;19:1030–1032. doi: 10.1096/fj.04-3213fje. [DOI] [PubMed] [Google Scholar]
- 45.Kuwano K., et al. P21Waf1/Cip1/Sdi1 and p53 expression in association with DNA strand breaks in idiopathic pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 1996;154:477–483. doi: 10.1164/ajrccm.154.2.8756825. [DOI] [PubMed] [Google Scholar]
- 46.Miller Y.E., et al. Induction of a high incidence of lung tumors in C57BL/6 mice with multiple ethyl carbamate injections. Cancer Lett. 2003;198:139–144. doi: 10.1016/s0304-3835(03)00309-4. [DOI] [PubMed] [Google Scholar]
- 47.Adams J.M., et al. The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318:533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 48.Corcoran L.M., Tawfilis S., Barlow L.J. Generation of B lymphoma cell lines from knockout mice by transformation in vivo with an Emu-myc transgene. J. Immunol. Methods. 1999;228:131–138. doi: 10.1016/s0022-1759(99)00094-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.