Abstract
TRAIL is a promising anticancer agent due to its ability to selectively induce apoptosis in established tumor cell lines but not nontransformed cells. Herein, we demonstrate a role for the apoptosis-inducing TRAIL receptor (TRAIL-R) as a metastasis suppressor. Although mouse models employing tumor transplantation have shown that TRAIL can reduce tumor growth, autochthonous tumor models have generated conflicting results with respect to the physiological role of the TRAIL system during tumorigenesis. We used a multistage model of squamous cell carcinoma to examine the role of TRAIL-R throughout all steps of tumor development. DMBA/TPA-treated TRAIL-R–deficient mice showed neither an increase in number or growth rate of benign papillomas nor an increase in the rate of progression to squamous cell carcinoma. However, metastasis to lymph nodes was significantly enhanced, indicating a role for TRAIL-R specifically in the suppression of metastasis. We also found that adherent TRAIL-R–expressing skin carcinoma cells were TRAIL resistant in vitro but were sensitized to TRAIL upon detachment by inactivation of the ERK signaling pathway. As detachment from the primary tumor is an obligatory step in metastasis, this provides a possible mechanism by which TRAIL-R could inhibit metastasis. Hence, treatment of cancer patients with agonists of the apoptosis-inducing receptors for TRAIL may prove useful in reducing the incidence of metastasis.
Introduction
Along with CD95 ligand (CD95L/FasL) and TNF, TNF-related apoptosis-inducing ligand (TRAIL) belongs to a subset of the TNF superfamily of cytokines known as “death ligands.” The members of this subfamily are capable of triggering the extrinsic apoptosis pathway by binding to their respective death domain–containing receptors. The intracellular death domain can recruit the adaptor molecule Fas-associated death domain protein (FADD), which upon ligand binding and activation of death-inducing signaling complex (DISC) binds to and activates caspases-8 and -10 and subsequently induces apoptosis (1). In human cells, TRAIL can induce apoptosis via 2 similar but distinct type I transmembrane receptors, TRAIL receptor 1 (TRAIL-R1/DR4) and TRAIL-R2 (KILLER/DR5/TRICK) (1). By contrast, there is only 1 apoptosis-inducing receptor for TRAIL in mice, TRAIL-R, which is equally related to human TRAIL-R1 and TRAIL-R2 (2). Due to their ability to selectively induce apoptosis in established tumor cell lines but not in normal cells in vitro and in vivo (3, 4), TRAIL and agonistic antibodies against the 2 apoptosis-inducing TRAIL receptors are promising anticancer therapeutics (5). Moreover, many studies have shown that TRAIL can have synergistic antitumor effects in combination with conventional DNA-damaging chemotherapeutics. These drugs usually activate the intrinsic mitochondrial apoptosis pathway, while TRAIL induces the extrinsic apoptosis pathway (6).
Tumor transplantation models in mice suggested that endogenous TRAIL expressed by NK cells plays an important role in tumor immune surveillance, as it can suppress tumor growth and experimental liver metastasis (7–11). However, in these studies TRAIL-sensitive cell lines were used, and it has now become clear that many primary tumor cells are TRAIL resistant (12–14). Furthermore, autochthonous tumor models using TRAIL- or TRAIL-R–deficient mice showed no increased susceptibility to epithelial tumorigenesis (15, 16), such as Her2/neu-initiated mammary tumors (16) or APCmin-induced intestinal tumors (15). In contrast, in nonepithelial tumors TRAIL can serve as a tumor suppressor, as TRAIL-deficient mice exhibit accelerated hematological malignancies when aged or in combination with p53 deficiency (16) and are more susceptible to methylcholanthrene-induced fibrosarcomas (9, 17). This difference might be due to genetic background, tumor-induction protocol, or a tissue-specific effect of the TRAIL/TRAIL-R pathway in tumor suppression. Of note, studies examining the role of the TRAIL system in epithelial tumorigenesis (15, 16) focused on early tumor stages but did not address later tumor stages or metastasis. Therefore, the physiological role of TRAIL and TRAIL-R in multistage tumorigenesis, particularly in the development of epithelial tumors, remains unclear. This is especially important because 90% of human cancers are of epithelial origin (18).
To examine the effect of TRAIL-R expression throughout the entire course of epithelial tumorigenesis, we applied a multistage model of skin cancer using TRAIL-R–deficient mice. The 7,12-dimethyl-benz-anthracene/12-O-tetradecanoylphorbol-13-acetate–induced (DMBA/TPA-induced) squamous cell carcinoma (SCC) model is a very well–characterized mouse model of epithelial neoplasia and mimics the multistage nature of human epithelial cancer development (19). The 2-stage chemical protocol involves treatment of mice with the carcinogen DMBA, followed by multiple applications of the tumor promoter TPA. This treatment induces benign squamous cell papillomas, nearly 100% of which have sustained a mutation in codon 61 of the Harvey ras oncogene (Ha-ras) (20), resulting in constitutive activation of Ras and of a number of signaling pathways (21). In most strains of mice, progression of benign papillomas to malignant SCCs is a rare and late event. These tumors break through the basement membrane and progressively invade the underlying dermis and subcutaneous tissues, infrequently metastasizing to regional and distant sites.
A unique advantage of this skin tumor model is that the different tumor stages — initiation of benign papilloma, tumor growth, progression to malignant carcinoma, and metastasis formation — can be readily assessed and quantified by visual inspection and histological analysis. Hence this carcinogen-induced tumor model is well suited to analyze the role of the TRAIL system in multistage tumorigenesis.
Results
Generation of TRAIL-R–deficient mice.
We initially generated mice conditionally deficient for the Trail-r gene by flanking exon 2 with loxP sites so that, upon expression of the Cre recombinase, a frame shift and an early stop codon was introduced (Figure 1A), resulting in a transcript devoid of TRAIL-binding cysteine-rich domains and an apoptosis-inducing death domain. Homologous recombination of the targeting construct in ES cells was tested by Southern blot analysis (Figure 1B). In order to generate constitutive TRAIL-R–deficient mice, conditional Trail-rflox/+ mice were crossed to mice expressing Cre recombinase in the germline. Constitutive germline deletion of exon 2 in offspring of these mice was verified by genomic PCR analysis of multiple tissues (Figure 1C). The absence of cell surface–expressed TRAIL-R was confirmed by staining of tumor cells derived from Trail-r–/– and Trail-r+/+ mice with an antibody specific for TRAIL-R and by flow cytometry analysis (Figure 2F). Constitutive Trail-r–/– mice were viable and did not show any reproductive or developmental defects (data not shown), consistent with previous reports of independently generated TRAIL-R–deficient mice (22, 23).
Figure 1. Generation of Trail-r–/– mice.
(A) Strategy to target exon 2 of the Trail-r gene. (i) Functional protein domains of TRAIL-R encoded by the Trail-r WT allele (ii). SP, signal peptide; CRD, cysteine-rich domain; TM, transmembrane domain; DD, death domain. Locations of the external (ext) 3′ and 5′ and the internal (int) probes and of genotyping primers mTR-a (a), mTR-b (b), and mTR-d (d) are indicated. (iii) Targeting construct. Open triangle, FRT sites; gray triangles, loxP sites; tkneo, selection cassette; DTA, diphtheria-toxin A-cassette. (iv) Null allele. (B) Southern blot analysis of targeted G418-selected ES cell clone after correct homologous recombination. (C) Constitutive deletion of exon 2 of Trail-r in different tissues. PCR analysis of genomic DNA of tail, lymph node, kidney, lung, heart, and brain of Trail-r–/–, Trail-r+/–, and Trail-r+/+ mice using primers a, b, and d is shown. The primers a and b amplified a 300-bp fragment of the Trail-r WT allele, and primers a and d amplified a 235-bp fragment of the Trail-r–null allele.
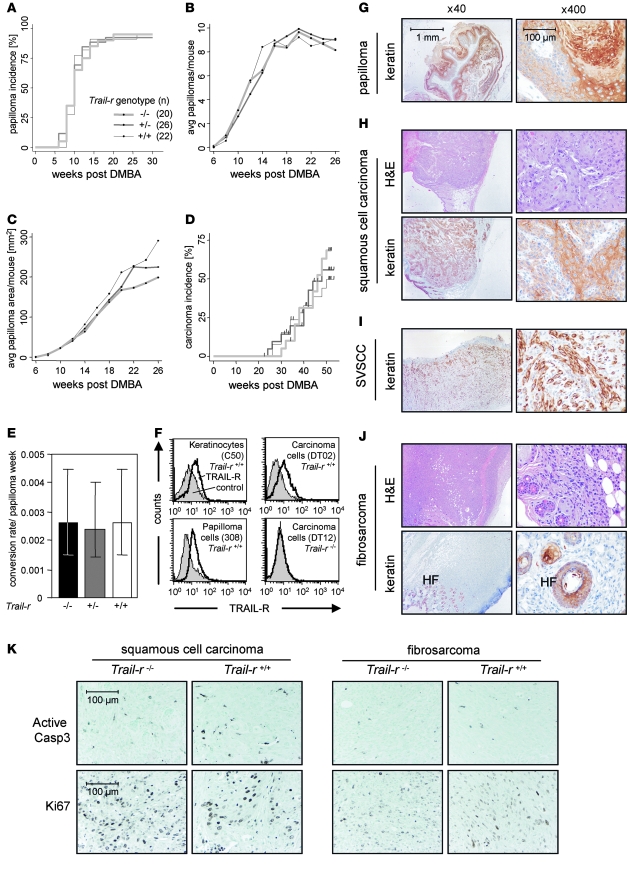
Figure 2. TRAIL-R does not influence primary skin tumorigenesis.
(A) Papilloma incidence rates. (B) Tumor initiation rates, as reflected in the average number of papillomas over time. (C) Tumor growth rates, represented by combined average papilloma area over time. (D) Carcinoma incidence rates. Vertical bars indicate deaths of mice with carcinomas. (E) Papilloma-to-carcinoma conversion rate per week of papilloma exposure. The 95% confidence intervals are displayed. (F) Cell surface–expressed TRAIL-R was examined using a TRAIL-R–specific antibody (MD5-1) on WT immortalized keratinocytes (cell line C-50), papilloma cells (cell line 308), and carcinoma cells (cell lines DT02 and DT12). Histological analysis (G–J) of tumor sections did not reveal any differences between the different Trail-r genotypes. Representative tissue sections (H&E staining and brown keratin staining) from Trail-r–/– mice are shown. (G) Keratin-positive papilloma. Among all genotypes, primary malignant tumors were (H) SCC, (I) spindle cell variant of SCC (SVSCC), or (J) fibrosarcomas, which is shown with keratin-positive hair follicles (HF). (K) Tissue sections of malignant carcinomas (fibrosarcoma or SCC) were stained with antibodies against active caspase-3 or Ki-67. Representative fields are shown (original magnification, ×400).
TRAIL-R deficiency does not enhance initiation, promotion, or malignant conversion of primary skin tumors.
To assess the role of TRAIL-R in multistage tumorigenesis, skin tumors were initiated in Trail-r–/–, Trail-r+/–, and Trail-r+/+ littermates by treatment with DMBA and TPA. We found that the number and rate of appearance (tumor incidence and initiation rate) of benign papillomas were nearly identical across the genotypes (Figure 2, A and B). Tumor growth rate also did not vary significantly (Figure 2C). Thus we concluded that TRAIL-R affects neither skin tumor initiation nor benign tumor growth. The next step in skin tumor development is conversion from benign papilloma to malignant carcinoma. The rates of appearance of carcinoma (Figure 2D) and papilloma-to-carcinoma conversion frequency were similar between the Trail-r genotypes (Figure 2E), indicating that TRAIL-R does not play a role in malignant invasion of DMBA/TPA-induced skin tumors. As determined by H&E analysis and keratin-specific staining, malignant tumors of all genotypes were either keratin-positive SCCs, spindle cell variants of SCCs, or keratin-negative fibrosarcomas (Figure 2, H–J). No apparent differences in morphology of the tumors were detected between the genotypes (data not shown). To determine whether TRAIL-R deficiency influenced apoptosis or proliferation of malignant carcinomas, tissue sections were stained with an antibody for active caspase-3 or Ki-67, respectively. However, no differences in apoptosis or proliferation were detected between primary carcinomas of Trail-r+/+ and Trail-r–/– mice (Figure 2K). To determine whether TRAIL-R expression was maintained throughout tumor development, we obtained immortalized keratinocytes and papilloma cell lines and established cell lines from skin carcinomas. While TRAIL-R was undetectable in Trail-r–/– cell lines, all Trail-r+/+ cell lines examined expressed TRAIL-R on the surface (Figure 2F and Supplemental Figure 1C; supplemental material available online with this article; doi:10.1172/JCI33061DS1). Thus although TRAIL-R is expressed in keratinocytes, benign papillomas, and malignant carcinomas, it does not appear to play a significant role in primary tumor development. Our finding is consistent with earlier studies in mice showing no role for TRAIL (16) or for TRAIL-R (15) in initiation of other epithelial tumors. This suggests that the TRAIL/TRAIL-R apoptosis system does not have a tumor suppressor function during early stages of primary tumorigenesis of many, if not all, epithelial tumors.
Increased lymph node metastasis in TRAIL-R–deficient mice.
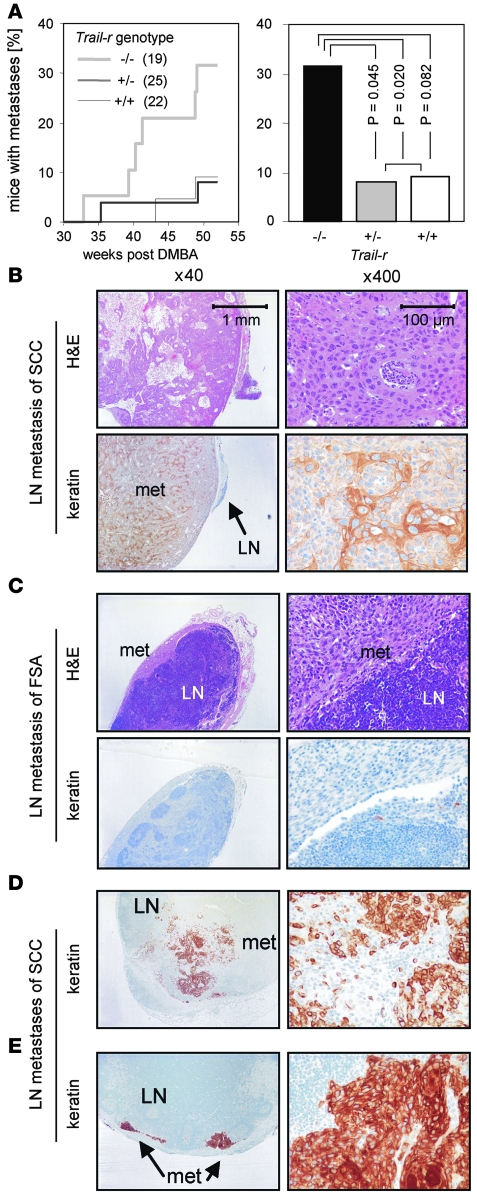
To investigate whether TRAIL-R expression influenced metastasis formation, tissue sections of enlarged inguinal and axial lymph nodes of carcinoma-bearing mice were stained with H&E and with a keratin-specific antibody (Figure 3, B–E). When TRAIL-R was absent, we detected metastasis formation earlier (Figure 3A) and at a significantly higher frequency: 32% of Trail-r–/– mice, in contrast to only 9% of Trail-r+/+ and 8% of Trail-r+/– mice, had developed lymph node metastases. This indicates that TRAIL-R plays a specific role in the suppression of metastasis without influencing the formation of primary tumors.
Figure 3. TRAIL-R suppresses lymph node metastases of skin carcinomas.
(A) Time course of detection of metastatic lymph nodes per total mice in study analyzed (left panel). Significantly higher incidence of lymph node metastases in TRAIL-R–deficient mice compared with TRAIL-R–expressing mice was detected upon necropsy (right panel), as determined by Pearson χ2 test. (B–E) Tissue sections were stained with H&E and a keratin-specific antibody. Lymph nodes of Trail-r–/– mice with keratin-positive metastatic lesions with SCC morphology (B, D, and E) or keratin-negative metastatic lesions (met) with fibrosarcoma (FSA) morphology (C). All stainings shown were from specimens obtained from tumors of Trail-r–/– mice.
Skin carcinoma cell lines are TRAIL-resistant but can be sensitized by proteasome inhibition.
To study the cellular mechanism of TRAIL-R–mediated metastasis suppression, we first compared the cell lines we had established from DMBA/TPA-induced skin carcinomas from Trail-r+/+ and Trail-r–/– mice. These cell lines displayed similar morphologies (Supplemental Figure 1A) and proliferated at similar rates in vitro (Supplemental Figure 1B), all Trail-r+/+ cell lines expressed comparable levels of TRAIL-R on the surface (Supplemental Figure 1C), and all tested cell lines (DT02, DT04, DT11, and DT12) contained the activating Ha-ras mutation in codon 61 (data not shown). To examine whether TRAIL-R expression results in increased migration, which could explain accelerated metastasis formation of skin carcinomas, we subjected Trail-r+/+, Trail-r–/–, and a WT cell line transfected with an siRNA knockdown construct for TRAIL-R in a wound-healing scratch assay. However, TRAIL-R expression did not impact cell migration (Supplemental Figure 1D).
We next examined sensitivity of skin carcinoma cells to TRAIL-induced cell death and found that all carcinoma cell lines were resistant to TRAIL-induced apoptosis when treated with 1 μg/ml TRAIL for 24 hours regardless of their Trail-r genotype (Figure 4A and Supplemental Figure 2A). This result is in line with the previously mentioned resistance of most early passage cells of primary tumors to TRAIL-induced apoptosis (12–14). As there was no difference in primary skin tumorigenesis between Trail-r+/+ and Trail-r–/– mice (Figure 2), a potential selection for TRAIL-resistant tumor cells was excluded. This indicated that TRAIL resistance of skin carcinoma cell lines was genuine. Recently, we and others have shown that the proteasome inhibitor bortezomib can sensitize tumor cells to TRAIL (12, 24, 25). Pretreatment with bortezomib sensitized most Trail-r+/+ carcinoma cells to TRAIL, while the Trail-r–/– cell lines remained resistant (Figure 4A and Supplemental Figure 2A). Thus although WT carcinoma cells were initially TRAIL resistant, the TRAIL-R apoptosis pathway was still functional in most of these cell lines.
Figure 4. Detachment sensitizes skin carcinoma cells specifically to TRAIL and CD95L.
(A) Subconfluent cells were incubated in the absence (control) or presence of TRAIL (iz-muTRAIL, 1 μg/ml) and/or bortezomib (20 nM) for 24 hours. Cell death was determined by PI staining and flow cytometric analysis. (B and C) Cells were trypsinized and cultured either on an uncoated (attached) or a poly-HEMA–coated surface (detached) in the absence or presence of TRAIL (B; 1 μg/ml) or at the indicated concentrations (C). (D) Subconfluent cells (DT02) were treated with or without 1 μM thapsigargin (Tgg), 30 μg/ml oxaliplatin (Oxp), 5 μg/ml cycloheximide (Chx), or 43°C heat shock (HS) or irradiated with 15 Gy (IR) and were incubated in the absence or presence of TRAIL (1 μg/ml) for 24 hours. (E) Attached (att) or detached (det) cells were incubated in the absence or presence of TRAIL (1 μg/ml) or CD95L (100 ng/ml) for 24 hours. Each data point represents one of the cell lines DT01–DT06 or DT11. The mean percentage of dead WT cells is indicated by a horizontal line. (F) Attached or detached cells were incubated in the absence or presence of TRAIL (1 μg/ml), CD95L (100 ng/ml), 5-FU (200 μg/ml), or etoposide (10 μM) for 24 hours. Standard deviation for duplicates or triplicates is shown. Results are representative of at least 2 independent experiments.
Detachment-induced sensitization of carcinoma cells.
TRAIL is known to be expressed on activated cells of the immune system, and T cell– and NK cell–derived TRAIL can play an important role in tumor immune surveillance (7, 8, 17, 26). For such TRAIL-induced tumor surveillance to be operative, there are 2 prerequisites: apart from expression of TRAIL on immune cells (27), tumor cells need to be TRAIL sensitive. Based on the increased susceptibility of Trail-r–/– mice to metastasis formation in our skin tumor model (Figure 3A), we hypothesized that TRAIL-resistant cells from the primary tumor may become TRAIL sensitive at some point during the metastatic process. An early obligatory step in metastasis formation is detachment from the primary tumor (28). As a consequence of detachment from the extracellular matrix, epithelial cells can undergo a form of cell death known as “anoikis” (28). In vitro, attachment of adherent cells to the cell culture dish can be prevented by coating cell culture dishes with poly[2-hydroxyethyl methacrylate] (poly-HEMA). To study whether detachment resulted in TRAIL-R–dependent apoptosis, we cultured Trail-r+/+ and Trail-r–/– carcinoma cells on poly-HEMA–coated plates. Irrespective of their Trail-r genotype, a similar percentage of cells of the different cell lines underwent anoikis (Figure 4B). This data indicates that TRAIL-R expression is not required for anoikis of skin carcinoma cells. Interestingly however, exogenous addition of TRAIL resulted in a dose-dependent increase in death of most of the detached Trail-r+/+ carcinoma cell lines tested, while detached Trail-r–/– cells remained TRAIL resistant (Figure 4, B, C, and E). Of note, the WT cell line DT06, which was not sensitized to TRAIL by bortezomib, also remained TRAIL resistant upon detachment. Thus loss of attachment resulted in sensitization to TRAIL/TRAIL-R–mediated apoptosis of most, but not all, previously TRAIL-resistant Trail-r+/+ skin carcinoma cell lines.
In order to examine whether detachment is a specific event to sensitize skin carcinoma cells to TRAIL-induced death, we subjected Trail-r+/+ skin carcinoma cells to various cellular stresses (Figure 4D). However, treatment of cells with heat shock, endoplasmic reticulum stress induced by thapsigargin, ionizing irradiation, and the DNA crosslinker oxaliplatin were unable to sensitize skin carcinoma cell lines to TRAIL. Only the well-known TRAIL-sensitizing translation-inhibiting agent cycloheximide, which we used as a control, sensitized the cells to TRAIL. This indicates that detachment is a specific event that renders these cells sensitive to TRAIL-induced apoptosis.
We next asked whether detachment-induced sensitization of skin carcinoma cells is specific to TRAIL or whether it can also sensitize these cells to other external death stimuli. We therefore treated the cells with CD95L or with the DNA-damaging agents 5-fluorouracil (5-FU) and etoposide, which activate the intrinsic apoptosis pathway. Interestingly, detached skin carcinoma cells were also sensitized to CD95L, including the TRAIL-R–deficient cell line DT11 (Figure 4, E and F), in a dose-dependent manner (Supplemental Figure 2B). In contrast, detachment did not increase or only marginally increased sensitivity to 5-FU and etoposide as compared with TRAIL or CD95L (Figure 4F). This result indicates that detachment sensitizes most skin carcinoma cell lines to inducers of the extrinsic but not intrinsic apoptosis pathway.
Detachment-induced inactivation of ERK sensitizes skin carcinoma cells to TRAIL-induced apoptosis.
We next investigated the molecular mechanism of detachment-induced TRAIL sensitization. To determine whether increased TRAIL and CD95L sensitivity was due to upregulation of the respective receptors, we compared death receptor expression of detached skin carcinoma cells with that of attached skin carcinoma cells (Figure 5A). Cell surface expression of TRAIL-R and CD95 remained unchanged upon detachment, suggesting that intracellular signaling events are responsible for detachment-induced sensitization to death ligands.
Figure 5. Detachment-induced inactivation of ERK sensitizes to TRAIL- and CD95L-induced apoptosis.
(A) Cells were incubated attached or detached for 12 hours. Cell surface–expressed TRAIL-R and CD95 were detected on the indicated skin carcinoma cell lines. Representative flow cytometry profiles of at least 2 independent experiments are shown. (B) DT02 cells and (C) DT01 cells were trypsinized (0 h) and incubated either detached or attached for the indicated times. Western blots were incubated with the indicated antibodies. In B, note that c-FLIPL, but not c-FLIPS, was detected. (D) DT02 cells were trypsinized and seeded into noncoated 24-well plates either with or without U0126 (40 μM) and TRAIL or CD95L at the indicated concentrations for 24 hours. Cell death was determined as described in Figure 4. Standard deviation for duplicates is shown. Results are representative of 3 independent experiments.
In many cells sensitivity to death ligands is regulated by the antiapoptotic caspase-8 modulator cellular FLICE-inhibitory protein (c-FLIP), which is expressed as a long (c-FLIPL) and a short (c-FLIPS) isoform. Therefore, we examined whether detachment could downregulate c-FLIP, which may explain the sensitization to TRAIL and CD95L (1). Whereas c-FLIPS was undetectable in the cells, c-FLIPL was increased when the cells were detached compared with attached TRAIL-resistant cells (Figure 5B). This indicates that c-FLIP cannot be responsible for detachment-induced sensitization to TRAIL.
We hypothesized that detachment of skin carcinoma cells could lead to inactivation of survival signaling pathways, thereby contributing to death ligand–mediated apoptosis. The molecular signature event in DMBA/TPA-induced skin tumors is constitutive activation of Ha-Ras, leading to activation of the Raf/MEK/ERK and the PI3K/AKT survival pathways in late papillomas and carcinomas, resulting in high ERK and AKT kinase activity (29). In epithelial cells, activation of the ERK and AKT survival pathways is often induced by attachment to the extracellular matrix via integrins, while upon detachment these kinases can become inactivated (30). To test whether inactivation of the ERK or AKT survival pathways could be responsible for detachment-induced sensitization to TRAIL in skin carcinoma cells, we examined phosphorylation of AKT and ERK in attached versus detached skin carcinoma cell lines, which reflects their activation status. Interestingly, phosphorylation of ERK was substantially decreased in detached DT01, DT02, and DT11 cells as compared with the respective attached cells (Figure 5C and data not shown). In contrast, AKT phosphorylation was even slightly increased in detached cells. These data show that inactivation of the ERK pathway but not the AKT pathway correlates with detachment-induced death ligand sensitization. To test whether specific inactivation of ERK is sufficient for sensitization of attached skin carcinoma cells to TRAIL- and CD95L-induced apoptosis, we treated skin carcinoma cells with the MEK1/2 inhibitor U0126. U0126 treatment resulted in complete ERK dephosphorylation but had no effect on the AKT phosphorylation status (Supplemental Figure 3A and data not shown), indicating specificity of U0126 for ERK in skin carcinoma cells. Treatment of attached cells with U0126 resulted in substantial sensitization to both TRAIL- and CD95L-induced cell death dependent on the dose of death ligand (Figure 5D) and U0126 (Supplemental Figure 3C). In contrast, inactivation of the AKT pathway with the PI3K inhibitor LY294002 did not sensitize skin carcinoma cells to TRAIL (Supplemental Figure 3, B and C). Together with the observed downregulation of ERK activity in detached cells, our data suggest that inactivation of the ERK survival pathway is responsible for detachment-induced sensitization of skin carcinoma cells to TRAIL and CD95L.
Discussion
TRAIL is an attractive candidate for tumor therapy due to its ability to selectively induce apoptosis in tumor cells but not in normal cells. However, the physiological role of the apoptosis-inducing TRAIL system in multistage tumorigenesis has not clearly been defined yet. Here we provide the first demonstration that TRAIL-R can selectively suppress metastasis of epithelial cancers without affecting growth of primary tumors.
Metastasis is infrequently observed in autochthonous mouse tumor models, which might be a reason why the role of the TRAIL system during metastasis formation of primary tumors has not yet been reported. Metastasis is often modeled by transplantation of tumor cells and is termed “experimental metastasis.” However, experimental metastasis models do not recapitulate several early steps necessary for metastasis formation, such as invasion, detachment from the primary tumor, and migration (31). Using TRAIL-sensitive cell lines, earlier studies had revealed that endogenous TRAIL could suppress the formation of multiple tumor foci in the liver (7–10), suggesting that TRAIL can suppress such experimental liver metastasis. However, upon local subcutaneous or intramammary fat pad injection of the same TRAIL-sensitive tumor cell lines, TRAIL was also shown to act as a tumor suppressor (9, 11). Hence these transplantation models were not able to differentiate between a tumor suppressor and a metastasis-specific suppressor function of the TRAIL system.
Here, using the autochthonous DMBA/TPA-induced skin carcinoma model, we monitored and analyzed all steps of primary tumor development up to metastasis formation. We detected that TRAIL-R deficiency leads to increased lymph node metastasis (Figure 3A), which had derived from primary epithelial skin tumors whose previous development in the very same mice had not been affected by absence of TRAIL-R (Figure 2). Primary tumor formation and metastasis are clearly distinct processes; genes that specifically regulate metastasis without affecting primary tumorigenesis are termed “metastasis suppressors” (32). Therefore, we provide here what we believe to be the first demonstration that Trail-r is a metastasis suppressor gene.
It is tempting to speculate that the TRAIL/TRAIL-R pathway may also specifically regulate metastasis formation in human tumors. If this proves to be the case, the presence of 2 death receptors for TRAIL in humans as compared with only 1 in mice might serve as an additional fail-safe mechanism to prevent metastasis. Indeed, there is evidence for selection pressure on inactivating apoptosis-inducing TRAIL receptors and increased TRAIL resistance specifically in established metastases of human epithelial tumors; for example, compared with primary tumors, increased mutation frequencies in TRAIL-R1– and TRAIL-R2–encoding genes and lower expression levels of TRAIL-R1 and TRAIL-R2 have been identified in metastases of mammary tumors (33) and melanomas (34), respectively. Furthermore, tumor cell lines derived from metastatic lymph nodes displayed a greater TRAIL resistance than cell lines derived from the respective primary oral tumors (35).
Previous studies have shown that not all tumors are TRAIL sensitive. About 50% of all examined human tumor cell lines were TRAIL resistant (4), as well as the majority of early passage tumor cells obtained from human neoplasias (12–14). We consistently found that cells isolated from benign papillomas as well as malignant carcinomas were also TRAIL resistant (Figure 4A and data not shown). How could the apoptosis-inducing TRAIL-R inhibit metastasis formation when no effect of TRAIL-R deficiency was seen on the primary tumors these metastases were derived from? We discovered that detachment of skin carcinoma cells from a solid surface sensitized them to TRAIL in a TRAIL-R–dependent manner (Figure 4, B and C). This is in line with a study in the human TRAIL-sensitive MCF-7 mammary carcinoma cell line, in which detachment resulted in further sensitization to TRAIL-induced apoptosis (36). TRAIL is known to be expressed on activated immune cells and to mediate tumor immune surveillance on TRAIL-sensitive tumor cells (7, 8, 17, 26). Detachment-induced TRAIL sensitization explains how TRAIL-mediated immune surveillance can specifically suppress metastases even if the primary tumors are TRAIL resistant.
Our finding that detachment also sensitized skin carcinoma cells to CD95L (Figure 4E) but not to stimuli of the intrinsic apoptosis pathway (Figure 4F) reveals a specific regulatory effect of loss of adhesion on the extrinsic apoptosis pathway. These data suggest that death receptor–mediated apoptosis may play a role in inhibiting metastases. In fact, besides TRAIL, CD95L can also be expressed on activated immune cells (37). In a mouse model of subcutaneously injected melanoma cells, CD95L was shown to specifically suppress spontaneous lung metastases without affecting tumor growth at the primary injection site (38). It will be interesting to test whether mice deficient for both TRAIL- and CD95L-induced apoptosis may present with a more striking phenotype with respect to metastasis formation in autochthonous tumor models. In this regard it is important to note that caspase-8, a common component of TRAIL-R and CD95 apoptosis pathways, is frequently deleted or inactivated in malignant disseminated childhood neuroblastoma (39–41). Moreover, caspase-8 has recently been identified as a metastasis suppressor in an experimental neuroblastoma metastasis model (42). It is possible that the metastasis suppressor function of caspase-8 is at least in part due to its function as a downstream mediator of death receptor–mediated apoptosis.
While detachment-induced TRAIL sensitization of early metastasizing skin carcinoma cells is a plausible cellular mechanism for inhibition of lymph node metastases in WT mice, it is also possible that other factors contribute to increased lymph node metastases in TRAIL-R–deficient mice. For example, disseminated tumor cells could be sensitized to TRAIL at the lymph node by certain cytokines (43). One might also speculate that the lack of TRAIL-R in nontumor tissue, for example in immune cells or endothelial cells, might contribute to increased lymph node metastasis, although we did not observe obvious differences in vascularization, inflammation, or macrophage distribution or unusual patterns of lymphocyte activation between the genotypes in the primary carcinomas or lymph nodes (data not shown). At this point, we also cannot formally rule out that absence of TRAIL-R could increase metastasis by altering homing properties of metastatic cancer cells independent of the apoptosis-inducing potential of TRAIL-R.
It will also be interesting to test which cells contribute to elimination of TRAIL-sensitized metastasizing tumor cells. Tumor models using mice conditionally deficient for TRAIL in certain immune cell populations such as NK cells, macrophages, or T and B lymphocytes could help to solve this question. However, TRAIL is a rather promiscuous ligand, which besides binding to TRAIL-R also binds to 3 other, non–apoptosis-inducing receptors in mice, namely the soluble osteoprotegerin (mOPG), or the membrane-bound receptors mTNFRH1 and -2 (44). The physiological relevance of their interactions with TRAIL is largely unknown, and deletion of TRAIL might lead to disruption of more pathways than disruption of TRAIL-R, for which TRAIL is the only known ligand.
With respect to the molecular mechanism responsible for death receptor sensitization by detachment, upregulation of TRAIL-R (Figure 5A), downregulation of FLIP (Figure 5B), and inactivation of the PI3K/AKT survival pathway did not correlate with detachment of skin carcinoma cells (Figure 5C). However, inactivation of the Raf/MEK/ERK prosurvival pathway did not only correlate with detachment, but also its targeted inactivation was sufficient for detachment-induced TRAIL and CD95L sensitization (Figure 5, C and D). Therefore, in skin carcinoma cells, detachment-induced TRAIL sensitization is likely to be achieved by detachment-induced inactivation of the ERK survival pathway. These data are in contrast to a recently published study that suggested that detachment of ovarian carcinoma cells leads to a PI3K/AKT-dependent sensitization to TRAIL, while no role of the MEK/ERK pathway was found (45). However, the authors excluded a role for the MEK/ERK pathway by using PD98059, a known low-affinity MEK inhibitor, which did not sensitize their cells to TRAIL. Of note, in contrast to the high-affinity MEK1/2 inhibitor U0126, PD98059 was also not able to sensitize attached cells to TRAIL in our skin carcinoma cell lines (Supplemental Figure 3C). In another study, sensitization of endothelial cells to CD95L by detachment was regulated by CD95 receptor upregulation and inactivation of the MEK/ERK pathway, leading to downregulation of FLIP expression (46). These data suggest that, depending on the cell type studied, detachment-induced sensitization to death receptor–mediated apoptosis may be regulated by different mechanisms.
Approximately 90% of human cancer deaths are caused by metastatic spread from primary tumors (28). Therefore, prevention of metastasis formation is an eminent goal in cancer research. Compared with canonical tumor suppressors, there is less selection pressure on metastasis suppressors or their signaling pathways during primary tumor formation, leaving metastasis suppressor pathways intact. In principle, by enhancing the function of metastasis suppressors, specific targeting of metastases could be achieved. Yet most metastasis suppressor genes identified thus far encode intracellular proteins (32), making them difficult to address therapeutically. In contrast, TRAIL receptors are expressed on the cell surface, and several TRAIL receptor agonists are currently in clinical trials (5). The application of TRAIL receptor agonists, either alone or in combination with TRAIL-sensitizing drugs, e.g., proteasome inhibitors or conventional chemotherapeutics, may prove particularly useful in targeting early-disseminating tumor cells in adjuvant cancer therapy.
Methods
Generation of Trail-r–/– mice.
A genomic BAC clone containing 10 kb of the Trail-r (Tnfrsf10b) gene (Figure 1A) was isolated from a genomic phage library derived from male ES cells of the mouse strain 129/Sv (Mobitec). The targeting construct for generation of conditional and constitutive Trail-r–/– mice (Figure 1A) was cloned by 2 rounds of ET cloning (RecE and RecT recombination) in E. coli, as previously described (47). The predicted polypeptide encoded by the null allele contains only the signal peptide, but did not encode the pre–ligand-binding assembly domain, the TRAIL-binding cysteine-rich domains, or the death domain (Figure 1A). The construct consisted of a 4.5-kb 5′ homology arm and a 3.1-kb 3′ homology arm, a tkneo selection cassette for positive selection by G418, and a diphtheria–toxin A (DTA) selection cassette for negative selection during homologous recombination at the 3′ end. Two loxP sites (recognized by the Cre recombinase) flanked a sequence containing exon 2 and the tkneo cassette, while the tkneo cassette alone was additionally flanked by 2 FRT sites (recognized by the Flp recombinase) (Figure 1A).
ES cells of the 129/Ola strain were transfected with the linearized targeting construct (Figure 1A). We isolated G418-resistant ES cell clones and confirmed homologous recombination by restriction digest of genomic DNA followed by Southern blot analysis using external 3′ and 5′ probes (3′-ext and 5′-ext) and an internal probe (Figure 1, A and B). In a correctly recombined ES cell clone, the tkneo selection cassette was excised by transfection with a eukaryotic expression vector encoding Flp recombinase (pCAGGS-FLP-e) (48) followed by negative selection with gancyclovir, which is toxic for cells expressing thymidine kinase. Correct excision of the selection cassette in ES cells was confirmed by PCR and Southern blot analysis (data not shown). ES cells containing the conditional Trail-rflox allele were injected into blastocysts, and chimeric mice were tested for germline transmission by PCR (data not shown). To generate the Trail-rnullallele, heterozygous conditional Trail-rflox/+ animals were crossbred to mice expressing the Cre recombinase under the control of the Nestin promoter, which is not only expressed in the brain but occasionally also in the germline (our unpublished observations). Constitutive germline deletion of exon 2 of Trail-r was determined by PCR genotyping analysis of genomic DNA of tail, lymph node, kidney, lung, heart, and brain (Figure 1C). RT-PCR analysis confirmed excision of exon 2 at the mRNA level (data not shown). Backcrossed mice were selected for loss of Cre recombinase.
DMBA/TPA-induced skin carcinogenesis.
Mice harboring the Trail-r–null allele were backcrossed for 2 generations from the Balb/c to the NIH/Ola background (Harlan Olac), which is susceptible to DMBA/TPA-induced skin carcinogenesis (49). Then, Trail-r+/– mice were intercrossed to generate the experimental cohort. The backs of 22 Trail-r+/+, 26 Trail-r+/–, and 20 Trail-r–/– 8-week-old littermates were shaved and topically treated with a single dose of DMBA (25 μg in 200 μl of acetone) and biweekly doses of TPA (200 μl of a 10–4 M solution in acetone) for 15 weeks, as previously described (49). The number and size range of tumors was recorded every other week by visual inspection. To determine papilloma area to assess tumor growth, we used the following formula: area [mm2] = A × (3/2)2À + B × (5/2)2À + C × (7/2)2À + D × (9/2)2À + E × (F/2)2À, where A is the number of tumors with a 2- to 4-mm diameter, B is the number of tumors with a 4- to 6-mm diameter, C is the number of tumors with a 6- to 8-mm diameter, D is the number of tumors with an 8- to 10-mm diameter, E is the number of tumors with a diameter greater than 10 mm, and F is the average of all tumors with diameters of 10 mm and greater. Papillomas were characterized by folded epidermal hyperplasia protruding from the skin surface, while carcinomas were usually endophytic tumors presenting as plaques with an ulcerated surface. Carcinomas break through the basement membrane and progressively invade the underlying dermis and subcutaneous tissues. The malignancy of all visually detected carcinomas was verified by histology upon necropsy. Mice were sacrificed if moribund because of excessive tumor load or 52 weeks after DMBA treatment. Tumors and lymph nodes were fixed in formalin. Lymph node metastases were assessed by histological analysis of all enlarged (≥ 2 mm) inguinal and axial lymph nodes of mice. All metastasis-containing lymph nodes were clearly enlarged (≥ 5 mm). A mouse was considered positive for metastasis when a coherent mass of cells of squamous or sarcomatoid morphology in at least 1 lymph node (as shown in Figure 3, B–E) was identified by 2 investigators. Care of experimental animals was in accordance with institutional guidelines. All animal experiments were approved by the IACUC of the Fred Hutchinson Cancer Research Center.
Cell lines.
Carcinoma cell lines were grown from malignant tumor explants obtained at necropsy. Tumors were excised, rinsed with 70% ethanol, cut into pieces, and incubated in culture medium (DMEM, 10% FCS, penicillin, and streptomycin) on cell culture dishes. The DMBA/TPA-induced carcinoma cell lines DT01, DT02, DT03, DT04, DT05, and DT06 were generated from malignant tumors of Trail-r+/+ mice, DT11 and DT12 from Trail-r–/– mice. The immortalized keratinocyte cell line C-50 and the papilloma cell line 308 were a kind gift from Susan Fischer (MD Anderson Cancer Research Center, Smithville, Texas, USA).
Detachment and apoptosis assays.
In order to prevent attachment of cells, freshly trypsinized cells were seeded into cell culture dishes that were coated with poly-HEMA according to the manufacturer’s instructions. Directly after seeding into coated 24-well plates in a volume of 2 ml, respectively, 500,000 cells/well were treated with isoleucine zipper murine TRAIL (iz-muTRAIL) and/or other agents at the indicated concentration for 24 hours. For attachment, cells were seeded into uncoated plates so that cells were at all times subconfluent. Heat shock was performed as described in ref. 50. Briefly, 37°C media was exchanged with 43°C media for 1 hour, followed by 24 hours of incubation at 37°C with or without TRAIL. Cell death was determined by loss of plasma membrane integrity as assessed by uptake of the DNA-intercalating dye propidium iodide (PI; 1 μg/ml) and flow cytometric analysis.
Western blot analysis.
Cells (500,000) were seeded in an uncoated 6-well dish when grown attached, or in a poly-HEMA–coated 24-well dish when grown detached. Attached cells were lysed in the dish, and suspended detached cells were collected and lysed in lysis buffer (30 mM Tris, 150 mM NaCl, 10% glycerol, 1% Triton X-100, 0.5% Igepal, supplemented with complete mini-protease inhibitor cocktail [Roche], 1 μM sodium fluoride, 1 μM sodium vanadate). Lysates were separated by standard SDS-polyacrylamide gel electrophoresis (12%), blotted onto PVDF membranes, and incubated with the indicated antibodies.
Reagents.
DMBA and TPA were from Sigma-Aldrich. Cell surface–expressed TRAIL-R was detected by hamster anti-mouse TRAIL-R antibody MD5-1 (eBioscience) or by mouse anti-mouse TRAIL-R antibody (mTR21), which was generated in Trail-r–/– mice immunized with an Ig-fusion protein containing the extracellular portion of TRAIL-R (mTRAIL-R-Fc). Anti-mCD95 (Jo2) was from Pharmingen. Anti–active caspase-3 and anti–Ki-67 antibodies were from Abcam. Cytokeratin was detected with polyclonal rabbit anti-keratin antibody (Cytokeratin WSS; Envision Plus). poly-HEMA and PI were from Sigma-Aldrich. Bortezomib was from Millennium Pharmaceuticals. iz-muTRAIL consisted of a synthetic isoleucine zipper motif fused to the extracellular domain of murine TRAIL (aa 99–291). Fc-CD95L was an Ig-fusion protein containing the extracellular portion of human CD95L. Cloning, production, and purification of iz-muTRAIL and Fc-CD95L was essentially done as described in ref. 51 and ref. 52, respectively. For western blot analysis, the following antibodies were used: anti–phospho-AKT (S473; catalog no. 9271), anti–phospho-ERK (catalog no. 9101), and anti-AKT (catalog no. 9272) from Cell Signaling and anti-ERK1 (C-16) and anti-actin (I-19) from Santa Cruz Biotechnology Inc. Anti-FLIP (Dave-2) was from Axxora.
Oligonucleotides.
For PCR genotyping of Trail-r, the following oligonucleotides were used: mTR-a, GAGCCATCTTTTTAAGTCTTGACTG; mTR-b, GACGATTATGGGCTGGGTTAGCTG; mTR-d, CGAACACAGCTGGTTTCCATGG.
Statistics.
Kaplan-Meier estimates were used to display papilloma (Figure 2A) and carcinoma (Figure 2D) incidence rates. Cox proportional hazard regression models were used to test for corresponding genotype effects, employing a single ordered genotype variable to test for gene dose effect and an indicator variable for 2 group comparisons. In Figure 2A, papilloma incidence rates are shown, with P = 0.96 across genotypes (trend test for genotype effect). In Figure 2D, carcinoma incidence rates are shown, with risk ratio (RR) = 1.3 (95% confidence interval, 0.62–2.5), P = 0.52 for Trail-r–/– versus Trail-r+/– and Trail- r+/+, and P = 0.69 across genotypes (trend test). In order to assess the effect of Trail-r genotype on tumor initiation (Figure 2B) and growth (Figure 2C), longitudinal observations of papilloma number and size, respectively, were analyzed using generalized estimating equations (GEEs) (53). The GEE regression approach accounted for the within-mouse correlation of tumor outcome measures over time, specified with a first-order autoregressive correlation structure. A test for a gene-dose effect across genotypes was accomplished by modeling genotype as a single-ordered covariate. Analyses of papilloma initiation and growth were restricted to observations during the first 26 weeks after DMBA treatment. In Figure 2B, tumor initiation rates are shown, with P = 0.83 (trend test). In Figure 2C, tumor growth rates are shown, with P = 0.37 (trend test) for growth rates between genotypes. Papilloma-to-carcinoma conversion rates (Figure 2E) were estimated in the context of Poisson regression models of carcinoma incidence on papilloma-time exposure. The number of papilloma-weeks exposure contributed by each animal was calculated from biweekly observations of tumor number. For the data shown in Figure 2E, P = 0.89 for Trail-r–/– versus Trail-r+/– and Trail-r+/+, and P = 0.99 across genotypes (trend test). For determination of percentage of mice with metastasis upon termination of the study at 52 weeks after DMBA treatment (Figure 3A), P = 0.020 for Trail-r–/– versus Trail-r+/+ and Trail-r+/–, P = 0.082 for Trail-r–/– versus Trail-r+/+, and P = 0.045 for Trail-r–/– mice versus Trail-r+/– mice (Pearson χ2). Group denominators included all mice in the study except for those with carcinoma whose lymph nodes could not be analyzed (25 Trail-r+/–, 19 Trail-r–/–, and 22 Trail-r+/+ mice were analyzed). The observed follow-up times for mice were similar across genotypes. All P values were based on 2-sided tests.
Supplementary Material
Acknowledgments
We thank Heidrun Kern, Sandra Fehsenfeld, and Claudia Rappl for excellent technical assistance, Johannes F. Coy for help in the isolation of the genomic Trail-r clone, Sue Knoblaugh and Denny Liggit for expert advice on histology analysis, Kay E. Gurley and Julie Randolph-Habecker for immunohistochemistry, Tobias Haas for generating the Fc-CD95L construct, and our colleagues at the FHCRC and DKFZ (especially Peter H. Krammer, Tim Wintermantel, Tom M. Ganten, Martin R. Sprick, Kay E. Gurley, Karen Kelly-Spratt, and Russell Moser) for helpful discussions during preparation of this work and manuscript. This work was supported by NIH research grants R01 CA070414 and R01 CA099517 to C.J. Kemp, a research fellowship from the DFG to A. Grosse-Wilde, and grants to H. Walczak from the BioFuture Program of the German Ministry of Education and Research and the Deutsche Krebshilfe.
Footnotes
Nonstandard abbreviations used: DMBA, 7,12-dimethyl-benz-anthracene; 5-FU, 5-fluorouracil; PI, propidium iodide; poly-HEMA, poly[2-hydroxyethyl methacrylate]; SCC, squamous cell carcinoma; TPA, 12-O-tetradecanoylphorbol-13-acetate; TRAIL, TNF-related apoptosis-inducing ligand; TRAIL-R, TRAIL receptor.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:100–110 (2008). doi:10.1172/JCI33061
Christopher J. Kemp and Henning Walczak contributed equally to this work.
See the related Commentary beginning on page 111.
References
- 1.Falschlehner C., Emmerich C.H., Gerlach B., Walczak H. TRAIL signalling: Decisions between life and death. Int. J. Biochem. Cell Biol. 2007;39:1462–1475. doi: 10.1016/j.biocel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 2.Wu G.S., Burns T.F., Zhan Y., Alnemri E.S., El-Deiry W.S. Molecular cloning and functional analysis of the mouse homologue of the KILLER/DR5 tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor. Cancer Res. 1999;59:2770–2775. [PubMed] [Google Scholar]
- 3.Ashkenazi A., et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J. Clin. Invest. 1999;104:155–162. doi: 10.1172/JCI6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walczak H., et al. Tumoricidal activity of tumor necrosis factor-related apoptosis- inducing ligand in vivo. Nat. Med. 1999;5:157–163. doi: 10.1038/5517. [DOI] [PubMed] [Google Scholar]
- 5.Koschny R., Walczak H., Ganten T.M. The promise of TRAIL-potential and risks of a novel anticancer therapy. J. Mol. Med. 2007;85:923–935. doi: 10.1007/s00109-007-0194-1. [DOI] [PubMed] [Google Scholar]
- 6.Yagita H., Takeda K., Hayakawa Y., Smyth M.J., Okumura K. TRAIL and its receptors as targets for cancer therapy. Cancer Sci. 2004;95:777–783. doi: 10.1111/j.1349-7006.2004.tb02181.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smyth M.J., et al. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) contributes to interferon gamma-dependent natural killer cell protection from tumor metastasis. J. Exp. Med. 2001;193:661–670. doi: 10.1084/jem.193.6.661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takeda K., et al. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in surveillance of tumor metastasis by liver natural killer cells. Nat. Med. 2001;7:94–100. doi: 10.1038/83416. [DOI] [PubMed] [Google Scholar]
- 9.Cretney E., et al. Increased susceptibility to tumor initiation and metastasis in TNF-related apoptosis-inducing ligand-deficient mice. J. Immunol. 2002;168:1356–1361. doi: 10.4049/jimmunol.168.3.1356. [DOI] [PubMed] [Google Scholar]
- 10.Sedger L.M., et al. Characterization of the in vivo function of TNF-alpha-related apoptosis-inducing ligand, TRAIL/Apo2L, using TRAIL/Apo2L gene-deficient mice. Eur. J. Immunol. 2002;32:2246–2254. doi: 10.1002/1521-4141(200208)32:8<2246::AID-IMMU2246>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 11.Takeda K., et al. Involvement of tumor necrosis factor-related apoptosis-inducing ligand in NK cell-mediated and IFN-gamma-dependent suppression of subcutaneous tumor growth. Cell. Immunol. 2001;214:194–200. doi: 10.1006/cimm.2001.1896. [DOI] [PubMed] [Google Scholar]
- 12.Koschny R., et al. Bortezomib sensitizes primary human astrocytoma cells of WHO grades I to IV for tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Clin. Cancer Res. 2007;13:3403–3412. doi: 10.1158/1078-0432.CCR-07-0251. [DOI] [PubMed] [Google Scholar]
- 13.Nguyen T., Zhang X.D., Hersey P. Relative resistance of fresh isolates of melanoma to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis. Clin. Cancer Res. 2001;7:966s–973s. [PubMed] [Google Scholar]
- 14.Eramo A., et al. Inhibition of DNA methylation sensitizes glioblastoma for tumor necrosis factor-related apoptosis-inducing ligand-mediated destruction. Cancer Res. 2005;65:11469–11477. doi: 10.1158/0008-5472.CAN-05-1724. [DOI] [PubMed] [Google Scholar]
- 15.Yue H.H., Diehl G.E., Winoto A. Loss of TRAIL-R does not affect thymic or intestinal tumor development in p53 and adenomatous polyposis coli mutant mice. Cell Death Differ. 2005;12:94–97. doi: 10.1038/sj.cdd.4401523. [DOI] [PubMed] [Google Scholar]
- 16.Zerafa N., et al. Cutting edge: TRAIL deficiency accelerates hematological malignancies. J. Immunol. 2005;175:5586–5590. doi: 10.4049/jimmunol.175.9.5586. [DOI] [PubMed] [Google Scholar]
- 17.Takeda K., et al. Critical role for tumor necrosis factor-related apoptosis-inducing ligand in immune surveillance against tumor development. J. Exp. Med. 2002;195:161–169. doi: 10.1084/jem.20011171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller S.J., Lavker R.M., Sun T.T. Interpreting epithelial cancer biology in the context of stem cells: tumor properties and therapeutic implications. Biochim. Biophys. Acta. 2005;1756:25–52. doi: 10.1016/j.bbcan.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 19.Kemp C.J. Multistep skin cancer in mice as a model to study the evolution of cancer cells. Semin. Cancer Biol. 2005;15:460–473. doi: 10.1016/j.semcancer.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 20.Quintanilla M., Brown K., Ramsden M., Balmain A. Carcinogen-specific mutation and amplification of Ha-ras during mouse skin carcinogenesis. Nature. 1986;322:78–80. doi: 10.1038/322078a0. [DOI] [PubMed] [Google Scholar]
- 21.Campbell S.L., Khosravi-Far R., Rossman K.L., Clark G.J., Der C.J. Increasing complexity of Ras signaling. Oncogene. 1998;17:1395–1413. doi: 10.1038/sj.onc.1202174. [DOI] [PubMed] [Google Scholar]
- 22.Diehl G.E., et al. TRAIL-R as a negative regulator of innate immune cell responses. Immunity. 2004;21:877–889. doi: 10.1016/j.immuni.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 23.Finnberg N., et al. DR5 knockout mice are compromised in radiation-induced apoptosis. Mol. Cell. Biol. 2005;25:2000–2013. doi: 10.1128/MCB.25.5.2000-2013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ganten T.M., et al. Proteasome inhibition sensitizes hepatocellular carcinoma cells, but not human hepatocytes, to TRAIL. Hepatology. 2005;42:588–597. doi: 10.1002/hep.20807. [DOI] [PubMed] [Google Scholar]
- 25.Sayers T.J., et al. The proteasome inhibitor PS-341 sensitizes neoplastic cells to TRAIL-mediated apoptosis by reducing levels of c-FLIP. Blood. 2003;102:303–310. doi: 10.1182/blood-2002-09-2975. [DOI] [PubMed] [Google Scholar]
- 26.Schmaltz C., et al. T cells require TRAIL for optimal graft-versus-tumor activity. Nat. Med. 2002;8:1433–1437. doi: 10.1038/nm1202-797. [DOI] [PubMed] [Google Scholar]
- 27.Smyth M.J., et al. Nature’s TRAIL--on a path to cancer immunotherapy. Immunity. 2003;18:1–6. doi: 10.1016/s1074-7613(02)00502-2. [DOI] [PubMed] [Google Scholar]
- 28.Mehlen P., Puisieux A. Metastasis: a question of life or death. Nat. Rev. Cancer. 2006;6:449–458. doi: 10.1038/nrc1886. [DOI] [PubMed] [Google Scholar]
- 29.Segrelles C., et al. Functional roles of Akt signaling in mouse skin tumorigenesis. Oncogene. 2002;21:53–64. doi: 10.1038/sj.onc.1205032. [DOI] [PubMed] [Google Scholar]
- 30.Gilmore A.P. Anoikis. Cell Death Differ. 2005;12(Suppl. 2):1473–1477. doi: 10.1038/sj.cdd.4401723. [DOI] [PubMed] [Google Scholar]
- 31.Khanna C., Hunter K. Modeling metastasis in vivo. Carcinogenesis. 2005;26:513–523. doi: 10.1093/carcin/bgh261. [DOI] [PubMed] [Google Scholar]
- 32.Rinker-Schaeffer C.W., O’Keefe J.P., Welch D.R., Theodorescu D. Metastasis suppressor proteins: discovery, molecular mechanisms, and clinical application. Clin. Cancer Res. 2006;12:3882–3889. doi: 10.1158/1078-0432.CCR-06-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin M.S., et al. Mutations of tumor necrosis factor-related apoptosis-inducing ligand receptor 1 (TRAIL-R1) and receptor 2 (TRAIL-R2) genes in metastatic breast cancers. Cancer Res. 2001;61:4942–4946. [PubMed] [Google Scholar]
- 34.Zhuang L., et al. Progression in melanoma is associated with decreased expression of death receptors for tumor necrosis factor-related apoptosis-inducing ligand. Hum. Pathol. 2006;37:1286–1294. doi: 10.1016/j.humpath.2006.04.026. [DOI] [PubMed] [Google Scholar]
- 35.Vigneswaran N., Wu J., Nagaraj N., Adler-Storthz K., Zacharias W. Differential susceptibility of metastatic and primary oral cancer cells to TRAIL-induced apoptosis. Int. J. Oncol. 2005;26:103–112. [PubMed] [Google Scholar]
- 36.Goldberg G.S., et al. Global effects of anchorage on gene expression during mammary carcinoma cell growth reveal role of tumor necrosis factor-related apoptosis- inducing ligand in anoikis. Cancer Res. 2001;61:1334–1337. [PubMed] [Google Scholar]
- 37.Li-Weber M., Krammer P.H. Function and regulation of the CD95 (APO-1/Fas) ligand in the immune system. Semin. Immunol. 2003;15:145–157. doi: 10.1016/s1044-5323(03)00030-7. [DOI] [PubMed] [Google Scholar]
- 38.Owen-Schaub L.B., van Golen K.L., Hill L.L., Price J.E. Fas and Fas ligand interactions suppress melanoma lung metastasis. J. Exp. Med. 1998;188:1717–1723. doi: 10.1084/jem.188.9.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teitz T., et al. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat. Med. 2000;6:529–535. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- 40.Teitz T., Lahti J.M., Kidd V.J. Aggressive childhood neuroblastomas do not express caspase-8: an important component of programmed cell death. J. Mol. Med. 2001;79:428–436. doi: 10.1007/s001090100233. [DOI] [PubMed] [Google Scholar]
- 41.Takita J., et al. Allelic imbalance on chromosome 2q and alterations of the caspase 8 gene in neuroblastoma. Oncogene. 2001;20:4424–4432. doi: 10.1038/sj.onc.1204521. [DOI] [PubMed] [Google Scholar]
- 42.Stupack D.G., et al. Potentiation of neuroblastoma metastasis by loss of caspase-8. Nature. 2006;439:95–99. doi: 10.1038/nature04323. [DOI] [PubMed] [Google Scholar]
- 43.Siegmund D., et al. Death receptor-induced signaling pathways are differentially regulated by gamma interferon upstream of caspase 8 processing. Mol. Cell. Biol. 2005;25:6363–6379. doi: 10.1128/MCB.25.15.6363-6379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schneider P., et al. Identification of a new murine tumor necrosis factor receptor locus that contains two novel murine receptors for tumor necrosis factor-related apoptosis-inducing ligand (TRAIL). J. Biol. Chem. 2003;278:5444–5454. doi: 10.1074/jbc.M210783200. [DOI] [PubMed] [Google Scholar]
- 45.Lane, D., Cartier, A., Rancourt, C., and Piché, A. 2007. Cell detachment modulates TRAIL resistance in ovarian cancer cells by downregulating the phosphatidylinositol 3-kinase/Akt pathway. Int. J. Gynecol. Cancer. doi:10.1111/j.1525-1438.2007.01062.x. [DOI] [PubMed] [Google Scholar]
- 46.Aoudjit F., Vuori K. Matrix attachment regulates Fas-induced apoptosis in endothelial cells: a role for c-flip and implications for anoikis. J. Cell Biol. 2001;152:633–643. doi: 10.1083/jcb.152.3.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wintermantel T.M., Mayer A.K., Schutz G., Greiner E.F. Targeting mammary epithelial cells using a bacterial artificial chromosome. Genesis. 2002;33:125–130. doi: 10.1002/gene.10097. [DOI] [PubMed] [Google Scholar]
- 48.Schaft J., Ashery-Padan R., van der Hoeven F., Gruss P., Stewart A.F. Efficient FLP recombination in mouse ES cells and oocytes. Genesis. 2001;31:6–10. doi: 10.1002/gene.1076. [DOI] [PubMed] [Google Scholar]
- 49.Kelly-Spratt K.S., Gurley K.E., Yasui Y., Kemp C.J. p19Arf suppresses growth, progression, and metastasis of Hras-driven carcinomas through p53-dependent and -independent pathways. PLoS Biol. 2004;2:E242. doi: 10.1371/journal.pbio.0020242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tu S., McStay G.P., Boucher L.M., Mak T., Beere H.M., Green D.R. In situ trapping of activated initiator caspases reveals a role for caspase-2 in heat shock-induced apoptosis. Nat. Cell Biol. 2006;8:72–77. doi: 10.1038/ncb1340. [DOI] [PubMed] [Google Scholar]
- 51.Ganten T.M., et al. Preclinical differentiation between apparently safe and potentially hepatotoxic applications of TRAIL either alone or in combination with chemotherapeutic drugs. Clin. Cancer Res. 2006;12:2640–2646. doi: 10.1158/1078-0432.CCR-05-2635. [DOI] [PubMed] [Google Scholar]
- 52.Holler N., et al. Two adjacent trimeric Fas ligands are required for Fas signaling and formation of a death-inducing signaling complex. Mol. Cell. Biol. 2003;23:1428–1440. doi: 10.1128/MCB.23.4.1428-1440.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zeger S.L., Liang K.Y. Longitudinal data analysis for discrete and continuous outcomes. Biometrics. 1986;42:121–130. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.