Abstract
The adult heart responds to excessive neurohumoral signaling and workload by a pathological growth response characterized by hypertrophy of cardiomyocytes and activation of a fetal program of cardiac gene expression. These responses culminate in diminished pump function, ventricular dilatation, wall thinning, and fibrosis, and can result in sudden death. Myocyte enhancer factor–2 (MEF2) transcription factors serve as targets of the signaling pathways that drive pathological cardiac remodeling, but the requirement for MEF2 factors in the progression of heart disease in vivo has not been determined. MEF2A and MEF2D are the primary MEF2 factors expressed in the adult heart. To specifically determine the role of MEF2D in pathological cardiac remodeling, we generated mice with a conditional MEF2D allele. MEF2D-null mice were viable, but were resistant to cardiac hypertrophy, fetal gene activation, and fibrosis in response to pressure overload and β-chronic adrenergic stimulation. Furthermore, we show in a transgenic mouse model that forced overexpression of MEF2D was sufficient to drive the fetal gene program and pathological remodeling of the heart. These results reveal a unique and important function for MEF2D in stress-dependent cardiac growth and reprogramming of gene expression in the adult heart.
Introduction
Acute or chronic injury to the adult heart activates a pathological response characterized by hypertrophic growth of cardiomyocytes, assembly of additional sarcomeres to enhance contractility, and activation of a fetal cardiac gene program, ultimately causing ventricular wall thinning, chamber dilation, heart failure, and sudden death (1–3). A variety of abnormalities cause pathological hypertrophy and heart failure, including hypertension, myocardial infarction, endocrine disorders, and inherited mutations in structural and contractile proteins of cardiomyocytes. These insults have been shown to activate a complex set of interwoven signaling pathways that culminate in the nucleus to activate various transcription factors, including nuclear factor of activated T cells (NFAT), GATA, and myocyte enhancer factor–2 (MEF2) factors, that drive the expression of fetal cardiac and stress response genes (3, 4). There has been great interest in deciphering the molecular mechanisms responsible for transcriptional reprogramming of the failing heart and in developing “transcriptional” therapies for enhancing cardiac function (4, 5).
Members of the MEF2 family of MADS (MCM1, agamous, deficiens, serum response factor) box transcription factors function as stress-dependent regulators of gene expression and have been implicated in multiple aspects of striated muscle development and disease (6). The 4 vertebrate MEF2 factors, MEF2A, -B, -C, and -D, display overlapping expression patterns in embryonic and adult tissues (7–9), and loss-of-function studies in mice have revealed distinct functions for the Mef2a and Mef2c genes in the developing heart. Mice lacking Mef2a survive until the perinatal period when they succumb to lethal cardiac arrhythmias accompanied by abnormalities in expression of contractile protein and metabolic genes (10). Mice lacking Mef2c die during early embryogenesis from a spectrum of cardiovascular defects (11). However, despite the key roles of MEF2 factors as regulators of cardiac gene expression during development, relatively little is known of their functions in the adult heart.
Using transgenic mice harboring a MEF2-dependent reporter gene, we showed previously that hypertrophic stimuli augment MEF2 activity in the adult heart (12). Stress-activated signaling pathways that drive pathological cardiac remodeling stimulate the transcriptional activity of MEF2 through multiple mechanisms. Activation of calcium/calmodulin-dependent protein kinase and protein kinase D stimulates MEF2 activity by promoting the phosphorylation and nuclear export of class II histone deacetylases, which function as MEF2 corepressors (13, 14). MAPK signaling leads to the phosphorylation of MEF2 and stimulation of transcriptional activity, and calcineurin promotes MEF2 activity through multiple mechanisms (15–17).
The major MEF2 isoforms in the adult heart are MEF2A and -D, which form heterodimers (10, 18). The lethal phenotype of Mef2a-null mice indicates that MEF2D alone cannot support normal postnatal cardiac function (10). Whether MEF2A can support cardiac function in the absence of MEF2D and whether MEF2D may also be essential in the adult heart has not been determined.
To define the potential functions of MEF2D in the heart, we generated mice with a Mef2d loss-of-function mutation. In contrast to the lethal cardiac phenotypes resulting from genetic deletion of Mef2a or Mef2c, we show that mice lacking Mef2d are viable, but display an impaired response to stress signals that normally lead to cardiac hypertrophy, fibrosis, and fetal gene activation. Conversely, forced overexpression of MEF2D in the heart is sufficient to evoke severe cardiomyopathy. These findings demonstrate that MEF2D plays a key role in mediating stress-dependent gene expression in the adult heart.
Results
Generation of Mef2d-null mice.
Because Mef2a- and Mef2c-null mice are not viable (10, 11), we generated a conditional Mef2d-null allele, called Mef2dloxP in order to determine the functions of Mef2d in the adult heart. The conditional allele contained loxP sites flanking the third exon, which encodes the MADS- and MEF2-specific domains that mediate DNA binding, dimerization, and cofactor interactions (Figure 1A). Deletion of the corresponding region between the loxP sites of Mef2a or Mef2c results in a null allele (10, 11).
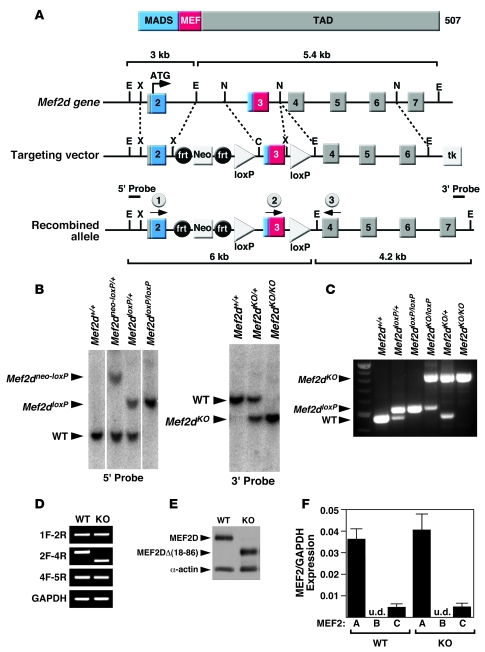
Figure 1. Generation of mice with a conditional Mef2d mutation.
(A) Schematic representation of the mouse Mef2d locus and targeting strategy. Positions of 3ι and 5ι probes used for Southern blots are shown. The positions of the PCR primers used for genotyping mutant alleles are marked with arrows (circles labeled 1, 2, and 3). C, ClaI; frt, FLP recombinase target; E, EcoRI; TAD, transactivation domain; MADS, MCM1, agamous, deficiens, serum response factor; N, NcoI; X, XhoI. (B) Southern blot analysis of Mef2d mutant alleles. Genomic DNA was digested with EcoRI and hybridized to a 5ι probe in the left panel and to a 3ι probe in the right panel. WT, wild-type allele; Mef2dneo-loxP, conditional allele; Mef2dloxP, conditional allele with the Neo cassette removed; Mef2dKO, null allele. (C) PCR genotyping to distinguish different Mef2d alleles. The positions of the primers that produce these PCR products are labeled (1, 2, and 3) and circled on A. All 3 primers were added to the PCR reactions and the PCR products were loaded into lanes 2–7. (D) Expression of wild-type and mutant Mef2d detected by RT-PCR. Mef2d mutant allele lacks exon 3, which encodes the MADS- and MEF2-specific domains. GAPDH was used as a loading control. Labels on the left side of the panel indicate exon location and direction of primers used for RT-PCR. (E) Western blot analysis to detect WT and mutant MEF2D (KO) proteins. The mutant MEF2D protein is truncated due to deletion of exon 3. α-Actin protein was used as a loading control. (F) Expression level of Mef2 detected by quantitative PCR. Error bars indicate ±SEM. u.d., undetectable.
To determine the complete loss-of-function phenotype of Mef2d, we initially deleted the gene using a Meox2-Cre transgene, which expresses Cre recombinase in epiblast-derived tissues as early as embryonic day 5 (19). Unexpectedly, mice homozygous for this deletion were viable and showed no obvious phenotypic abnormalities. Complete deletion of the genomic region between the 2 loxP sites was confirmed by Southern blot and PCR of genomic DNA (Figure 1, B and C). By RT-PCR of RNA from adult heart, we found that exon 2 was spliced to exon 4 in the mutant allele, resulting in an in-frame deletion within the coding region of the Mef2d mRNA (Figure 1D). Western blot analysis of cardiac extracts confirmed that the mutant MEF2D protein lacking the MADS and MEF2 domains was expressed at a level comparable to the wild-type protein (Figure 1E).
To confirm that the residual MEF2 protein did not function as a dominant-negative mutant, we coexpressed it with the wild-type protein in transfected COS-7 cells and monitored expression of a MEF2-dependent luciferase reporter. As shown in Supplemental Figure 1 (supplemental material available online with this article; doi:10.1172/JCI33255DS1), the MEF2DΔ(18-86) protein was devoid of transcriptional activity and showed no dominant-negative effect on wild-type MEF2A protein.
We analyzed numerous adult tissues, including skeletal muscle, heart, liver, thymus, spleen, and brain of Mef2d mutant mice and observed no histological abnormalities. Mef2d mutant mice also appeared normal by other criteria, including behavior, weight, fertility, and lifespan. We conclude that MEF2D is not required for viability in mice. Quantification of Mef2a and Mef2c mRNA expression in Mef2d mutant hearts showed no compensatory upregulation of these genes (Figure 1F).
Mef2d-null mice are resistant to cardiac remodeling and fibrosis in response to pressure overload.
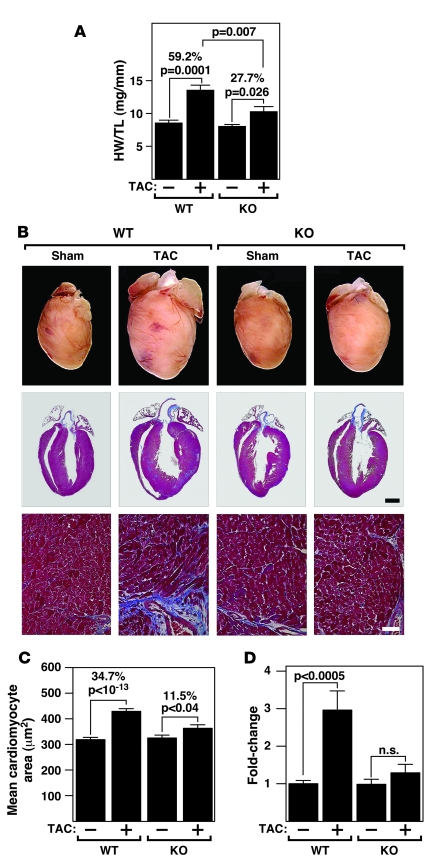
To investigate whether MEF2D was required for pathological remodeling of the heart in response to pressure overload, we performed thoracic aortic constriction (TAC) on adult mice. As shown in Figure 2A, the hearts of wild-type and Mef2d–/– mice were comparable in size at baseline. Twenty-one days following TAC, wild-type mice showed a 59.2% increase in heart weight/tibia length (HW/TL). In contrast, Mef2d mutant mice showed only a 27.7% increase in HW/TL (Figure 2A). Based on histological sections, less cardiac remodeling was observed in Mef2d mutant mice compared with wild-type mice (Figure 2B). Consistent with these findings, following pressure overload, we observed a 34.7% ± 0.25% increase in cross-sectional area of cardiomyocytes in wild-type mice, in comparison to only a 11.5% ± 0.40% increase in mutant mice (Figure 2C).
Figure 2. Blunted hypertrophy of Mef2d mutant mice following TAC.
(A) HW/TL ratios (±SEM) of WT and Mef2d mutant mice were determined 21 days after TAC. (B) Hearts from WT and Mef2d mutant mice subjected to either a sham operation or pressure overload (TAC) are shown at the top. Histological sections stained with Masson’s trichrome are shown on the bottom. Masson’s trichrome staining of WT and Mef2d mutant hearts indicates lack of fibrosis in Mef2d–/– hearts in response to pressure overload by TAC. Scale bars: 1 mm (middle panel); 40 μm (bottom panel). (C) Mean cross-sectional area of cardiomyocytes (±SEM) in WT and Mef2d mutant mice was measured 21 days after TAC. (D) Using morphometric analysis of picrosirius red stained heart sections, the amount of myocardial fibrosis was assessed. Fibrosis was apparent in the hearts of WT mice following TAC. In contrast, virtually no fibrosis was detected in the hearts of Mef2d mutant mice. Values indicate fold changes of fibrosis in each group compared with a group of sham-operated WT mice (±SEM).
Pressure overload hypertrophy in wild-type mice was accompanied by extensive fibrosis of the ventricular wall as detected by Masson’s trichrome staining (Figure 2B). Remarkably, there was virtually no ventricular fibrosis in Mef2d mutant mice. These findings were corroborated by morphometric analysis of picrosirius red stained sections of wild-type and mutant hearts showing that Mef2d mutant mice had much less fibrosis after pressure overload compared with wild-type mice (Figure 2D).
Functional analysis of hearts of Mef2d-null mice.
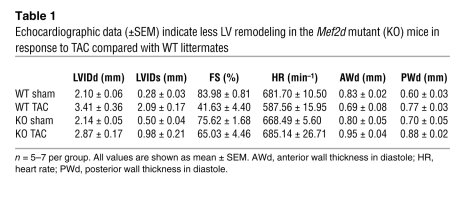
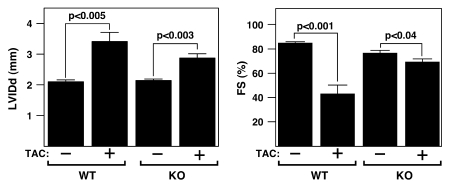
The response to cardiac stress of Mef2d mice was assessed by echocardiography. At baseline, there were no substantial differences seen in the LV end-diastolic diameter (LVIDd) or heart rate (HR), and only minimal differences seen in the LV end-systolic diameter (LVIDs) and fractional shortening (FS) in wild-type and mutant Mef2d mice (Table 1 and Supplemental Table 1). Three weeks after TAC surgery, wild-type mice showed a dramatic increase in LVIDd accompanied by a pronounced reduction in cardiac contractility, as indicated by a decreased FS. In contrast, Mef2d mutant animals were largely resistant to LV dilation and its concomitant decrease in contractility (Figure 3). Additionally, compared with Mef2d mutant animals, wild-type animals experienced a significant reduction in HR following TAC, which is indicative of the pathological state of the heart. These data demonstrate that in response to pressure overload Mef2d mutant hearts are resistant to cardiac remodeling and functional deterioration.
Table 1 .
Echocardiographic data (±SEM) indicate less LV remodeling in the Mef2d mutant (KO) mice in response to TAC compared with WT littermates
Figure 3. Echocardiographic analysis of Mef2d mutant mice following TAC.
Mef2d mutant mice display less LV dilation during diastole and a less pronounced decrease in FS in response to TAC than WT mice.
Fetal gene activation is blunted in Mef2d-null mice in response to pressure overload.
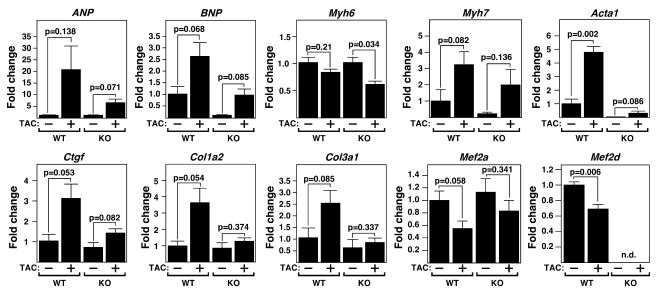
MEF2D was required not only for hypertrophic growth and fibrosis in response to pressure overload, but was also essential for fetal gene activation, a prototypical consequence of cardiac stress. As shown in Figure 4, upregulation of the hypertrophic gene markers atrial natriuretic peptide (ANP), brain natriuretic peptide (BNP), myosin, heavy polypeptide 7, cardiac muscle, β (Myh7), and actin, α 1, skeletal muscle (Acta1) was severely blunted in mutant mice. Also, induction of connective tissue growth factor (Ctgf), procollagen, type I, α 2 (Col1a2), and procollagen, type III, α 1 (Col3a1), which are upregulated during cardiac fibrosis, was compromised in Mef2d–/– mice. Baseline expression of several fetal cardiac genes was also diminished in Mef2d–/– mice, suggesting that MEF2D is required for their normal expression in the absence of stress (Figure 4 and Supplemental Table 1). These data are consistent with Masson’s trichrome and picrosirius red staining, and support the conclusion that cardiac hypertrophy and fibrosis induced by pressure overload are suppressed in Mef2d–/– mice.
Figure 4. Resistance of Mef2d mutant mice to fetal gene activation following TAC.
Expression of cardiac hypertrophic markers, fibrosis markers, Mef2a, and Mef2d in WT and Mef2d mutant hearts was detected by quantitative PCR 21 days after TAC (n = 3–5 per group). Values indicate expression level relative to a WT sham-operated group (±SEM). ANP, atrial natriuretic peptide; BNP, brain natriuretic peptide; Myh6, myosin, heavy polypeptide 6, cardiac muscle, alpha; Myh7, myosin, heavy polypeptide 7, cardiac muscle, beta; Acta1, actin, alpha 1, skeletal muscle; Ctgf, connective tissue growth factor; Col1a2, procollagen, type I, alpha 2; Col3a1, procollagen, type III, alpha 1; n.d., non-determined.
We observed a decrease in Mef2a and Mef2d expression level following TAC; however, previous reports showed that MEF2 activity is increased following cardiac remodeling (12, 20).We believe that the decrease in MEF2 expression may represent a compensatory mechanism to diminish pathological signaling via MEF2 in the heart.
Mef2d-null mice are resistant to cardiac remodeling in response to β-adrenergic stimulation.
We also investigated whether MEF2D was necessary for cardiac remodeling in response to chronic β-adrenergic stimulation. Isoproterenol (ISO), a β-adrenergic receptor agonist, was administered over 7 days to mice using osmotic minipumps (8.8 mg/kg/day). Compared with wild-type mice, which showed a 25.9% increase in HW/TL (P = 0.0008), mutant mice showed an insignificant in HW/TL (Figure 5, A and B). This finding suggests that MEF2D acts within signaling pathways activated not only by pressure overload, but also β-adrenergic stimulation.
Figure 5. Decreased hypertrophic response of Mef2d mutant mice following β-adrenergic stimulation.
(A) WT and Mef2d mutant mice were chronically infused with either saline vehicle or ISO (8.8 mg/kg/day) for 7 days and sacrificed for assessment of cardiac remodeling. HW/TL ratios (±SEM) are shown as bar graphs (n = 7–9 per group). n.s., non-significant. (B) Whole hearts of WT and Mef2d mutant mice treated with either saline vehicle or ISO (8.8 mg/kg/day) are shown at the top. Hearts were sectioned and stained with H&E as seen on bottom. Scale bar: 1 mm. (C) Representative bar graphs indicating less LV dilation and a decrease in FS in Mef2d mutant mice compared with WT mice in response to ISO treatment.
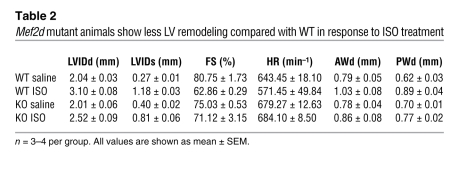
Echocardiography confirmed that ISO administration resulted in a more pronounced increase in LV wall thickness in wild-type mice compared with Mef2d mutant mice, indicative of cardiac hypertrophy (Table 2). In addition, the wild-type animals experienced enhanced LV remodeling in response to ISO, as shown by more LV dilation and a decrease in contractility (Table 2 and Figure 5C).
Table 2 .
Mef2d mutant animals show less LV remodeling compared with WT in response to ISO treatment
Forced expression of MEF2D causes severe cardiomyopathy.
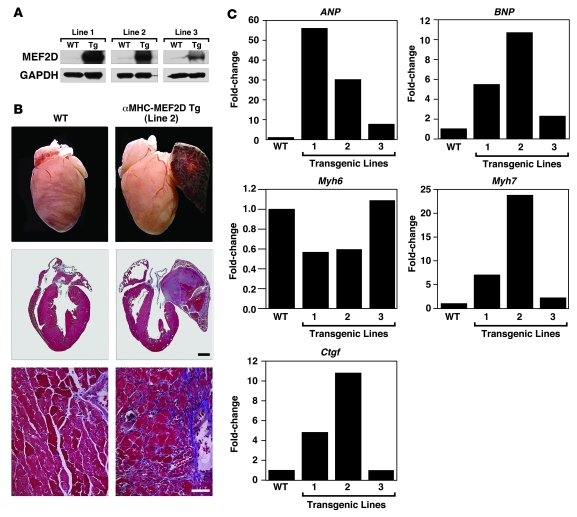
To determine whether MEF2D is sufficient to induce pathological cardiac remodeling, we generated transgenic mice in which the α–myosin heavy chain (α-MHC) transgene directed cardiac-specific expression of MEF2D. Three independent stable transgenic mouse lines (lines 1, 2, and 3) were established that, based on Western blot analysis, expressed MEF2D in the heart approximately 18-, 15-, and 2.5-fold higher, respectively, than the wild-type animals. We focused on transgenic line 2, which showed clear signs of cardiomyopathy — extensive fibrosis, atrial enlargement, and thrombi as well as severely congested lungs and liver — at 8 weeks of age.
Fetal cardiac genes as well as connective tissue growth factor (Ctgf), a marker of fibrosis, were dramatically upregulated in transgenic lines 1 and 2 and were upregulated to a lesser extent in line 3, consistent with the lower level of MEF2D expression in this line (Figure 6C). MEF2D overexpression in transgenic lines 1 and 2 also caused a diminution in expression of myosin, heavy polypeptide 6, cardiac muscle, α (Myh6) and upregulation of Myh7, a hallmark of pathological cardiac remodeling and reduced cardiac contractility. We conclude that MEF2D is sufficient to induce cardiac remodeling and cardiomyopathy.
Figure 6. Pathological cardiac remodeling induced by overexpression of MEF2D.
(A) Proteins were extracted from ventricles of α-MHC-MEF2D transgenic mouse lines at 6 weeks to 8 weeks of age for Western blot analysis using an antibody against MEF2D. GAPDH was used as a loading control. (B) Hearts were dissected from α-MHC-MEF2D male mice (line 2) and WT littermates at 8 weeks of age. Note the enlarged left atrium in the α-MHC-MEF2D heart. The lower panels are histological sections stained with Masson’s trichrome to detect fibrosis. Scale bar: 1 mm (middle panel); 40 μm (bottom panel). (C) Expression level of various hypertrophy and fibrosis markers was evaluated by quantitative PCR in each transgenic line. Note the correlation between expression level of MEF2D (A) and that of hypertrophy and fibrosis markers.
Discussion
The results of this study demonstrate that MEF2D plays a unique and important role as a mediator of stress-dependent pathological remodeling of the adult heart. In the absence of MEF2D hypertrophy, chamber dilatation, fibrosis, and fetal gene activation were profoundly blunted following pressure overload or chronic β-adrenergic stimulation. Conversely, elevating MEF2D activity by transgenic overexpression was sufficient to substitute for stress signals and drive pathological cardiac remodeling. The viability of Mef2d-null mice contrasts with the early embryonic lethality of Mef2c-null mice (11) and perinatal lethality of Mef2a-null mice (10), pointing to distinct functions for each MEF2 isoform at different stages of heart development.
Signaling pathways leading to cardiac remodeling.
Our results indicate that MEF2D is a critical component of the signaling pathways through which pressure overload and adrenergic signaling drive cardiac remodeling and fetal gene activation. In this regard, prior studies showed that MEF2 regulates an array of genes involved in ion handling, extracellular matrix remodeling, and metabolism during pathological cardiac remodeling (21, 22). Some of these genes are direct targets of MEF2, whereas others are regulated indirectly by MEF2 through protein-protein interactions with other transcription factors and through subordinate MEF2-regulated transcription factors. Recent studies pointing to the importance of stress-inducible microRNAs in the cardiac remodeling response reveal an additional layer of potential regulation by MEF2 (23, 24).
Forced expression of MEF2A or -C in transgenic mice has been shown previously to cause dilated cardiomyopathy and fetal gene activation with little or no hypertrophy (21, 22). Overexpression of MEF2 or expression of a superactive MEF2-VP16 chimeric protein in primary cardiomyocytes also evokes a phenotype of myocyte elongation and sarcomere disorganization (21, 22). Similarly, our results show that overexpression of MEF2D causes ventricular dilation and fibrosis in transgenic mice.
Expression of a dominant-negative MEF2 mutant protein in the adult heart also reduces chamber dilation and improves contractility in the presence of constitutive calcineurin activation (22). Notably, dominant-negative MEF2 results in a minimal block to hypertrophy, suggesting that MEF2 function is required primarily for late stages of pathological cardiac remodeling. Since MEF2 associates with NFAT and GATA factors, both of which have been shown to mediate pathological cardiac signaling (17, 25–27), the blockade of cardiac remodeling by dominant-negative MEF2 likely involves the inhibition of multiple downstream transcriptional effectors in addition to endogenous MEF2. The diminution of cardiac remodeling in Mef2d mutant mice therefore provides a more direct illustration of the specific contribution of this MEF2 isoform to the cardiac stress response. It will be of interest to examine mice with a cardiomyocyte-specific deletion of Mef2d to determine whether the reduction of cardiac remodeling in Mef2d mutant mice results exclusively from the lack of MEF2D in cardiomyocytes.
While hypertrophy, fetal gene activation, and fibrosis were substantially blunted in Mef2d mutant mice, we observed residual hypertrophy and fetal gene activation in these animals. This likely reflects redundant functions of MEF2A, the other major MEF2 factor expressed in the adult heart, as well as other stress-responsive transcription factors such as NFAT, GATA, and CAMTA (25–27).
Myriad functions of Mef2 genes in vivo.
The phenotypes of Mef2a, -c, and -d mutant mice reveal unique roles for each of these MEF2 factors at different steps in cardiac development: MEF2C regulates early cardiac morphogenesis (11), MEF2A regulates genes involved in contractility and energy metabolism after birth (10), and MEF2D serves as a mediator of stress-dependent gene expression in the adult heart. These unique functions are likely to reflect the distinct expression patterns of the different Mef2 genes and, possibly, intrinsic differences in the transcriptional activities and protein-protein interactions of the MEF2 factors due to their amino acid sequence differences.
In addition to their functions in the heart, MEF2 factors are expressed in most or all tissues, and are likely to play key roles in diverse gene programs for cell differentiation and stress responsiveness. Conditional gene deletion of Mef2c, for example, has revealed important roles for this gene in regulation of skeletal muscle fiber type and resistance to fatigue, as well as in endochondral bone development (28, 29). MEF2 factors also participate in neural and vascular development (30–32) and, recently, have been implicated in coronary artery disease, cancer and hepatic fibrosis (33–36). Thus, strategies to modulate the MEF2 activity in adult tissues may be therapeutically beneficial in the settings of a variety of diseases.
Methods
Generation of Mef2d mutant mice.
The mouse Mef2d genomic locus was isolated from a 129SvEv mouse BAC library (BACPAC Resource Center at CHORI, Oakland, California, USA). A 1.3-kb genomic DNA fragment (short arm) that includes exon 2 and flanking introns was digested with XhoI and EcoRI restriction enzymes. The EcoRI restriction site of this genomic DNA fragment was blunted and subcloned into the XhoI site located upstream of the FLP recombinase target–flanked (FRT-flanked) neomycin resistance cassette of the pDelboy targeting vector. A 1.2-kb genomic DNA fragment (the deleted region) that encoded exon 3 was digested by NcoI restriction enzyme, blunted and inserted in between loxP sites of the targeting vector via ClaI and XhoI restriction sites. A 3.3-kb genomic fragment (long arm) that encodes exons 4–6 was digested and blunt-end cloned into the EcoRI restriction site downstream of the loxP sites in the targeting vector. This EcoRI site was not destroyed by the blunt cloning and was used for screening for positive recombination by Southern blot analysis (Figure 1A).
The Mef2d targeting vector was linearized by digestion with NotI and electroporated into 129SvEv-derived embryonic stem cells. Following positive-negative selection with G418 and FIAU, resistant colonies were screened by Southern blot analysis of EcoRI digested DNA using 5ι and 3ι probes (Figure 1, A and B). Three correctly targeted clones were expanded and injected into C57BL/6 blastocysts, which were transferred to the uterus of pseudopregnant females. High-percentage chimeric male mice (Mef2dneo-loxP/+) were bred into a C57BL/6 background to obtain germline transmission. Heterozygous Mef2dneo-loxP mice were intercrossed with hACTB:FLPe transgenic mice (37) to remove the neomycin resistance cassette. Southern blot analysis and PCR were performed to confirm the removal of the neomycin resistance cassette in the genome of Mef2dloxP/+ mice. To obtain mice with a Mef2d-null allele, the intercross of Mef2dloxP/+ mice to Meox2-Cre transgenic mice (19) was performed. Southern blot analysis and PCR were performed to confirm mice with Mef2d heterozygous- or homozygous-null (Mef2dKO) alleles (Figure 1, B and C). All experiments on mice were conducted in 129SvEv/C57BL/6 mixed background. All animal studies were approved by the IACUC of the University of Texas Southwestern Medical Center.
Southern blot analysis and PCR genotyping.
DNA sequences for PCR genotyping primers are as follows: 5′-TTCTGCAGACCATGAACTGATCACTAGT-3′; 5′-TAGAGCAAGTACCAGAATAGCCAGG-3′; and 5′-ACAGGAAAGGACAATACAAGAAAGCAC-3′. Southern blot probes were generated by PCR using the following primer sets: 5′ probe forward, 5′-AAGCTTAGAATTTACAGCCTGCAGCGCTGA-3', reverse, 5′-GGATCCGAGCTAAGGTTTTGTTAAGTTTCC-3′; and 3′ probe forward, 5′-CTCGAGTCATGCATCCTGTGTGAA-3′; reverse, 5′-GGATGACTTTGTTTAGGCTGTTGC-3'. In brief, tail genomic DNA was digested with EcoRI and analyzed using a standard Southern blot protocol. The 25 μl PCR genotyping reaction contained 1 μl of tail DNA as template, 2.5 U of Taq polymerase (Promega) in 0.5 μl, and a final concentration of 2 mM MgCl2. An annealing temperature of 70°C was used and the PCR products were electrophoresed on a 2% agarose gel.
RNA analysis.
Total RNA was purified from isolated heart ventricles with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. Two micrograms of RNA was used as template to synthesize cDNA using random hexamers. Primer sequences used for RT-PCR are as follows: 1 foward, 5′-TTCGCTGTTTCCCGTCGGAG-3′; 2 reverse, 5′-TGGCGGTTCCGTTCATCAGTG-3′, forward, 5′-GTCCCCAGACATTCATGAGC-3′; 4 reverse, 5′-GCTCTGCTCCAGTGAGTCCT-3′, forward, 5′-CCCTGAGGAAGAAGGGTTTC-3′; 5 reverse, 5′-CAGGGAAGGAGTGACCAGAG-3′; GAPDH forward, 5′-AGACAGCCGCATCTTCTTGT-3′, reverse, 5′-TGATGGCAACAATGTCCACT-3′. Quantitative PCR was performed using the following Taqman probes purchased from ABI: Acta1, Mm00808218_g1; ANP, Mm01255748_g1; BNP, Mm00435304_g1; Col1a2, Mm00483888_m1; Col3a1, Mm00802331_m1; Ctgf, Mm00515790_g1; Mef2a, Mm01318991_m1; Mef2d, Mm00504931_m1; Myh6, Mm00440354_m1; Myh7, Mm01319006_g1.
Histology.
Hearts were fixed in 4% paraformaldehyde in phosphate-buffered saline, embedded in paraffin, and sectioned at 5-μm intervals. H&E staining, Masson’s trichrome staining, and picrosirius red staining were performed using standard procedures.
Measurements of mean cardiomyocyte area and myocardial fibrosis.
Cardiomyocyte size was assessed on H&E-stained sections using ImageJ software (http://rsb.info.nih.gov/ij/) (NIH). About 100–150 randomly chosen cardiomyocytes from each group (n = 2–3) were analyzed to measure cross-sectional cardiomyocyte area. ImageJ software (NIH) was used to measure the amount of myocardial fibrosis on picrosirius red stained sections. About 30–50 randomly chosen frames from each group (n = 2–3) were analyzed.
Transthoracic echocardiography.
Cardiac function and heart dimensions were evaluated by 2-dimensional echocardiography on conscious mice using a Vingmed System (GE Vingmed Ultrasound) and a 11.5-MHz linear array transducer. M-mode tracings were used to measure anterior and posterior wall thicknesses at end diastole and end systole. LV internal diameter (LVID) was measured as the largest anteroposterior diameter in either diastole (LVIDd) or systole (LVIDs). The data were analyzed by a single observer blinded to mouse genotypes. LV FS was calculated according to the following formula: FS (%) = [(LVIDd – LVIDs)/LVIDd] × 100.
Statistics.
Gene expression was normalized to GAPDH expression level and calculated as relative change. Differences in morphological, physiological, and biochemical parameters between groups were analyzed by 2-sided Student’s t test using Microsoft Excel 2002. Non-significant (n.s.) difference indicates a P value greater than 0.05.
TAC and ISO administration.
Male mice (8- to 12-week-old) either underwent a sham operation or were subjected to TAC as described (38). Perioperative mortalities (within 24 hours of surgery) were similar (about 10%) in wild-type and mutant mice subjected to TAC. There was no excess mortality during follow-up in any of the 4 groups (Sham or TAC, wild-type or mutant mice). Hearts were isolated 21 days following TAC for assessment of cardiac hypertrophy. ISO (Sigma-Aldrich) (8.8 mg/kg/day) or saline vehicle was administered using miniosmotic pumps (Model 2001; Alzet) subcutaneously implanted on the back of 8- to 10-week-old male mice. Cardiac hypertrophy was evaluated by measuring HW/TL of sacrificed mice 7 days after pump implantation.
Generation of transgenic mice.
A cDNA encoding mouse FLAG-tagged MEF2D was cloned into a plasmid containing the α-myosin heavy chain transgene and human growth hormone (hGH) poly(A)+ signal (39), and transgenic mice were generated by standard techniques. Genotyping was performed by PCR analysis using genomic DNA, and transgenic expression of MEF2D in the heart was confirmed by Western blot analysis using mouse anti-MEF2D antibody (1:1,000; BD Transduction Laboratories) and mouse anti-GAPDH antibody (1:5,000; Chemicon).
Tissue culture and transfection.
Standard procedures were used and described in detail in Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Alisha Tizenor for graphics, Jennifer Brown for editorial assistance, and Yongli Kong, Cheryl Nolen, John Shelton, Evelyn Tennison, and Jonathan Guevara for technical support. We are also grateful to Jens Fielitz for helpful discussions. D. Phan was supported by a NIH postdoctoral fellowship. This work was supported by grants from the NIH and the Donald W. Reynolds Cardiovascular Clinical Research Center to J.A. Hill and E.N. Olson and the Robert A. Welch Foundation to E.N. Olson.
Footnotes
Nonstandard abbreviations used: FS, fractional shortening; HW/TL, heart weight/tibia length; ISO, isoproterenol; LVIDd, LV end-diastolic diameter; LVIDs, LV end-systolic diameter; MEF2, myocyte enhancer factor–2; MHC, myosin heavy chain; NFAT, nuclear factor of activated T cells; TAC, thoracic aortic constriction.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 118:124–132 (2008). doi:10.1172/JCI33255
Dillon Phan’s present address is: Gilead Colorado Inc., Westminster, Colorado, USA.
Da-Zhi Wang’s present address is: Carolina Cardiovascular Biology Center, Department of Cell and Developmental Biology, University of North Carolina, Chapel Hill, North Carolina, USA.
Yuri Kim and Dillon Phan contributed equally to this work.
References
- 1.Czubryt M.P., Olson E.N. Balancing contractility and energy production: the role of myocyte enhancer factor 2 (MEF2) in cardiac hypertrophy. Recent Prog. Horm. Res. 2004;59:105–124. doi: 10.1210/rp.59.1.105. [DOI] [PubMed] [Google Scholar]
- 2.Frey N., Katus H.A., Olson E.N., Hill J.A. Hypertrophy of the heart: a new therapeutic target? Circulation. 2004;109:1580–1589. doi: 10.1161/01.CIR.0000120390.68287.BB. [DOI] [PubMed] [Google Scholar]
- 3.Frey N., Olson E.N. Cardiac hypertrophy: the good, the bad, and the ugly. Annu. Rev. Physiol. 2003;65:45–79. doi: 10.1146/annurev.physiol.65.092101.142243. [DOI] [PubMed] [Google Scholar]
- 4.McKinsey T.A., Olson E.N. Toward transcriptional therapies for the failing heart: chemical screens to modulate genes. J. Clin. Invest. 2005;115:538–546. doi: 10.1172/JCI24144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Backs J., Olson E.N. Control of cardiac growth by histone acetylation/deacetylation. Circ. Res. 2006;98:15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 6.Black B.L., Olson E.N. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu. Rev. Cell Dev. Biol. 1998;14:167–196. doi: 10.1146/annurev.cellbio.14.1.167. [DOI] [PubMed] [Google Scholar]
- 7.Molkentin J.D., et al. MEF2B is a potent transactivator expressed in early myogenic lineages. Mol. Cell. Biol. 1996;16:3814–3824. doi: 10.1128/mcb.16.7.3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edmondson D.G., Lyons G.E., Martin J.F., Olson E.N. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120:1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- 9.Martin J.F., et al. A Mef2 gene that generates a muscle-specific isoform via alternative mRNA splicing. Mol. Cell. Biol. 1994;14:1647–1656. doi: 10.1128/mcb.14.3.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naya F.J., et al. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat. Med. 2002;8:1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 11.Lin Q., Schwarz J., Bucana C., Olson E.N. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Passier R., et al. CaM kinase signaling induces cardiac hypertrophy and activates the MEF2 transcription factor in vivo. J. Clin. Invest. 2000;105:1395–1406. doi: 10.1172/JCI8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKinsey T.A., Zhang C.L., Lu J., Olson E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature. 2000;408:106–111. doi: 10.1038/35040593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vega R.B., et al. Protein kinases C and D mediate agonist-dependent cardiac hypertrophy through nuclear export of histone deacetylase 5. Mol. Cell. Biol. 2004;24:8374–8385. doi: 10.1128/MCB.24.19.8374-8385.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang S.H., Galanis A., Sharrocks A.D. Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol. Cell. Biol. 1999;19:4028–4038. doi: 10.1128/mcb.19.6.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu H., et al. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J. 2001;20:6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Molkentin J.D., et al. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Molkentin J.D., Black B.L., Martin J.F., Olson E.N. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- 19.Tallquist M.D., Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26:113–115. doi: 10.1002/(sici)1526-968x(200002)26:2<113::aid-gene3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 20.Zhang C.L., et al. Class II histone deacetylases act as signal-responsive repressors of cardiac hypertrophy. Cell. 2002;110:479–488. doi: 10.1016/s0092-8674(02)00861-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu J., et al. Myocyte enhancer factors 2A and 2C induce dilated cardiomyopathy in transgenic mice. J. Biol. Chem. 2006;281:9152–9162. doi: 10.1074/jbc.M510217200. [DOI] [PubMed] [Google Scholar]
- 22.van Oort R.J., et al. MEF2 activates a genetic program promoting chamber dilation and contractile dysfunction in calcineurin-induced heart failure. Circulation. 2006;114:298–308. doi: 10.1161/CIRCULATIONAHA.105.608968. [DOI] [PubMed] [Google Scholar]
- 23.van Rooij E., et al. A signature pattern of stress-responsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Rooij E., et al. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 25.Morin S., Charron F., Robitaille L., Nemer M. GATA-dependent recruitment of MEF2 proteins to target promoters. EMBO J. 2000;19:2046–2055. doi: 10.1093/emboj/19.9.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang Q., et al. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J. Biol. Chem. 2001;276:30245–30253. doi: 10.1074/jbc.M102174200. [DOI] [PubMed] [Google Scholar]
- 27.Song K., et al. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell. 2006;125:453–466. doi: 10.1016/j.cell.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 28.Arnold M.A., et al. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev. Cell. 2007;12:377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Potthoff M.J., et al. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J. Clin. Invest. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shalizi A., et al. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 31.Flavell S.W., et al. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 32.Lin Q., et al. Requirement of the MADS-box transcription factor MEF2C for vascular development. Development. 1998;125:4565–4574. doi: 10.1242/dev.125.22.4565. [DOI] [PubMed] [Google Scholar]
- 33.Du Y., Spence S.E., Jenkins N.A., Copeland N.G. Cooperating cancer-gene identification through oncogenic-retrovirus-induced insertional mutagenesis. Blood. 2005;106:2498–2505. doi: 10.1182/blood-2004-12-4840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L., Fan C., Topol S.E., Topol E.J., Wang Q. Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science. 2003;302:1578–1581. doi: 10.1126/science.1088477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., et al. Regulation of hepatic stellate cell activation and growth by transcription factor myocyte enhancer factor 2. Gastroenterology. 2004;127:1174–1188. doi: 10.1053/j.gastro.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Bai X., et al. Overexpression of myocyte enhancer factor 2 and histone hyperacetylation in hepatocellular carcinoma. J. Cancer Res. Clin. Oncol. 2007;134:83–91. doi: 10.1007/s00432-007-0252-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez C.I., et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 38.Hill J.A., et al. Cardiac hypertrophy is not a required compensatory response to short-term pressure overload. Circulation. 2000;101:2863–2869. doi: 10.1161/01.cir.101.24.2863. [DOI] [PubMed] [Google Scholar]
- 39.Gulick J., Subramaniam A., Neumann J., Robbins J. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J. Biol. Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.