Abstract
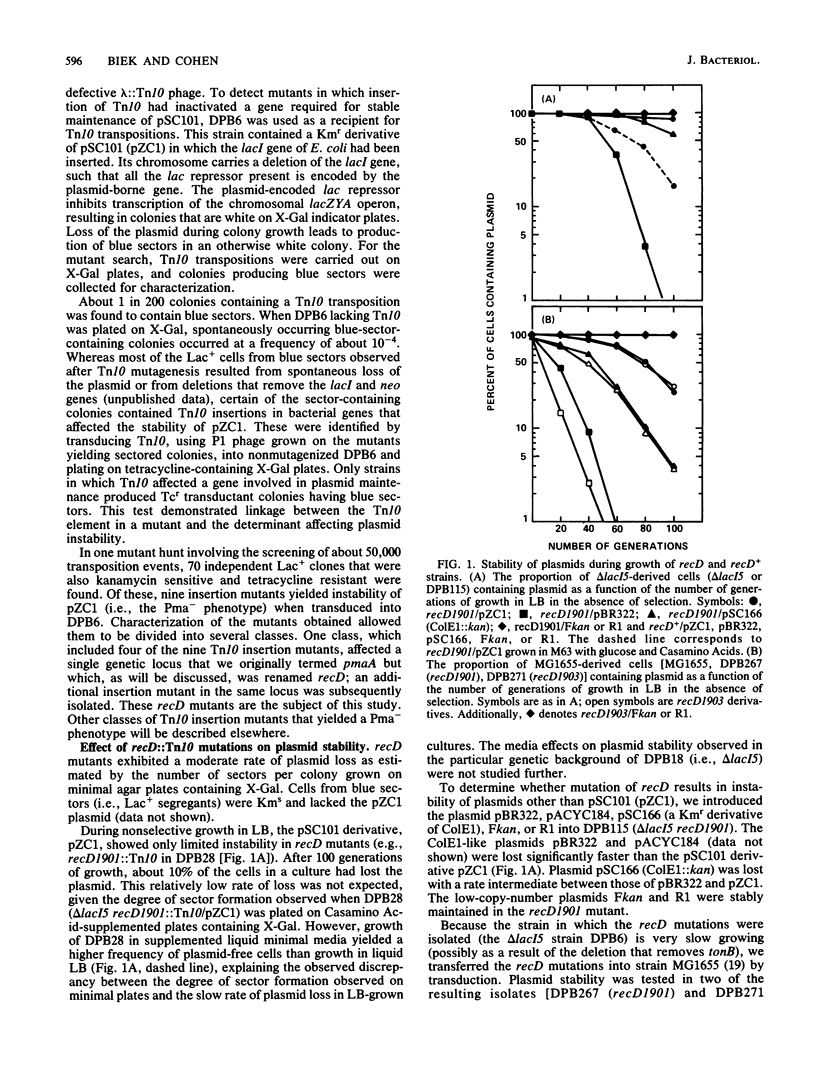
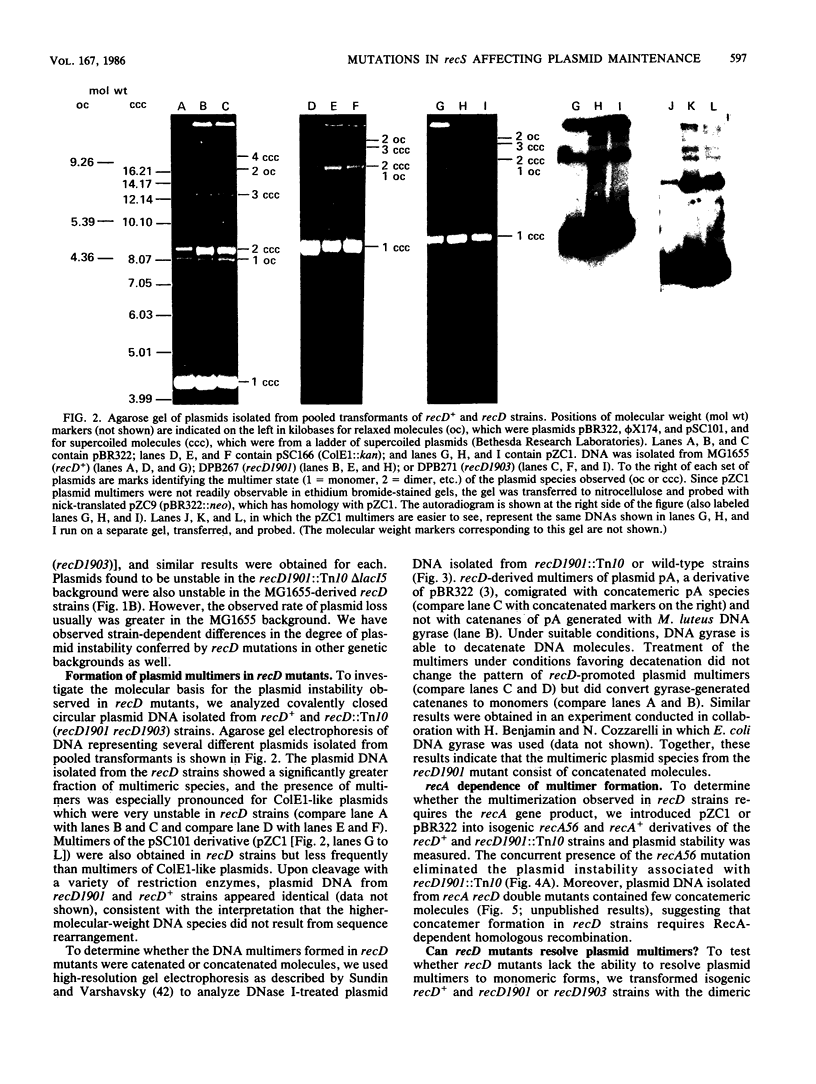
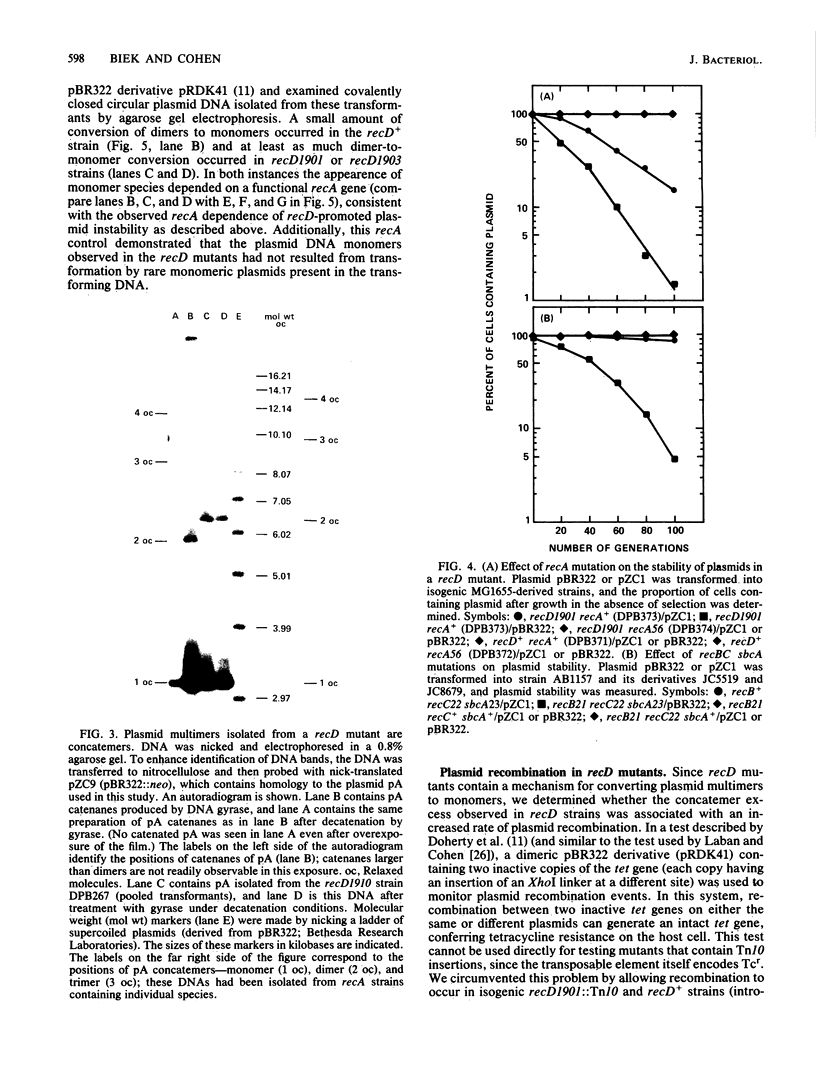
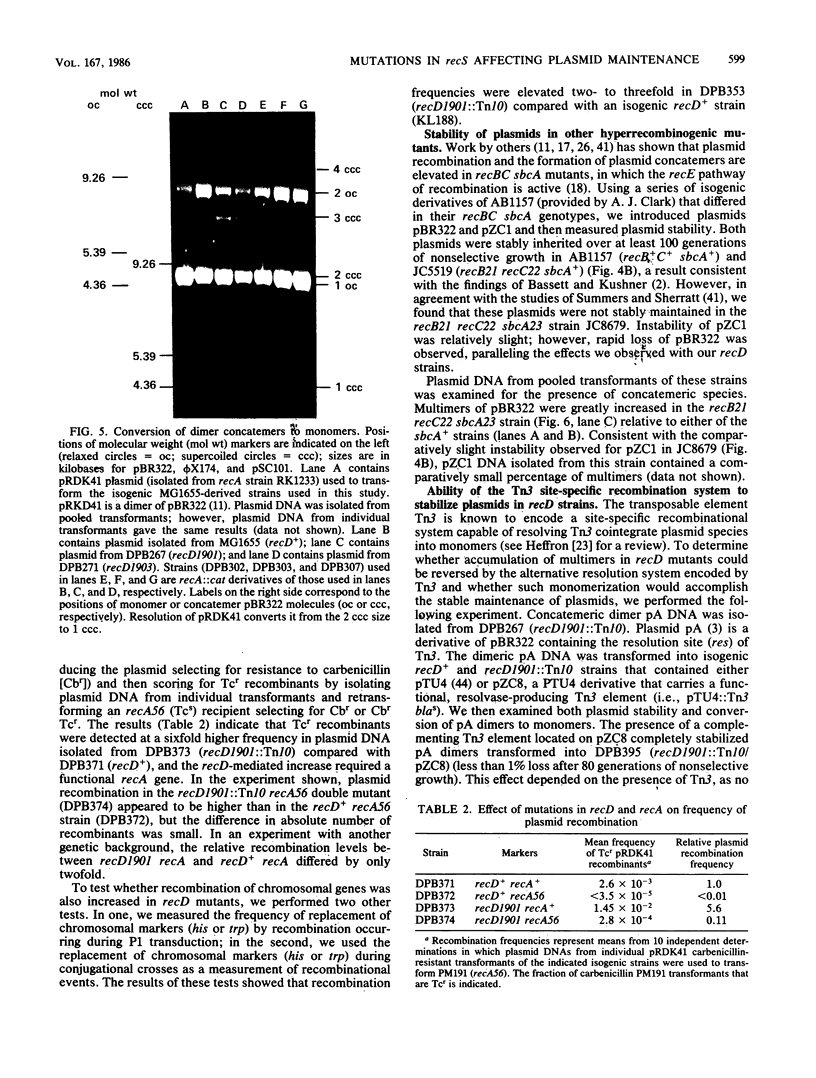
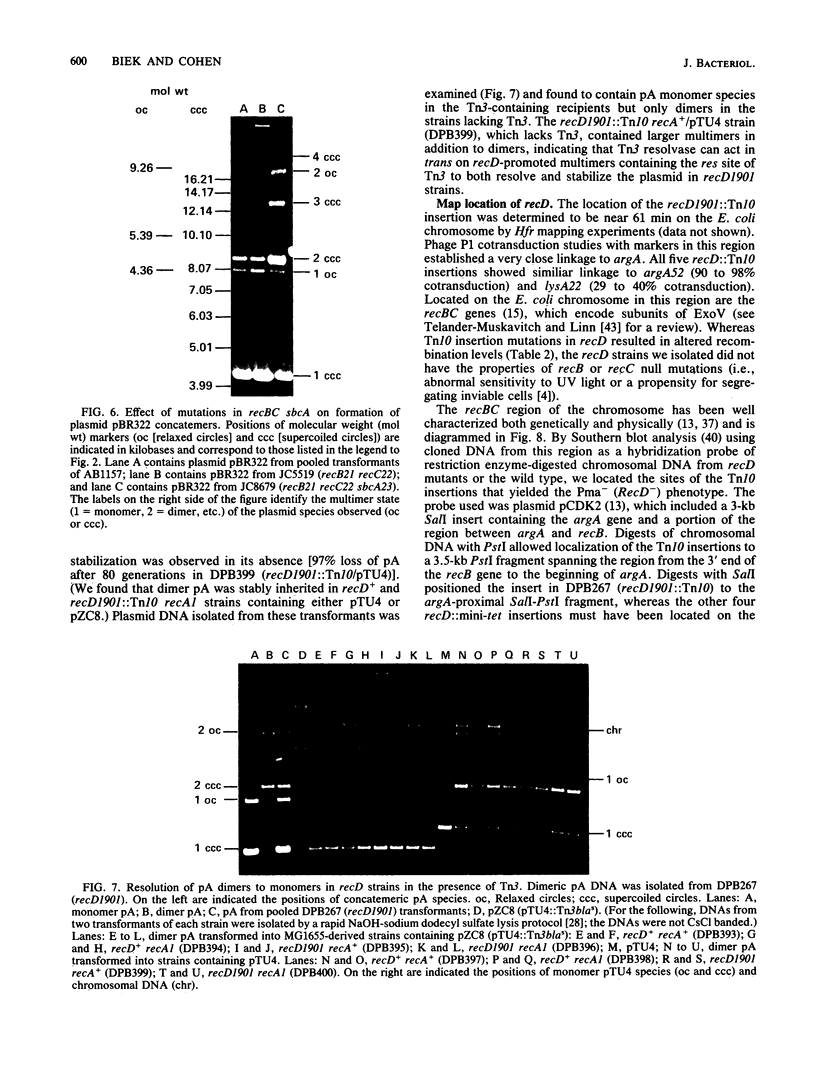
We isolated mutations that reduce plasmid stability in dividing cell populations and mapped these mutations to a previously undescribed gene, recD, that affects recombination frequency and consequently the formation of plasmid concatemers. Insertions of the transposable element Tn10 into recD resulted in increased concatemerization and loss of pSC101 and ColE1-like replicons during nonselective growth. Both concatemer formation and plasmid instability in recD mutants require a functional recA gene. Mutations in recD are recessive to recD+ and map to a small region of the Escherichia coli chromosome located between recB and argA. Although the recD locus is distinct from loci encoding the two previously identified subunits of the RecBC enzyme, mutations in recD appear to affect the exonuclease activity of this enzyme.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin S., Abeles A. Partition of unit-copy miniplasmids to daughter cells. I. P1 and F miniplasmids contain discrete, interchangeable sequences sufficient to promote equipartition. J Mol Biol. 1983 Sep 15;169(2):353–372. doi: 10.1016/s0022-2836(83)80055-2. [DOI] [PubMed] [Google Scholar]
- Austin S., Ziese M., Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell. 1981 Sep;25(3):729–736. doi: 10.1016/0092-8674(81)90180-x. [DOI] [PubMed] [Google Scholar]
- Bassett C. L., Kushner S. R. Exonucleases I, III, and V are required for stability of ColE1-related plasmids in Escherichia coli. J Bacteriol. 1984 Feb;157(2):661–664. doi: 10.1128/jb.157.2.661-664.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamin H. W., Matzuk M. M., Krasnow M. A., Cozzarelli N. R. Recombination site selection by Tn3 resolvase: topological tests of a tracking mechanism. Cell. 1985 Jan;40(1):147–158. doi: 10.1016/0092-8674(85)90318-6. [DOI] [PubMed] [Google Scholar]
- Capaldo-Kimball F., Barbour S. D. Involvement of recombination genes in growth and viability of Escherichia coli K-12. J Bacteriol. 1971 Apr;106(1):204–212. doi: 10.1128/jb.106.1.204-212.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A. M., Smith G. R. A new class of Escherichia coli recBC mutants: implications for the role of RecBC enzyme in homologous recombination. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7850–7854. doi: 10.1073/pnas.81.24.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Cohen A., Clark A. J. Synthesis of linear plasmid multimers in Escherichia coli K-12. J Bacteriol. 1986 Jul;167(1):327–335. doi: 10.1128/jb.167.1.327-335.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C. Revised interpretation of the origin of the pSC101 plasmid. J Bacteriol. 1977 Nov;132(2):734–737. doi: 10.1128/jb.132.2.734-737.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty M. J., Morrison P. T., Kolodner R. Genetic recombination of bacterial plasmid DNA. Physical and genetic analysis of the products of plasmid recombination in Escherichia coli. J Mol Biol. 1983 Jul 5;167(3):539–560. doi: 10.1016/s0022-2836(83)80097-7. [DOI] [PubMed] [Google Scholar]
- Dykstra C. C., Palas K. M., Kushner S. R. Purification and characterization of exonuclease V from Escherichia coli K-12. Cold Spring Harb Symp Quant Biol. 1984;49:463–467. doi: 10.1101/sqb.1984.049.01.052. [DOI] [PubMed] [Google Scholar]
- Dykstra C. C., Prasher D., Kushner S. R. Physical and biochemical analysis of the cloned recB and recC genes of Escherichia coli K-12. J Bacteriol. 1984 Jan;157(1):21–27. doi: 10.1128/jb.157.1.21-27.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely S., Wright A. Maintenance of plasmid pSC101 in Escherichia coli requires the host primase. J Bacteriol. 1985 Oct;164(1):484–486. doi: 10.1128/jb.164.1.484-486.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emmerson P. T., Howard-Flanders P. Cotransduction with thy of a gene required for genetic recombination in Escherichia coli. J Bacteriol. 1967 May;93(5):1729–1731. doi: 10.1128/jb.93.5.1729-1731.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enquist L. W., Skalka A. Replication of bacteriophage lambda DNA dependent on the function of host and viral genes. I. Interaction of red, gam and rec. J Mol Biol. 1973 Apr 5;75(2):185–212. doi: 10.1016/0022-2836(73)90016-8. [DOI] [PubMed] [Google Scholar]
- Fishel R. A., James A. A., Kolodner R. recA-independent general genetic recombination of plasmids. Nature. 1981 Nov 12;294(5837):184–186. doi: 10.1038/294184a0. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Rasmussen P. B., Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A. 1986 May;83(10):3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillen J. R., Willis D. K., Clark A. J. Genetic analysis of the RecE pathway of genetic recombination in Escherichia coli K-12. J Bacteriol. 1981 Jan;145(1):521–532. doi: 10.1128/jb.145.1.521-532.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyer M. S., Reed R. R., Steitz J. A., Low K. B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harb Symp Quant Biol. 1981;45(Pt 1):135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- Hakkaart M. J., van den Elzen P. J., Veltkamp E., Nijkamp H. J. Maintenance of multicopy plasmid Clo DF13 in E. coli cells: evidence for site-specific recombination at parB. Cell. 1984 Jan;36(1):203–209. doi: 10.1016/0092-8674(84)90090-4. [DOI] [PubMed] [Google Scholar]
- Hasunuma K., Sekiguchi M. Effect of dna mutations on the replication of plasmid pSC101 in Escherichia coli K-12. J Bacteriol. 1979 Mar;137(3):1095–1099. doi: 10.1128/jb.137.3.1095-1099.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasunuma K., Sekiguchi M. Replication of plasmid pSC101 in Escherichia coli K12: requirement for dnaA function. Mol Gen Genet. 1977 Sep 9;154(3):225–230. doi: 10.1007/BF00571277. [DOI] [PubMed] [Google Scholar]
- Jaffé A., Ogura T., Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985 Sep;163(3):841–849. doi: 10.1128/jb.163.3.841-849.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karu A. E., Sakaki Y., Echols H., Linn S. The gamma protein specified by bacteriophage gamma. Structure and inhibitory activity for the recBC enzyme of Escherichia coli. J Biol Chem. 1975 Sep 25;250(18):7377–7387. [PubMed] [Google Scholar]
- Laban A., Cohen A. Interplasmidic and intraplasmidic recombination in Escherichia coli K-12. Mol Gen Genet. 1981;184(2):200–207. doi: 10.1007/BF00272905. [DOI] [PubMed] [Google Scholar]
- Lieberman R. P., Oishi M. The recBC deoxyribonuclease of Escherichia coli: isolation and characterization of the subunit proteins and reconstitution of the enzyme. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4816–4820. doi: 10.1073/pnas.71.12.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meacock P. A., Cohen S. N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Miller C. A., Tucker W. T., Meacock P. A., Gustafsson P., Cohen S. N. Nucleotide sequence of the partition locus of Escherichia coli plasmid pSC101. Gene. 1983 Oct;24(2-3):309–315. doi: 10.1016/0378-1119(83)90091-4. [DOI] [PubMed] [Google Scholar]
- Nordström K., Molin S., Aagaard-Hansen H. Partitioning of plasmid R1 in Escherichia coli. I. Kinetics of loss of plasmid derivatives deleted of the par region. Plasmid. 1980 Sep;4(2):215–227. doi: 10.1016/0147-619x(80)90011-6. [DOI] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983 Feb;32(2):351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- Oliver D. B., Goldberg E. B. Protection of parental T4 DNA from a restriction exonuclease by the product of gene 2. J Mol Biol. 1977 Nov;116(4):877–881. doi: 10.1016/0022-2836(77)90276-5. [DOI] [PubMed] [Google Scholar]
- Peterson B. C., Hashimoto H., Rownd R. H. Cointegrate formation between homologous plasmids in Escherichia coli. J Bacteriol. 1982 Sep;151(3):1086–1094. doi: 10.1128/jb.151.3.1086-1094.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss B., Sprengel R., Schaller H. Protein fusions with the kanamycin resistance gene from transposon Tn5. EMBO J. 1984 Dec 20;3(13):3317–3322. doi: 10.1002/j.1460-2075.1984.tb02297.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Fujiyoshi T., Shimada K., Takagi Y. Fine structure of the recB and recC gene region of Escherichia coli. Biochem Biophys Res Commun. 1982 Nov 30;109(2):414–422. doi: 10.1016/0006-291x(82)91737-5. [DOI] [PubMed] [Google Scholar]
- Schmeissner U., Ganem D., Miller J. H. Genetic studies of the lac repressor. II. Fine structure deletion map of the lacI gene, and its correlation with the physical map. J Mol Biol. 1977 Jan 15;109(2):303–326. doi: 10.1016/s0022-2836(77)80036-3. [DOI] [PubMed] [Google Scholar]
- Scott J. R. Regulation of plasmid replication. Microbiol Rev. 1984 Mar;48(1):1–23. doi: 10.1016/b978-0-12-048850-6.50006-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Summers D. K., Sherratt D. J. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984 Apr;36(4):1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- Sundin O., Varshavsky A. Arrest of segregation leads to accumulation of highly intertwined catenated dimers: dissection of the final stages of SV40 DNA replication. Cell. 1981 Sep;25(3):659–669. doi: 10.1016/0092-8674(81)90173-2. [DOI] [PubMed] [Google Scholar]
- Tu C. P., Cohen S. N. Translocation specificity of the Tn3 element: characterization of sites of multiple insertions. Cell. 1980 Jan;19(1):151–160. doi: 10.1016/0092-8674(80)90396-7. [DOI] [PubMed] [Google Scholar]
- Tucker W. T., Miller C. A., Cohen S. N. Structural and functional analysis of the par region of the pSC 10 1 plasmid. Cell. 1984 Aug;38(1):191–201. doi: 10.1016/0092-8674(84)90540-3. [DOI] [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]