Abstract
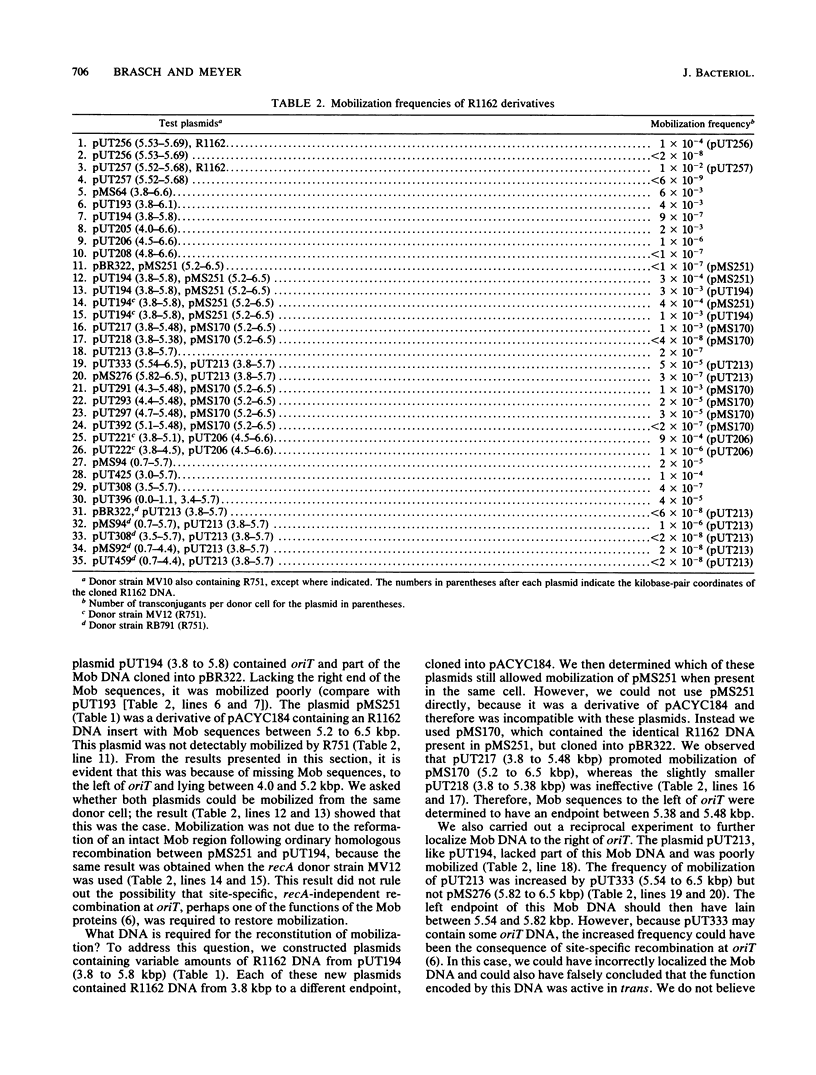
DNA involved in the mobilization of broad-host-range plasmid R1162 was localized to a region of 2.7 kilobases within coordinates 3.4 to 6.1 kilobases on the R1162 map. By examining the transfer properties of plasmids containing cloned fragments of DNA from within this region, we showed that at least four trans-active products and a cis-active site (oriT) were involved in mobilization. A cloned DNA fragment of 155 base pairs was capable of providing full oriT activity. This fragment was located within 600 base pairs of DNA containing the origin of replication of R1162, and its nucleotide sequence and that of neighboring DNA were determined. Activation of oriT required R1162-encoded, trans-acting products. Deletions which resulted in the loss of one or more of these had a variable effect on transfer efficiency and indicated the presence of both essential and nonessential Mob products. Regions encoding these products flanked oriT and in one case appeared to overlap a gene essential for plasmid replication. The implications of these findings with respect to the broad host range of R1162 are discussed.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amann E., Brosius J., Ptashne M. Vectors bearing a hybrid trp-lac promoter useful for regulated expression of cloned genes in Escherichia coli. Gene. 1983 Nov;25(2-3):167–178. doi: 10.1016/0378-1119(83)90222-6. [DOI] [PubMed] [Google Scholar]
- Barth P. T., Grinter N. J. Comparison of the deoxyribonucleic acid molecular weights and homologies of plasmids conferring linked resistance to streptomycin and sulfonamides. J Bacteriol. 1974 Nov;120(2):618–630. doi: 10.1128/jb.120.2.618-630.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastia D. Determination of restriction sites and the nucleotide sequence surrounding the relaxation site of ColE1. J Mol Biol. 1978 Oct 5;124(4):601–639. doi: 10.1016/0022-2836(78)90174-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Boulnois G. J., Wilkins B. M., Lanka E. Overlapping genes at the DNA primase locus of the large plasmid ColI. Nucleic Acids Res. 1982 Feb 11;10(3):855–869. doi: 10.1093/nar/10.3.855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan G., Sherratt D. The transposon Tn1 as a probe for studying ColE1 structure and function. Mol Gen Genet. 1977 Mar 7;151(2):151–160. doi: 10.1007/BF00338689. [DOI] [PubMed] [Google Scholar]
- Everett R., Willetts N. Characterisation of an in vivo system for nicking at the origin of conjugal DNA transfer of the sex factor F. J Mol Biol. 1980 Jan 15;136(2):129–150. doi: 10.1016/0022-2836(80)90309-5. [DOI] [PubMed] [Google Scholar]
- Figurski D., Meyer R., Miller D. S., Helinski D. R. Generation in vitro of deletions in the broad host range plasmid RK2 using phage Mu insertions and a restriction endonuclease. Gene. 1976;1(1):107–119. doi: 10.1016/0378-1119(76)90010-x. [DOI] [PubMed] [Google Scholar]
- Grinter N. J., Barth P. T. Characterization of SmSu plasmids by restriction endonuclease cleavage and compatibility testing. J Bacteriol. 1976 Oct;128(1):394–400. doi: 10.1128/jb.128.1.394-400.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiney D. G., Helinski D. R. Relaxation complexes of poasmid DNA and protein. III. Association of protein with the 5' terminus of the broken DNA strand in the relaxed complex of plasmid ColE1. J Biol Chem. 1975 Nov 25;250(22):8796–8803. [PubMed] [Google Scholar]
- Guiney D. G., Helinski D. R. The DNA-protein relaxation complex of the plasmid RK2: location of the site-specific nick in the region of the proposed origin of transfer. Mol Gen Genet. 1979 Oct 3;176(2):183–189. doi: 10.1007/BF00273212. [DOI] [PubMed] [Google Scholar]
- Hershfield V., Boyer H. W., Yanofsky C., Lovett M. A., Helinski D. R. Plasmid ColEl as a molecular vehicle for cloning and amplification of DNA. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3455–3459. doi: 10.1073/pnas.71.9.3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Inselburg J., Applebaum B. Proteins synthesized in minicells containing plasmid ColE1 and its mutants. J Bacteriol. 1978 Mar;133(3):1444–1451. doi: 10.1128/jb.133.3.1444-1451.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobanputra R. S., Datta N. Trimethoprim R factors in enterobacteria from clinical specimens. J Med Microbiol. 1974 May;7(2):169–177. doi: 10.1099/00222615-7-2-169. [DOI] [PubMed] [Google Scholar]
- Lanka E., Barth P. T. Plasmid RP4 specifies a deoxyribonucleic acid primase involved in its conjugal transfer and maintenance. J Bacteriol. 1981 Dec;148(3):769–781. doi: 10.1128/jb.148.3.769-781.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanka E., Lurz R., Kröger M., Fürste J. P. Plasmid RP4 encodes two forms of a DNA primase. Mol Gen Genet. 1984;194(1-2):65–72. doi: 10.1007/BF00383499. [DOI] [PubMed] [Google Scholar]
- Lin L. S., Meyer R. J. Nucleotide sequence and functional properties of DNA encoding incompatibility in the broad host-range plasmid R1162. Mol Gen Genet. 1984;194(3):423–431. doi: 10.1007/BF00425554. [DOI] [PubMed] [Google Scholar]
- Lovett M. A., Helinski D. R. Relaxation complexes of plasmid DNA and protein. II. Characterization of the proteins associated with the unrelaxed and relaxed complexes of plasmid ColE1. J Biol Chem. 1975 Nov 25;250(22):8790–8795. [PubMed] [Google Scholar]
- Marko M. A., Chipperfield R., Birnboim H. C. A procedure for the large-scale isolation of highly purified plasmid DNA using alkaline extraction and binding to glass powder. Anal Biochem. 1982 Apr;121(2):382–387. doi: 10.1016/0003-2697(82)90497-3. [DOI] [PubMed] [Google Scholar]
- Messing J., Vieira J. A new pair of M13 vectors for selecting either DNA strand of double-digest restriction fragments. Gene. 1982 Oct;19(3):269–276. doi: 10.1016/0378-1119(82)90016-6. [DOI] [PubMed] [Google Scholar]
- Meyer R. J., Lin L. S., Kim K., Brasch M. A. Broad host-range plasmid R1162: replication, incompatibility, and copy-number control. Basic Life Sci. 1985;30:173–188. doi: 10.1007/978-1-4613-2447-8_16. [DOI] [PubMed] [Google Scholar]
- Meyer R., Hinds M., Brasch M. Properties of R1162, a broad-host-range, high-copy-number plasmid. J Bacteriol. 1982 May;150(2):552–562. doi: 10.1128/jb.150.2.552-562.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris C. F., Hershberger C. L., Rownd R. Strand-specific nick in open circular R-factor deoxyribonucleic acid: attachment of the linear strand to a proteinaceous cellular component. J Bacteriol. 1973 Apr;114(1):300–308. doi: 10.1128/jb.114.1.300-308.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordheim A., Hashimoto-Gotoh T., Timmis K. N. Location of two relaxation nick sites in R6K and single sites in pSC101 and RSF1010 close to origins of vegetative replication: implication for conjugal transfer of plasmid deoxyribonucleic acid. J Bacteriol. 1980 Dec;144(3):923–932. doi: 10.1128/jb.144.3.923-932.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. J., Saul M. W., Sherratt D. J. ColE1 plasmid mobility: essential and conditional functions. Mol Gen Genet. 1979 Feb 16;170(1):103–107. doi: 10.1007/BF00268585. [DOI] [PubMed] [Google Scholar]
- Warren G. J., Twigg A. J., Sherratt D. J. ColE1 plasmid mobility and relaxation complex. Nature. 1978 Jul 20;274(5668):259–261. doi: 10.1038/274259a0. [DOI] [PubMed] [Google Scholar]
- Willetts N., Skurray R. The conjugation system of F-like plasmids. Annu Rev Genet. 1980;14:41–76. doi: 10.1146/annurev.ge.14.120180.000353. [DOI] [PubMed] [Google Scholar]
- Willetts N., Wilkins B. Processing of plasmid DNA during bacterial conjugation. Microbiol Rev. 1984 Mar;48(1):24–41. doi: 10.1128/mr.48.1.24-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E. M., Muesing M. A., Polisky B. Temperature-sensitive copy number mutants of CoIE1 are located in an untranslated region of the plasmid genome. Proc Natl Acad Sci U S A. 1982 Jun;79(11):3570–3574. doi: 10.1073/pnas.79.11.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]



