Abstract
The principal cause of bacterial resistance to penicillin and other β-lactam antibiotics is the acquisition of plasmid-encoded β-lactamases, enzymes that catalyze hydrolysis of the β-lactam bond and render these antibiotics inactive. Clavulanic acid is a potent inhibitor of β-lactamases and has proven clinically effective in combating resistant infections. Although clavulanic acid and penicillin share marked structural similarities, the biosyntheses of their bicyclic nuclei are wholly dissimilar. In contrast to the efficient iron-mediated oxidative cyclization of a tripeptide to isopenicillin N, the critical β-lactam ring of clavulanic acid is demonstrated to form by intramolecular closure catalyzed by a new type of ATP/Mg2+-dependent enzyme, a β-lactam synthetase (β-LS). Insertional inactivation of its encoding gene in wild-type Streptomyces clavuligerus resulted in complete loss of clavulanic acid production and the accumulation of N2-(carboxyethyl)-l-arginine (CEA). Chemical complementation of this blocked mutant with authentic deoxyguanidinoproclavaminic acid (DGPC), the expected product of the β-LS, restored clavulanic acid synthesis. Finally, overexpression of this gene gave the β-LS, which was shown to mediate the conversion of CEA to DGPC in the presence of ATP/Mg2+. Primary amino acid sequence comparisons suggest that this mode of β-lactam formation could be more widely spread in nature and mechanistically related to asparagine synthesis.
Keywords: β-lactam antibiotics/biosynthesis/clavulanic acid/amidotransferases
Penicillin and related β-lactam antibiotics have been a mainstay in the treatment of infections for 50 years. Their effectiveness in medicine, however, like other classes of known antibiotics, has come under increasing challenge from the rise of multiply drug-resistant pathogenic bacteria (1, 2). Prominant among the resistance mechanisms to penicillin and other β-lactams are the β-lactamases, which hydrolyze these antibiotics and disarm their ability to inhibit bacterial cell wall biosynthesis. Clavulanic acid is a naturally occurring inhibitor of β-lactamase enzymes, whose value in combating antibiotic resistance has been demonstrated clinically (3). A new mechanism of β-lactam biosynthesis has been identified in the clavulanate pathway, which may play an analogous role in the formation of the carbapenems, another clinically important family of these antibiotics. Application of these enzymes in the preparation of these and other antibiotics may be envisioned.
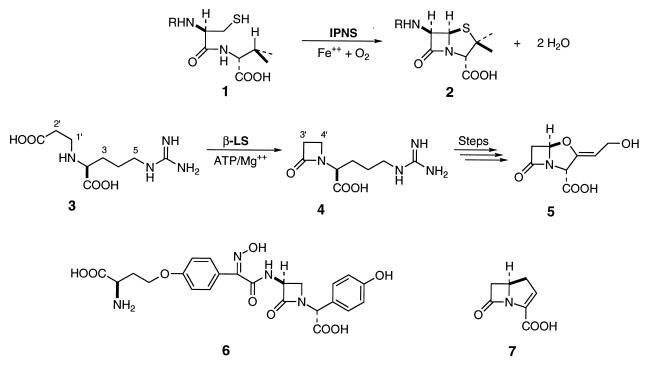
As largely the products of fermentation, mechanistic and practical interests have given impetus to the study and genetic manipulation of antibiotic biosynthesis (4–7). Great effort has been focused on understanding the formation of these important natural products and to identifying the genes that encode their biosynthetic pathways. The β-lactam antibiotic family can be divided into at least four known groups, of which the biosynthetic study of penicillin N (Fig. 1, 2) is by far the most advanced. A tripeptide precursor 1 is produced by a nonribosomal peptide synthetase (8, 9), which is cyclized with impressive efficiency by isopenicillin N synthase in the presence of ferrous ion and molecular oxygen to isopenicillin N (Fig. 1, 2; subsequent boldface numbers will refer to corresponding sections of figure). Research has culminated recently in the x-ray crystal structure of isopenicillin N synthase with iron and substrate bound (10). The structural features of this complex, in addition to a vast base of earlier experimental data, have led to the proposal of a detailed catalytic mechanism involving successive oxidative cyclization reactions mediated by iron and oxygen resulting in the formation of two rings and two molecules of water (10, 11).
Figure 1.
Reaction of isopenicillin N synthase, an overview of clavulanic acid biosynthesis and representative members of the β-lactam antibiotic classes. In penicillin biosynthesis, δ-(l-aminoadipoyl)-l-cysteinyl-d-valine (ACV) (1) is cyclized to isopenicillin N (2) by isopenicillin N synthase in the presence of ferrous ion and molecular oxygen. Clavulanic acid biosynthesis differs in that N2-(2-carboxyethyl)arginine (CEA) (3) cyclizes first to the monocyclic β-lactam deoxyguanidinoproclavaminate (DGPC) (4) mediated by β-lactam synthetase (β-LS) and ATP/Mg2+. Several subsequent transformations of this molecule are required to form clavulanic acid (5). The penam/cephem group is represented here by isopenicillin N (2), the clavams by clavulanic acid (5), the monocyclic β-lactams by nocardicin A (6), and the carbapenems by carbapenem-3-carboxylic acid (7).
The highly efficacious β-lactamase inhibitor clavulanic acid (5) bears strong structural similarity to isopenicillin N (2), but its primary modes of biochemical activity and synthesis differ sharply (3). Unlike the economical two-step biosynthesis of isopenicillin N (2), the fused bicyclic motif in clavulanic acid is assembled in at least eight steps from the building blocks of primary metabolism. The earliest proposed intermediate in clavulanic acid biosynthesis is CEA (3) (12, 13). An isotopically labeled sample of this material was shown to give an intact, albeit low, incorporation into 5 when administered to a fermentation of Streptomyces clavuligerus. Similarly, the monocyclic β-lactam 4 (DGPC) was prepared in labeled form and analogously incorporated into clavulanic acid, suggesting a precursor relationship of the former to the latter.
In this paper we describe the first evidence that the transformation of CEA (3) to DGPC (4) is catalyzed by a single enzyme in the presence of ATP/Mg2+. This intramolecular amide bond formation is previously unknown and has led to the designation of a β-LS. The β-LS is encoded by orf3, a component of the clavulanic acid gene cluster (14) having deduced amino acid similarity to a subset of the amidotransferases (15). It is located approximately 1 kb upstream of orf5, which encodes clavaminate synthase (16), a later enzyme in the pathway responsible for formation of the bicyclic nucleus of clavulanic acid (17). Targeted disruption of orf3 in the wild-type S. clavuligerus led to complete blocking of clavulanic acid production and the accumulation of CEA. Chemical complementation of orf3 mutants with authentic DGPC was shown to restore clavulanic acid production. Finally, separate overexpression of orf3 afforded β-LS, which was demonstrated to catalyze the in vitro conversion of CEA (3) to DGPC (4) in the presence of ATP/Mg2+.
EXPERIMENTAL PROCEDURES
Gene Disruption.
All restriction enzymes were purchased from New England Biolabs. Plasmid pLRF16 contained a 2.0-kb blunt-ended KpnI–BglII S. clavuligerus chromosomal fragment, comprising the full orf3 gene with a 5′ flanking region, in EcoRV digested pKC1139 (18). The thiostrepton resistance gene (tsr) in pIJ680 (19) was excised as a 1.08-kb BclI–BclI fragment and inserted into the unique internal NcoI site of orf3 by blunt-ended ligation. Plasmid pLRF25, used for the disruption of orf3 gene, was prepared by insertion of the inactivated orf3 into the replicationally unstable vector pLRF66 derived from pIJ680. The resulting plasmid was used to transform S. clavuligerus protoplasts. The transformants containing pLRF25 were subjected to two rounds of sporulation in the absence of antibiotic selection. These spores were grown on thiostrepton-containing plates and then replicated on neomycin-containing plates. The resulting TsrR NeoS colonies were presumptive gene replacement integrants.
Southern Hybridization.
Genomic DNA was completely digested with SphI, BclI, or KpnI–SacI and the fragments, separated by agarose gel electrophoresis, were transferred to nylon membranes. [α-32P]dCTP was used to label DNA probes by the random priming method. Hybridization was carried out for 16 hr at 55°C in a solution containing 5× SSC buffer, 50% formamide, 5× Denhardt’s solution, and 1% SDS (20).
Analysis of Antibiotics.
Bioassays were conducted by the method of agar plate diffusion with appropriate indicator organisms seeded on nutrient agar. Clavulanic acid was detected by β-lactamase inhibition by using Klebsiella pneumoniae subsp. pneumoniae (ATCC 29665) and benzylpenicillin (21), whereas penicillin, cephalosporin, and cephamycin were detected with Escherichia coli SC 12155 (22). Clavulanic acid was also detected by reaction with imidazole, as described (23, 24).
Isolation of CEA.
A 0.5 liter culture of mutant RFL35 was grown in SA medium (25) containing 10 μg/ml thiostrepton for 100 hr. The fermentation broth was centrifuged to remove cells, lyophilized, and taken up in 75% ethanol to remove high molecular weight biomolecules. The extract was then passed through an Amberlite IRA-68 (Aldrich) column to remove anionic impurities followed by lyophilization. The cationic compounds in this lyophilizate were then isolated by solid phase extraction over SCX preparative filter columns (Varian). The extract was purified further by C-18 HPLC chromatography by using a Prodigy 5μ ODS(3) column (250 × 10 mm; Phenomenex, Torrance, CA) with water as eluant.
Sakaguchi Analysis.
Guanidino-containing compounds were detected in cultures of mutants and during product isolation by employing modified Sakaguchi conditions (12, 26). Fifty microliters of sample was incubated with 50 ml of 0.1% 8-hydroxyquinoline for one min. Fifty microliters of 0.2% bromine/0.5 N NaOH was added, followed immediately by 350 ml distilled, deionized H2O. Absorbance at 500 nm was correlated with the presence of the guanidino function by construction of a standard curve by using arginine.
Overproduction of β-LS.
β-LS was cloned into the pET-24a(+) (Novagen) vector and was used to transform BL21(DE3) cells. Cell paste was disrupted by French press and DNA was precipitated by the addition of KCl and MnCl2 to final concentrations of 100 mM and 30 mM, respectively. After dialysis, the cell extract was purified by Q-Sepharose (Sigma) chromatography by using a 0–500 mM KCl gradient. β-LS activity in column fractions was assayed by monitoring ATP hydrolysis in the presence of CEA as described below. Fractions containing β-LS activity eluted at 280 mM KCl and corresponded to the induced protein by SDS/PAGE. Pooled fractions were concentrated to 3 ml in an Amicon ultrafiltration cell fitted with a PM10 membrane.
ATP hydrolysis was monitored by using the EnzCheck pyrophosphate assay kit (Molecular Probes). In this assay, PPi is converted to Pi by inorganic pyrophosphatase. 2-Amino-6-mercapto-7-methylpurine ribonucleoside is enzymatically phosphorylated to the spectrophotometrically distinct 2-amino-6-mercapto-7-methylpurine, resulting in a shift of the absorbance maximum from 330 to 360 nm.
In Vitro Reaction.
Enzymatic reactions of CEA [1H NMR (400 MHz, D2O) δ 1.4–1.6 (sym m, 2H, H-4), 1.83 (m, 2H, H-3), 2.69 (t, J = 6.7 Hz, 2H, C-2′), 3.06 (t, J = 7.3 Hz, 2H, H-5), 3.20 (t, J = 6.7 Hz, 2H, C-1′), 3.80 (dd, J = 5.0, 7.2 Hz, 1H, H-2)] and partially purified β-LS were carried out in 100 mM Pipes buffer (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 10 mM ATP, and 6 mM CEA. After 3 hr reaction, the enzyme was removed by centrifugal filtration (UltraFree 10,000 MWCO; Millipore) and purified by C-18 HPLC on a Phenomenex Prodigy 5μ ODS(3) column (250 × 10 mm) with 50 mM ammonium bicarbonate as eluant (5 ml/min). DGPC (27) had a retention time of 13.75 min under these conditions.
RESULTS
Construction of Disruption Mutant.
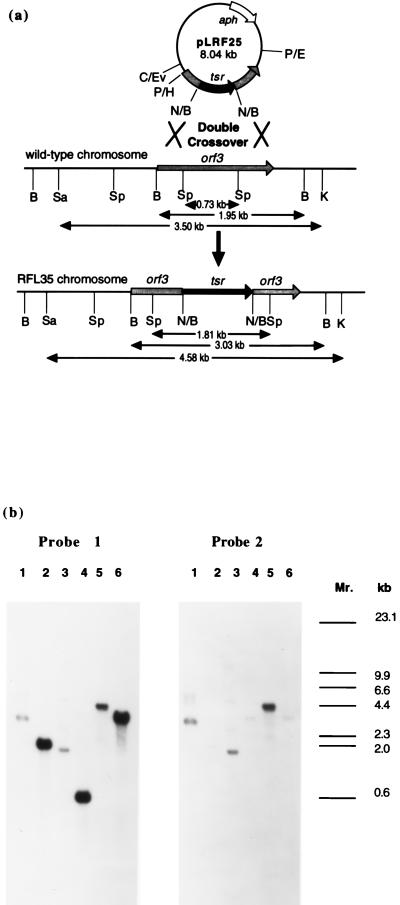
To access the role of Orf 3 in clavulanic acid biosynthesis, the corresponding gene was disrupted by inserting the thiostrepton resistance gene (tsr) into the unique internal NcoI site. S. clavuligerus was transformed with the corresponding plasmid, pRFL25, which also conferred neomycin resistance. Only colonies found to be resistant to thiostrepton and sensitive to neomycin were studied as potential double crossover products between the chromosomal orf3 and the disrupted copy from pLRF25. Successful gene replacement in one of the TsrR NeoS integrant strains, S. clavigerus RFL35, was confirmed by Southern hybridization analysis (Fig. 2). Orf3 probes consistently showed hybridization to fragments from digested RFL35 DNA, which were 1.1 kb longer than those observed in screening wild-type digestions indicating insertion of the tsr gene. The tsr probe selected for fragments of exactly the same size as those obtained with the orf3 probe, but tsr hybridizing fragments were absent in the wild-type chromosomal screening. These results demonstrated that in RFL35 the native orf3 had been replaced by the disrupted copy in a double crossover event.
Figure 2.
(a) Restriction map showing the predicted result of a double crossover between homologous regions of orf3 in pLRF25 and the S. clavuligerus chromosome. Solid box represents the tsr gene, open box represents the neomycin resistance gene, and the crosshatched box represents orf3. Abbreviations: B, BclI; C, ClaI; E, EcoRI; Ev, EcoRV; H, HindIII; K, KpnI; N, NcoI; P, PvuII; Sa, SacI; Sp, SphI. (b) Southern hybridization of S. clavuligerus chromosomal DNA from RFL35 and wild type with orf3-specific probe (probe 1) and tsr-specific probe (probe 2, after striping gel of probe 1). Lanes: 1, RFL35/BclI; 2, WT/BclI; 3, RFL35/SphI; 4, WT/SphI; 5, RFL35/KpnI–SacI; 6, WT/KpnI–SacI; Mr, λ/HindIII.
Analysis of Mutant RFL35.
Cultures of S. clavuligerus RFL35 were grown in SA fermentation medium (25) and analyzed for the production of clavulanic acid both by bioassay (28) and by reaction with imidazole (23, 24). No clavulanic acid was detectable in the supernatants of RFL35 cultures. To ensure that the double crossover mutation had no deleterious effect on other aspects of secondary metabolism, bioassay with the β-lactam supersensitive E. coli SC 12155 (22) was performed. This experiment showed that RFL35 exhibited the same levels of penicillin and cephamycin production as did the wild type, confirming that the disruption of orf3 had no effect on their production.
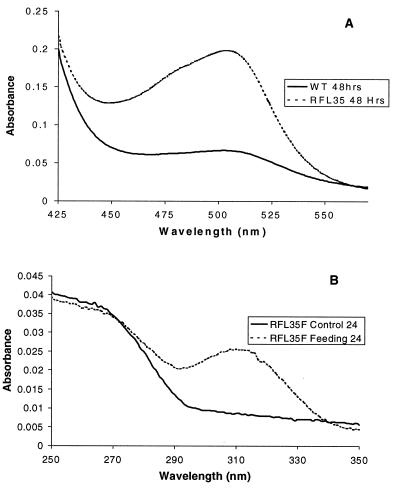
To establish the role of the orf3 gene product in clavulanic acid biosynthesis, chemical complementation of orf3 disruption mutants with DGPC (4) was carried out. Synthetic DGPC, the hypothetical product of Orf 3, was fed to 24-hr cultures of RFL35 at a final concentration of 2 mM. Clavulanic acid production was then followed over the course of several days by imidazole assay and bioassay to determine whether the addition of DGPC restored the clavulanic acid pathway. After 24 hr, clavulanic acid (5) was clearly seen in the supplemented RFL35 and absent in a RFL35 control culture (Fig. 3). Indeed, at no time was any clavulanic acid production detected in the control cultures. However, the control cultures of RFL35 were observed to accumulate large quantities (1.8–2.8 mM) of a metabolite containing a guanidino functional group, as determined by a modified Sakaguchi color reaction (12, 26). The level of production of Sakaguchi-positive materials was at least three times higher than in the wild type. Because DGPC (4) was presumably not present in the cells of RFL35, as shown by the chemical supplementation experiments, the accumulated metabolite was believed to be CEA (3). In an attempt to confirm this proposal, an extract was prepared from a 60-hr fermentation of S. clavuligerus RFL35. The extract was purified further by IRA 68 anion exchange and Varian SCX cation exchange chromatography, followed by reversed-phase HPLC purification on a C18 column. By comparison with an authentic sample, 1H NMR spectroscopy showed the major metabolite to be CEA. Although ion exchange conditions do not preclude the possibility that the isolated CEA arose from hydrolysis of DGPC, our chemical complementation experiments would suggest that this is not the case.
Figure 3.
Colorimetric assays. Accumulated intermediates in RFL35 vs. wild-type (WT) strains are evident by Sakaguchi color reaction in A. (B) Chemical complementation of RFL35 with deoxyguanidinoproclavaminate, visible by imidazole assay in as little as 24 hr.
Expression of β-LS and Characterization.
Orf3 was cloned into the pET-24a(+) expression vector and transformed into E. coli BL21(DE3). The overproduced protein was substantially purified by Q-Sepharose ion exchange chromatography. Owing to its homology to Class B asparagine synthases (AS-B, vide infra), activity of the partially purified protein was assayed for the ability to hydrolyze ATP in the presence of CEA (3) by using reaction conditions modeled on those of AS-B (29). For this purpose, the EnzCheck pyrophosphate assay kit (Molecular Probes) was used. Greater than 50 times the background rate of ATP hydrolysis was observed in the presence of CEA. Omission of inorganic pyrophosphatase in this assay gave only basal levels of phosphate, indicating that the protein overproduced from orf3 catalyzed the hydrolysis of ATP to specifically release pyrophosphate in the presence of the putative substrate, CEA. ATP hydrolysis was shown to be dependent on CEA concentration and independent of other added cofactors. These experiments established that β-LS uses CEA as a substrate in the presence of ATP/Mg2+, and supported the proposed role of the enzyme in the biosynthetic pathway.
To establish unambiguously the identity of the product, the in vitro reaction of overproduced β-LS with CEA in the presence of ATP/Mg2+ was followed by reversed-phase HPLC chromatography on a C18 column. Compared with controls, a new peak was observed, which increased in a time-dependent manner and coeluted with a standard sample of DGPC. 1H-NMR analysis of this new product showed it to correspond to DGPC (4) (27).
DISCUSSION
Orf3 encodes a 54.5 kDa protein whose amino acid sequence analysis shows up to 33% identity and 49% similarity (blast 2) with the primary metabolic class of amidotransferases (15) known as the asparagine synthetases, Class B (AS-B) (30). AS-B catalyzes the conversion of aspartic acid to asparagine in the presence of ATP/Mg2+, transferring a nitrogen from either glutamine or ammonia. The mechanism of glutamine nitrogen transfer mediated by asparagine synthetase has not been fully elucidated, but it is clear that the reaction pathway proceeds through a β-aspartyl-AMP intermediate (31), which subsequently interacts with the glutamyl nitrogen in an undetermined fashion distinct from glutamine phosphoribosylpyrophosphate amidotransferase (32). A covalently linked glutamine adduct has been proposed as a hypothetical intermediate (33). Highly conserved N-terminal cysteine residues, shown to be essential for glutamine-dependent asparagine synthetase activity, have been postulated to attack the carboxyamide to generate a nucleophilic amino group for transfer and yield a thioacyl enzyme intermediate, which subsequently reacts with water to give glutamate and the resting state of the enzyme (15, 33). In the event, because the conversion of CEA to DGPC could be considered an intramolecular amide-forming reaction, the homology of the protein encoded by orf3 (β-LS) to asparagine synthetase suggested its possible role in clavulanic acid biosynthesis. β-LS showed similarity in several regions to asparagine synthetase, particularly in the ATP and putative glutamine-binding domains. Notably, however, the strictly conserved N-terminal cysteine residue characteristic of AS-B enzymes is missing in Orf 3 (Fig. 4).
Figure 4.
Alignment of homologous regions of AS and β-LS. Heavy shading is used to indicate conserved residues and light shading is used to indicate similarity. The first and second regions are closely linked to glutamine-dependent activity in AS, and the third region is associated with ATP binding. Human AS (human AS P08243), Rice AS (rice AS U55873), Myco tuber. AS (Mycobacterium tuberculosis), Eco K12 AS-B (Q10374 E. coli P22106), β-LS (β-LS from S. clavuligerus), CarA (CarA from Erwinia cartovora).
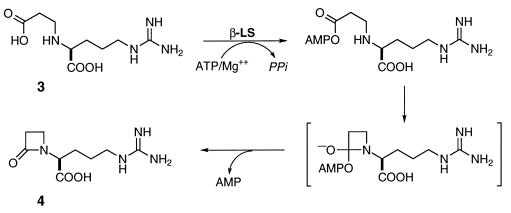
To investigate the possibility of an analogous intramolecular amide bond-forming transformation in clavulanic acid (5) biosynthesis, orf3 was disrupted by homologous recombination in S. clavuligerus. Clavulanic acid synthesis was completely lost and comparatively high concentrations of CEA (3) were observed to accumulate. Clavulanic acid production could be regained by reintroduction of an undamaged copy of orf3 (data not shown) and, importantly, by chemical complementation with DGPC (4), the putative product of the defective enzyme. Subsequent overexpression of orf3 gave the β-LS, whose conversion of CEA to DGPC (Fig. 1) was shown to require ATP/Mg2+ and proceed with loss of pyrophosphate. In light of these findings and the amino acid sequence similarity between Orf 3 and AS-Bs, we propose a mechanism for the conversion of CEA (3) to DGPC (4). In this mechanism, CEA is first activated by reaction with ATP/Mg2+, and the β-lactam ring is then formed by the intramolecular attack of the β-nitrogen on the activated carboxyl group (Fig. 5). This process may resemble the attack of the glutamyl nitrogen on the aspartyladenylate in the AS-B catalyzed reaction. It should be noted that the proposed mechanism is functionally the reverse of the reaction catalyzed by β-lactamases.
Figure 5.
Proposed mechanism of β-LS based on analogy to AS-B and β-lactamase.
The formation of the β-lactam ring in clavulanic acid takes place in the closure of CEA (3) to DGPC (4). Despite the apparent structural similarity between clavulanic acid (5) and isopenicillin N (2), the modes of azetidinone synthesis are strikingly different. The strained bicyclic nucleus of penicillin is generated in two sequential oxidative cyclization reactions catalyzed by ferrous isopenicillin N synthase. The overall thermodynamic cost of this process is paid by the concommitant reduction of a single molecule of dioxygen to two molecules of water. The cyclization of CEA to DGPC is similarly contrathermodynamic, but made favorable by intermediate acyl activation through reaction with ATP/Mg2+. Although this thermodynamic solution is shared with ribosomal and nonribosomal peptide synthetases and the genetically related asparagine synthetases, the interaction of an internal nucleophile is an unprecedented mode of amide bond synthesis and a new mechanism of β-lactam ring formation.
Whereas this and other biochemical evidence suggests that different biosynthetic strategies to β-lactam antibiotic formation have evolved (16), β-LS shows 26% identity and 46% similarity (blast 2) to the protein encoded by carA, a gene recently identified in E. cartovora and thought to be involved in the synthesis of carbapenem-3-carboxylic acid (7) (34). The extent of primary sequence similarity between these two proteins and their mutual lack of an N-terminal cysteine raise the intriguing possibility of an analogous role for proteins of this class in the biosynthesis of carbapenem antibiotics. The possible broader occurrence of this reaction motif poses renewed questions about the evolution of β-lactam antibiotic biosynthesis. The latitude these enzymes exhibit with respect to substrate specificity and their ability to catalyze stereospecific ring-forming reactions are of mechanistic interest, especially as a variant of AS-B enzymes. The practical value of such reactions is apparent.
Acknowledgments
We are grateful to Drs. T.-K. Wu and J. Pitlik for preliminary experiments that led to the research described in this paper, and to Dr. T. S. Hitchman for his critical comments during preparation of the manuscript. We thank Prof. C. R. Hutchinson (University of Wisconsin) for providing strains and vectors. We also thank the National Institutes of Health for financial support (Grant AI 14937).
ABBREVIATIONS
- β-LS
β-lactam synthetase
- CEA
N2-(carboxyethyl)-l-arginine
- DGPC
deoxyguanidinoproclavaminic acid
- AS-B
asparagine synthetase, Class-B
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF071051).
References
- 1.Service R F. Science. 1995;270:724–727. doi: 10.1126/science.270.5237.724. [DOI] [PubMed] [Google Scholar]
- 2.Neu H C. Science. 1992;257:1064–1073. doi: 10.1126/science.257.5073.1064. [DOI] [PubMed] [Google Scholar]
- 3.Baggaley K H, Brown A G, Schofield C J. Nat Prod Rep. 1997;14:309–333. doi: 10.1039/np9971400309. [DOI] [PubMed] [Google Scholar]
- 4.Marsden A F A, Wilkinson B, Cortes J, Dunster N J, Staunton J, Leadlay P F. Science. 1998;279:199–202. doi: 10.1126/science.279.5348.199. [DOI] [PubMed] [Google Scholar]
- 5.Jacobsen J R, Hutchinson C R, Cane D E, Khosla C. Science. 1997;277:367–369. doi: 10.1126/science.277.5324.367. [DOI] [PubMed] [Google Scholar]
- 6.Katz L, Donadio S. Annu Rev Microbiol. 1993;47:875–912. doi: 10.1146/annurev.mi.47.100193.004303. [DOI] [PubMed] [Google Scholar]
- 7.Hutchinson C R, Fujii I. Annu Rev Microbiol. 1995;49:201–238. doi: 10.1146/annurev.mi.49.100195.001221. [DOI] [PubMed] [Google Scholar]
- 8.Marahiel M A, Stachelhaus T, Mootz H D. Chem Rev. 1997;97:2651–2673. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- 9.von Döhren H, Keller U, Vater J, Zocher R. Chem Rev. 1997;97:2675–2705. doi: 10.1021/cr9600262. [DOI] [PubMed] [Google Scholar]
- 10.Roach P L, Clifton I J, Hensgens C M H, Shibata N, Schofield C J, Hajdu J, Baldwin J E. Nature (London) 1997;387:827–830. doi: 10.1038/42990. [DOI] [PubMed] [Google Scholar]
- 11.Townsend C A. Chem Biol. 1997;4:721–730. doi: 10.1016/s1074-5521(97)90310-0. [DOI] [PubMed] [Google Scholar]
- 12.Elson, S. W., Baggaley, K. H., Fulston, M., Nicholson, N. H., Tyler, J. W., Edwards, J., Holms, H., Hamilton, I. & Mousdale, D. M. (1993) J. Chem. Soc. Chem. Commun. 1211–1212.
- 13.Elson, S. W., Baggaley, K. H., Davidson, M., Fulston, M., Nicholson, N. H., Risbridger, G. D. & Tyler, J. W. (1993) J. Chem. Soc. Chem. Commun. 1212–1214.
- 14.Ward J M, Hodgson J E. FEMS Microbiol Lett. 1993;110:239–242. doi: 10.1111/j.1574-6968.1993.tb06326.x. [DOI] [PubMed] [Google Scholar]
- 15.Zalkin H. Adv Enzymol Relat Areas Mol Biol. 1993;66:203–309. doi: 10.1002/9780470123126.ch5. [DOI] [PubMed] [Google Scholar]
- 16.Marsh E N, Chang M D-T, Townsend C A. Biochemistry. 1992;31:12648–12657. doi: 10.1021/bi00165a015. [DOI] [PubMed] [Google Scholar]
- 17.Busby R W, Townsend C A. Bioorg Med Chem. 1996;4:1059–1064. doi: 10.1016/0968-0896(96)00088-0. [DOI] [PubMed] [Google Scholar]
- 18.Bierman M, Logan R, O’Brian K, Seno E T, Nagaraja Rao R, Schoner B I. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 19.Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M. Genetic Manipulation of Streptomyces: A Laboratory Manual. Norwich, U.K.: John Innes Foundation; 1985. [Google Scholar]
- 20.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, Albright L M, Coen D M, Varki A. Current Protocols in Molecular Biology. New York: Wiley; 1995. [Google Scholar]
- 21.Bailey C R, Butler M J, Normansell I D, Rowland R T, Winstanley D J. Bio/Technology. 1984;2:808–811. [Google Scholar]
- 22.Aoki H, Kubochi Y, Iguchi E, Imanaka H. J Antibiot. 1976;29:492–500. [Google Scholar]
- 23.Bird A E, Bellis J M, Gasson B C. Analyst. 1982;107:1241–1245. [Google Scholar]
- 24.Salowe S P, Marsh E N, Townsend C A. Biochemistry. 1990;29:6499–6508. doi: 10.1021/bi00479a023. [DOI] [PubMed] [Google Scholar]
- 25.Paradkar A S, Jensen S E. J Bacteriol. 1995;177:1307–1314. doi: 10.1128/jb.177.5.1307-1314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jepson J B, Smith I. Nature (London) 1953;172:1100–1101. doi: 10.1038/1721100b0. [DOI] [PubMed] [Google Scholar]
- 27.Wu T K, Busby R W, Houston T A, McIlwaine D B, Egan L A, Townsend C A. J Bacteriol. 1995;177:3714–3720. doi: 10.1128/jb.177.13.3714-3720.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero J, Liras P, Martín J F. Appl Microbiol Biotechnol. 1984;20:318–324. [Google Scholar]
- 29.Sheng S, Moraga-Amador D A, van Heeke G, Allison R D, Richards N G J, Schuster S M. J Biol Chem. 1993;268:16771–16780. [PubMed] [Google Scholar]
- 30.Scofield M A, Lewis W S, Schuster S M. J Biol Chem. 1990;265:12895–12902. [PubMed] [Google Scholar]
- 31.Cedar H, Schwartz J H. J Biol Chem. 1969;244:4112–4121. [PubMed] [Google Scholar]
- 32.Krahn J M, Kim J H, Burns M R, Parry R J, Zalkin H, Smith J L. Biochemistry. 1997;36:11061–11068. doi: 10.1021/bi9714114. [DOI] [PubMed] [Google Scholar]
- 33.Stoker P W, O’Leary M H, Boehlein S K, Schuster S M, Richards N G J. Biochemistry. 1996;35:3024–3030. doi: 10.1021/bi952504t. [DOI] [PubMed] [Google Scholar]
- 34.McGowan S J, Sebaihia M, Porter L E, Stewart G S A B, Williams P, Bycroft B W, Salmond G P C. Mol Microbiol. 1996;22:415–426. [PubMed] [Google Scholar]