Abstract
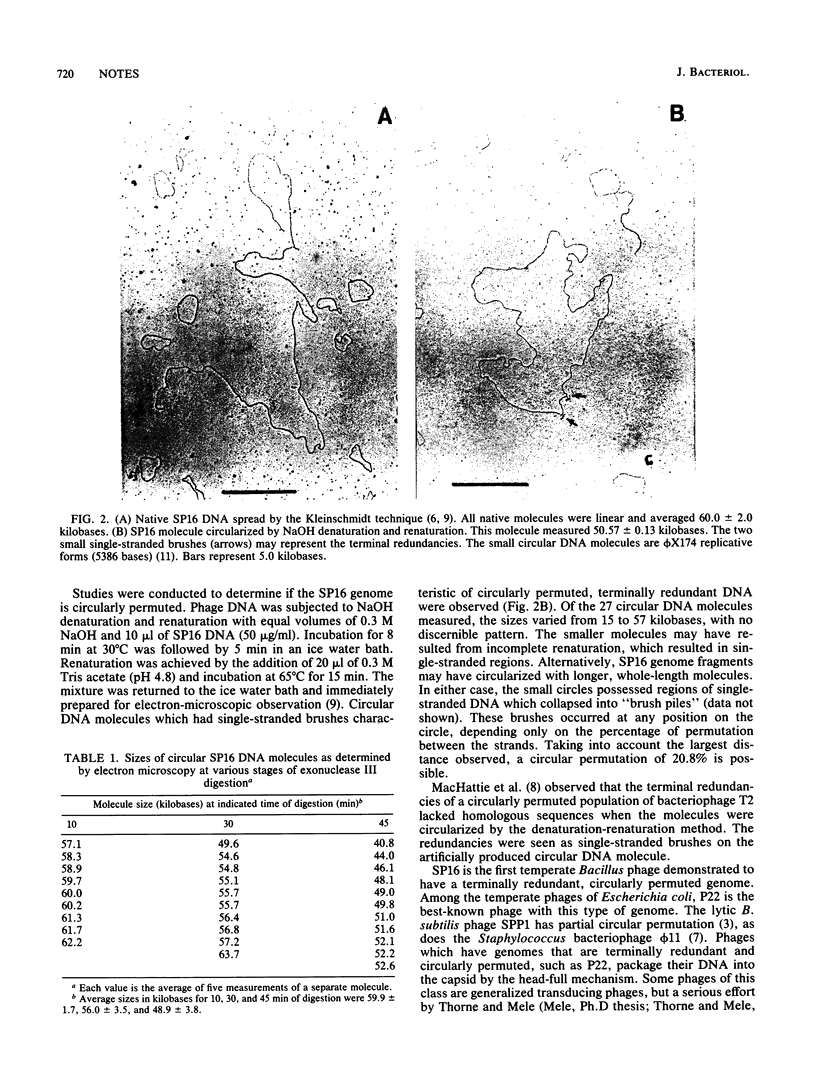
The physical nature of temperate Bacillus bacteriophage SP16 DNA was analyzed by electron microscopy, exonuclease digestion, denaturation-renaturation experiments, and restriction enzyme analysis. The SP16 genome is a linear molecule 60.0 +/- 2.0 kilobases in length without cohesive ends. Electron micrographs of denatured and renatured SP16 DNA showed that the DNA is circularly permuted. The genome possesses terminal redundancy, as demonstrated by electron microscopy of exonuclease III-digested DNA.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ansevin A. T., Vizard D. L., Mandel M. L., Dean D. H. Similarity of genetic distance determined from DNA thermal denaturation profiles to standard estimates of bacteriophage relatedness. Biochim Biophys Acta. 1983 Jun 24;740(2):127–133. doi: 10.1016/0167-4781(83)90069-6. [DOI] [PubMed] [Google Scholar]
- Deichelbohrer I., Messer W., Trautner T. A. Genome of Bacillus subtilis Bacteriophage SPP1: Structure and Nucleotide Sequence of pac, the Origin of DNA Packaging. J Virol. 1982 Apr;42(1):83–90. doi: 10.1128/jvi.42.1.83-90.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelson J. E., Wu R. Nucleotide sequence analysis of deoxyribonucleic acid. VII. Characterization of Escherichia coli exonuclease 3 activity for possible use in terminal nucleotide sequence analysis of duplex deoxyribonucleic acid. J Biol Chem. 1972 Jul 25;247(14):4661–4668. [PubMed] [Google Scholar]
- Helling R. B., Goodman H. M., Boyer H. W. Analysis of endonuclease R-EcoRI fragments of DNA from lambdoid bacteriophages and other viruses by agarose-gel electrophoresis. J Virol. 1974 Nov;14(5):1235–1244. doi: 10.1128/jvi.14.5.1235-1244.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löfdahl S., Zabielski J., Philipson L. Structure and restriction enzyme maps of the circularly permuted DNA of staphylococcal bacteriophage phi 11. J Virol. 1981 Feb;37(2):784–794. doi: 10.1128/jvi.37.2.784-794.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacHattie L. A., Ritchie D. A., Thomas C. A., Jr, Richardson C. C. Terminal repetition in permuted T2 bacteriophage DNA molecules. J Mol Biol. 1967 Feb 14;23(3):355–363. doi: 10.1016/s0022-2836(67)80110-4. [DOI] [PubMed] [Google Scholar]
- Rudinski M. S., Dean D. H. Evolutionary considerations of related Bacillus subtilis temperate phages phi 105, rho 14, rho 10, and rho 6 as revealed by heteroduplex analysis. Virology. 1979 Nov;99(1):57–65. doi: 10.1016/0042-6822(79)90036-9. [DOI] [PubMed] [Google Scholar]
- Sanger F., Coulson A. R., Friedmann T., Air G. M., Barrell B. G., Brown N. L., Fiddes J. C., Hutchison C. A., 3rd, Slocombe P. M., Smith M. The nucleotide sequence of bacteriophage phiX174. J Mol Biol. 1978 Oct 25;125(2):225–246. doi: 10.1016/0022-2836(78)90346-7. [DOI] [PubMed] [Google Scholar]
- Scher B. M., Dean D. H., Garro A. J. Fragmentation of Bacillus bacteriophage phi105 DNA by complementary single-stranded DNA in the cohesive ends of the molecule. J Virol. 1977 Aug;23(2):377–383. doi: 10.1128/jvi.23.2.377-383.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THORNE C. B. Transduction in Bacillus subtilis. J Bacteriol. 1962 Jan;83:106–111. doi: 10.1128/jb.83.1.106-111.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]




