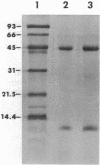
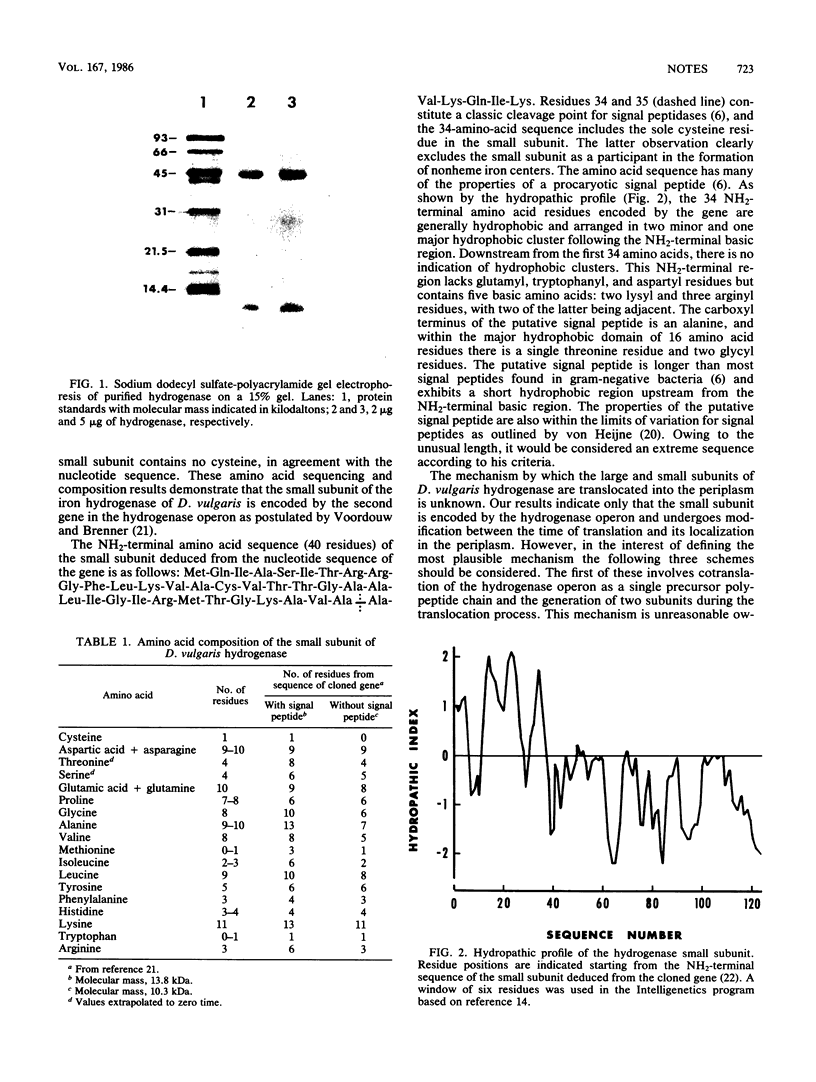
Abstract
We sequenced the NH2 terminus of the large and small subunits of the periplasmic hydrogenase from the sulfate-reducing bacterium Desulfovibrio vulgaris (Hildenborough) and found that the small subunit lacks a region of 34 NH4-terminal amino acids coded by the gene for the small subunit (G. Voordouw and S. Brenner, Eur. J. Biochem. 148:515-520, 1985). We suggest that this region constitutes a signal peptide based on comparison with known procaryotic signal peptides.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ackrell B. A., Asato R. N., Mower H. F. Multiple forms of bacterial hydrogenases. J Bacteriol. 1966 Oct;92(4):828–838. doi: 10.1128/jb.92.4.828-838.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. R., LeGall L., Peck H. D. Evidence for the periplasmic location of hydrogenase in Desulfovibrio gigas. J Bacteriol. 1974 Nov;120(2):994–997. doi: 10.1128/jb.120.2.994-997.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn M., Andreesen J. R., Gottschalk G. Fumarate reductase of Clostridium formicoaceticum. A peripheral membrane protein. Arch Microbiol. 1978 Oct 4;119(1):7–11. doi: 10.1007/BF00407920. [DOI] [PubMed] [Google Scholar]
- Hatchikian E. C., Bruschi M., Le Gall J. Characterization of the periplasmic hydrogenase from Desulfovibrio gigas. Biochem Biophys Res Commun. 1978 May 30;82(2):451–461. doi: 10.1016/0006-291x(78)90896-3. [DOI] [PubMed] [Google Scholar]
- Hunkapiller M. W., Lujan E., Ostrander F., Hood L. E. Isolation of microgram quantities of proteins from polyacrylamide gels for amino acid sequence analysis. Methods Enzymol. 1983;91:227–236. doi: 10.1016/s0076-6879(83)91019-4. [DOI] [PubMed] [Google Scholar]
- Huynh B. H., Czechowski M. H., Krüger H. J., DerVartanian D. V., Peck H. D., Jr, LeGall J. Desulfovibrio vulgaris hydrogenase: a nonheme iron enzyme lacking nickel that exhibits anomalous EPR and Mössbauer spectra. Proc Natl Acad Sci U S A. 1984 Jun;81(12):3728–3732. doi: 10.1073/pnas.81.12.3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kröger A., Dorrer E., Winkler E. The orientation of the substrate sites of formate dehydrogenase and fumarate reductase in the membrane of Vibrio succinogenes. Biochim Biophys Acta. 1980 Jan 4;589(1):118–136. doi: 10.1016/0005-2728(80)90136-x. [DOI] [PubMed] [Google Scholar]
- Krüger H. J., Huynh B. H., Ljungdahl P. O., Xavier A. V., Der Vartanian D. V., Moura I., Peck H. D., Jr, Teixeira M., Moura J. J., LeGall J. Evidence for nickel and a three-iron center in the hydrogenase of Desulfovibrio desulfuricans. J Biol Chem. 1982 Dec 25;257(24):14620–14623. [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Matsubara H., Sasaki R. M. High recovery of tryptophan from acid hydrolysates of proteins. Biochem Biophys Res Commun. 1969 Apr 29;35(2):175–181. doi: 10.1016/0006-291x(69)90263-0. [DOI] [PubMed] [Google Scholar]
- Meek R. L., Walsh K. A., Palmiter R. D. The signal sequence of ovalbumin is located near the NH2 terminus. J Biol Chem. 1982 Oct 25;257(20):12245–12251. [PubMed] [Google Scholar]
- Odom J. M., Peck H. D., Jr Localization of dehydrogenases, reductases, and electron transfer components in the sulfate-reducing bacterium Desulfovibrio gigas. J Bacteriol. 1981 Jul;147(1):161–169. doi: 10.1128/jb.147.1.161-169.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenkamp D. J., Peck H. D., Jr Proton translocation associated with nitrite respiration in Desulfovibrio desulfuricans. J Biol Chem. 1981 Jun 10;256(11):5450–5458. [PubMed] [Google Scholar]
- Voordouw G., Brenner S. Nucleotide sequence of the gene encoding the hydrogenase from Desulfovibrio vulgaris (Hildenborough). Eur J Biochem. 1985 May 2;148(3):515–520. doi: 10.1111/j.1432-1033.1985.tb08869.x. [DOI] [PubMed] [Google Scholar]
- Voordouw G., Walker J. E., Brenner S. Cloning of the gene encoding the hydrogenase from Desulfovibrio vulgaris (Hildenborough) and determination of the NH2-terminal sequence. Eur J Biochem. 1985 May 2;148(3):509–514. doi: 10.1111/j.1432-1033.1985.tb08868.x. [DOI] [PubMed] [Google Scholar]
- Yamamoto I., Ishimoto M. Hydrogen-dependent growth of Escherichia coli in anaerobic respiration and the presence of hydrogenases with different functions. J Biochem. 1978 Sep;84(3):673–679. doi: 10.1093/oxfordjournals.jbchem.a132172. [DOI] [PubMed] [Google Scholar]
- de Vries W., Niekus H. G., van Berchum H., Stouthamer A. H. Electron transport-linked proton translocation at nitrite reduction in Campylobacter sputorum subspecies bubulus. Arch Microbiol. 1982 Mar;131(2):132–139. doi: 10.1007/BF01053995. [DOI] [PubMed] [Google Scholar]
- de Vries W., van Berchum H., Stouthamer A. H. Localization of hydrogenase and nitrate reductase in Campylobacter sputorum subsp. bubulus. Antonie Van Leeuwenhoek. 1984;50(1):63–73. doi: 10.1007/BF00404908. [DOI] [PubMed] [Google Scholar]
- van der Westen H. M., Mayhew S. G., Veeger C. Separation of hydrogenase from intact cells of Desulfovibrio vulgaris. Purification and properties. FEBS Lett. 1978 Feb 1;86(1):122–126. doi: 10.1016/0014-5793(78)80112-4. [DOI] [PubMed] [Google Scholar]
- von Heijne G. Signal sequences. The limits of variation. J Mol Biol. 1985 Jul 5;184(1):99–105. doi: 10.1016/0022-2836(85)90046-4. [DOI] [PubMed] [Google Scholar]