Abstract
Interaction between Agrobacterium tumefaciens and plants provides a unique example of interkingdom gene transfer. Agrobacterium, a plant pathogen, is capable to stably transform the plant cell with a segment of its own DNA called T-DNA (transferred DNA). This process depends, among others, on the specialized bacterial virulence proteins VirD1 and VirD2 that excise the T-DNA from its adjacent sequences. Subsequent to transfer to the plant cell, the virulence protein VirD2, through its nuclear localization signal (NLS), is believed to guide the T-DNA to the nucleus. The T-DNA then is integrated into the plant genome. Although both of these proteins are essential for bacterial virulence, physical interaction of them has not been analyzed so far. We studied associations between these proteins by expressing them in mammalian cells and by testing for intracellular localization and colocalization. When expressed in human cells [HeLa, human embryo kidney (HEK) 293], the VirD2 protein homogeneously distributed over the nucleoplasm. The presence of any of two NLSs, on the N and C termini of VirD2, was sufficient for its efficient nuclear localization whereas deletion of both NLSs rendered the protein cytoplasmic. However, this double NLS mutant was translocated to the nucleus in the presence of wild-type VirD2 protein, implying VirD2–VirD2 interaction. The VirD1 protein, by itself localized in the cytoplasm, moved to the nucleus when coexpressed with the VirD2 protein, suggesting VirD1–VirD2 interaction. This interaction was confirmed by coimmunoprecipitation tests. Of interest, both proteins coimported to the nucleus showed a similar, peculiar sublocalization. The data are discussed in terms of functions of the VirD proteins. In addition, coimport of proteins into nuclei is suggested as a useful system in studying individual protein–protein interactions.
Keywords: nuclear import/virulence proteins/protein–protein interaction
Agrobacterium tumefaciens is a soil bacterium that produces tumors on dicotyledonous plants. The bacterium transfers a segment of a large plasmid (Ti plasmid) to the plant nucleus, where it is integrated stably. The transferred region (T-DNA) contains genes that direct the production of special compounds called opines, which the bacteria use, as well as enzymes involved in plant hormone synthesis, a cause of tumor formation. Transformed plant cells therefore grow into tumors secreting nutrients for the use of the plant pathogen. The excision and transfer of T-DNA is performed by virulence proteins, also encoded by genes situated on the Ti plasmid (1). The T-DNA in the bacterium is flanked by 24-bp border repeats that are recognized by two Agrobacterium proteins, VirD1 and VirD2. These proteins are introducing specific cuts into one strand of the border sequences. The single-stranded T-DNA then is displaced by DNA replication. Processing of T-DNA also has been documented in heterologous systems: VirD1 and VirD2 proteins overexpressed in Escherichia coli were shown to be functional (2, 3); proteins produced in transiently transfected plant cells were able to process correctly a cotransfected T-DNA (4, 5). Furthermore, efficient in vitro T-DNA processing was demonstrated with the use of purified VirD1 and VirD2 proteins (6). Although absolutely required for the nicking reaction, the precise role of VirD1 is still unknown. The T-DNA, covalently linked to VirD2, is transported from the bacterium to the plant cell and from there to the plant nucleus (7–9). The fact that any DNA that is located between the borders is transferred to the plant and is integrated in its genome makes Agrobacterium a unique tool for plant transformation.
Import of proteins into nuclei is mediated by nuclear localization signals (NLSs) that are categorized mainly as monopartite, SV40 large T-antigen type, possessing a single short region of basic amino acids, and bipartite, composed of two clusters of basic amino acids separated by a spacer (10). The VirD2 protein bears two NLSs, a monopartite one located in the N-terminal part of the protein and a bipartite one located close to the C terminus, which are functional in both plant (11, 12, 13) and yeast cells (12). Their functions have been characterized as ability to target a reporter protein fused to the N-terminal part of VirD2 (11), to the N-terminal NLS alone (12), or to the C-terminal NLS of VirD2 (12, 13), to the nucleus of transiently transfected cells. Recent studies suggested the activity of VirD2 NLSs to be universal because the recombinant protein accumulated in the nuclei of microinjected Xenopus oocytes and Drosophila embryos (14). Although activity of both VirD2 NLSs was documented in the studies listed above, only the C-terminal NLS has been found to be involved in tumorigenicity of Agrobacterium (15–18).
We studied subcellular localization of VirD2 and derivatives deleted in one or both NLSs in HeLa and HEK 293 cells. As a reporter gene, the green fluorescent protein (GFP) was used, which has been shown to be useful not only in determination of subcellular localization of proteins (19) but also in studies of protein–protein interaction (20). We focused our attention on the use of this system to monitor interaction of VirD2 with VirD2. Furthermore, because VirD2 is acting together with VirD1 in processing the T-DNA in the bacterium, we have asked whether these proteins interact.
Our results show that the VirD2 protein localized exclusively in the nuclei when overexpressed in mammalian cells. The presence of either the N- or C- terminal NLS of VirD2 was sufficient for efficient nuclear localization whereas deletion of both NLS sequences rendered the protein cytoplasmic. However, this double NLS mutant was translocated to the nucleus in the presence of wild-type VirD2 protein, suggesting VirD2–VirD2 interaction. Evidence for VirD1–VirD2 interaction was obtained by using the same experimental system. This was further confirmed by coimmunoprecipitation experiments.
MATERIALS AND METHODS
Construction of Plasmids.
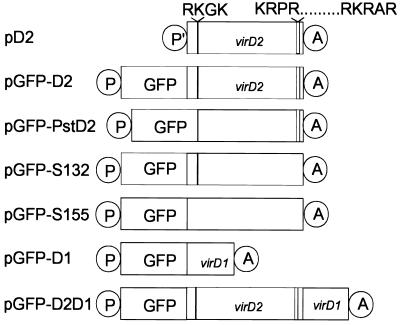
Plasmids used for monitoring subcellular localization of proteins are schematically depicted in Fig. 1. Plasmid pD2 contains the coding region of virD2, amplified by PCR from plasmid pVCK225 (21) whereas two modifications were introduced: A SmaI restriction site and the “Kozak” sequence ACCACC (22) were added upstream of the native ATG codon, and an XbaI cloning site was introduced after the native stop codon. The SmaI/XbaI virD2-containing fragment was cloned in EcoRV/XbaI sites of the pcDNA3 expression vector (Invitrogen). The correctness of the clone was verified by sequencing.
Figure 1.
Schematic representation of the constructs: P′-cytomegalovirus promoter, P-β-actin promoter, GFP-sequence coding for the green fluorescent protein, virD1- sequence coding for the VirD1 protein, virD2- sequence coding for the VirD2 protein, and A-simian virus 40 (SV40) polyadenylation signal. Regions coding for N- and C-terminal NLSs are represented by filled and opened bars, respectively. The corresponding amino acid sequences are indicated. pD2, wild-type virD2; pGFP-D2, wild-type virD2 fused to gfp; pGFP-PstD2, virD2 mutant, lacking the N-terminal NLS coding sequence and fused to gfp; pGFP-S132, virD2 mutant, lacking the C-terminal NLS coding sequence and fused to gfp; pGFP-S155, virD2 mutant, lacking both N- and C-terminal NLS coding sequences and fused to gfp; pGFP-D1, wild-type virD1 fused to gfp; pGFP-D2D1, three genes fused in frame: gfp, wild-type virD2, and wild-type virD1.
pGFP-D1, pGFP-D2, pGFP-S132, and pGFP-S155 are GFP fusions of VirD1, VirD2, VirD2 C-terminal NLS mutant, and VirD2 double NLS mutant, respectively, cloned as SpeI/EcoRV PCR products in the corresponding sites of pβactinNGFP (23). pVCK225 was used as a template for PCR for the virD1 and virD2 sequences, pVD44 (16) was used for the VirD2 C-terminal NLS mutant, and pS155 was used for the VirD2 double NLS mutant. pS155 is pVD44 missing the N-terminal NLS sequence coding for the four amino acids essential for NLS activity (S. Kocher, personal communication). The primers for all reactions were lacking native start and stop codons of the amplified genes.
In pGFP-PstD2, the N-terminal NLS coding sequence of virD2 was deleted by cloning a PstI/EcoRI fragment of pVD43 (16) into pβactinNGFP. In order for the two ORFs to be in frame, the ends of the PstI/EcoRI fragment were repaired by T4 polymerase and were ligated to the blunted SpeI site of the vector.
pGFP-D2D1 was made from pGFP-D2 that was opened by EcoRV and was ligated in frame to the EcoRV PCR fragment containing the virD1 ORF lacking the start codon. pVCK225 was used as a template in the PCR reaction.
The hemagglutinin (HA) epitope tagged construct pHA-D1 was prepared by ligating an HA epitope encoding oligonucleotide to the 5′-end of the PCR virD1 product described above, in-frame with the initiator methionine, in the mammalian expression vector pcDNA3.
Cell Culture.
HeLa cells and HEK 293 cells were maintained in DMEM supplemented with 10% fetal calf serum (Life Technologies, Grand Island, NY) at 37°C in an atmosphere containing 5% CO2. Cells seeded at a density of 106 and 0.5 × 106 per 10-cm and 6-cm dish, respectively, were transfected the following day with 1–2 μg/ml plasmid DNA by using a modified calcium phosphate method (24). The transfection mixture was removed after 16 hr of incubation, and the cells were analyzed for expression 24 hr later.
Indirect Immunofluorescence Staining.
Cells were plated and transfected on sterile coverslips. Fixation of cells was performed with 3.7% paraformaldehyde and permeabilization with 0.2% Triton X-100 (25). Then, 1:50 diluted anti-HA epitope 12CA5 mAb (Boehringer Mannheim), 1:1000 diluted rabbit polyclonal anti-VirD2 serum (a gift of Erich Lanka, Max-Planck-Institut, Berlin), or 1:400 diluted rabbit anti-GFP serum (Molecular Probes) was applied for 1 hr at 37°C. The cells subsequently were washed with PBS and were incubated with 1:100 diluted fluorescein isothiocyanate- or 1:50 diluted tetramethylrhodamine isothiocyanate-conjugated anti-mouse or anti-rabbit IgG (Sigma), supplemented with 4′,6-diamidino-2-phenylindole or propidium iodide, for thirty minutes. The coverslips were washed with PBS and were mounted on glass slides by using Gelvatol (or airvol; Air Products and Chemicals, Allentown, PA). HeLa cells grown on ellocate coverslips (Eppendorf) were microinjected by using an Eppendorf 5171 micromanipulator and an Eppendorf 5242 microinjection device. After 12 hr, the cells were immunostained as described above. All images were collected on a Leica TCS 4D microscope (Deerfield, IL).
Immunoprecipitation.
Cells were washed, were collected by centrifugation in ice-cold PBS, and were extracted in lysis buffer containing 50 mM Tris⋅HCl (pH 7.5) and 1% wt/vol Nonidet P-40 supplemented with complete protease inhibitor mixture (Boehringer Mannheim). Lysates were centrifuged for 15 min at 12,000 × g and were precleared with protein A Sepharose (Pharmacia) for 1 hr. The VirD2 protein was immunoprecipitated from 50% of the cell lysate from each plate with rabbit polyclonal anti-VirD2 serum. Immune complexes were collected by using protein A Sepharose. The immune complexes on the beads were washed three times with lysis buffer and were resuspended in 1× SDS sample buffer.
Immunoblot Analysis.
Cell extracts and immunoprecipitates were resolved by 15% SDS/PAGE and were transferred to Immobilon P membranes (Millipore). The filters were blocked for 30 min with 5% skimmed milk in 1× Tris-buffered saline (150 mM NaCl/50 mM Tris⋅HCl, pH 7.4), 0.2% Triton X-100, and 0.1% Tween-20 followed by 2-hr incubation with a rabbit polyclonal anti-VirD2 serum (diluted 1:5000) or with the anti HA-epitope 12CA5 mAb (diluted 1:100) in the same blocking solution. The secondary antibody was 1000-fold diluted alkaline-phosphatase conjugated anti-mouse Ig (Southern Biotechnology Associates), or, alternatively, alkaline-phosphatase conjugated protein A (Sigma). Detection was performed by using the AP color development reagents from Bio-Rad.
RESULTS
Nuclear Localization of VirD2 in Mammalian Cells Depends on the Presence of Either NLS.
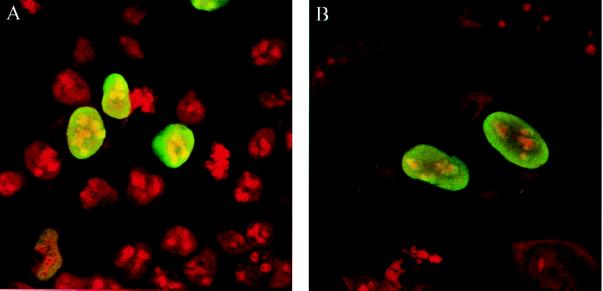
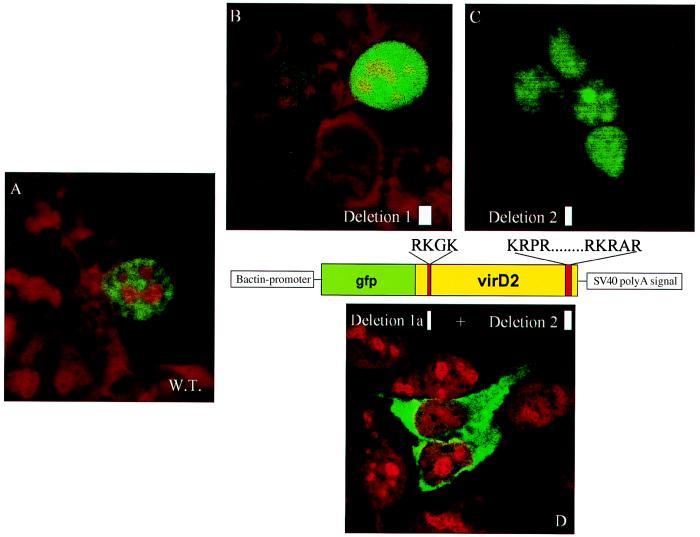
To monitor the subcellular localization of the VirD2 protein in human cells, the translational start site of the bacterial VirD2 protein was modified to conform to Kozak’s consensus sequence for eukaryotic translational initiation, and the gene was cloned in a mammalian expression vector (see Materials and Methods; Fig. 1). Protein localization in transfected cells was observed by indirect immunofluorescence with an anti-VirD2 antibody (Fig. 2). In both HEK 293 (Fig. 2A) and HeLa cells (Fig. 2B), the VirD2 protein was exclusively nuclear. The same results were obtained when VirD2 protein was fused, at its N terminus, to GFP (Fig. 3A). Deletion of the N-terminal (pGFP-PstD2) or C-terminal (pGFP-S132) NLS of VirD2 did not change the protein localization (Fig. 3 B and C, respectively). However, a mutant VirD2 protein lacking both NLSs (pGFP-S155) remained exclusively in the cytoplasm (Fig. 3D). Taken together, these results show that nuclear localization of VirD2 in mammalian cells depends on NLSs operational in plant cells and that any of the two NLSs, monopartite or bipartite, is sufficient to target the protein to the nucleus of a mammalian cell.
Figure 2.
Nuclear localization of the VirD2 protein in human cells. (A) HEK 293 cells. (B) HeLa cells. The cells were transfected transiently (HEK 293) or were microinjected (HeLa) with pD2. VirD2 protein was visualized by indirect immunofluorescence using anti-VirD2 antibody and fluorescein isothiocyanate-conjugated anti-rabbit IgG antibody. The cells were fixed and stained with propidium iodide.
Figure 3.
Subcellular localization of the VirD2 NLS mutant proteins fused to GFP, in HEK 293 cells. (A) Wild-type VirD2 protein fused to GFP (cells were transfected transiently with pGFP-D2). (B) N-terminal NLS mutant VirD2 protein (cells were transfected transiently with pGFP-PstD2. (C) C-terminal NLS mutant VirD2 protein (cells were transfected transiently with pGFP-S132). (D) Mutant VirD2 protein lacking both N- and C-terminal NLS (cells were transfected transiently with pGFP-S155). Cells were fixed and stained with propidium iodide.
Coimport to the Nucleus of GFP-VirD2 Double NLS Mutant Protein by the Wild-Type VirD2 Protein.
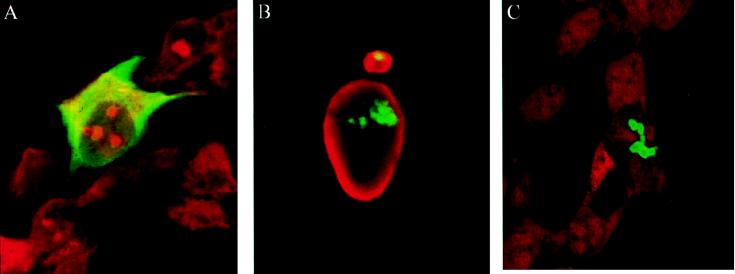
Interaction of VirD2 with VirD2 was tested by coexpression of (GFP-VirD2 NLS double mutant (pGFP-S155) with wild-type VirD2 (pD2) in HEK 293 cells. In the presence of wild-type VirD2 protein, a mutant cytoplasmic protein was translocated to the cell nucleus (Fig. 4A). As a control, the cells were cotransfected with GFP and VirD2. The presence of VirD2 did not affect the native localization of GFP (results not shown). Of interest, coimported GFP-S155 protein was not distributed evenly through the nucleoplasm but was concentrated in several subnuclear regions (Fig. 4B).
Figure 4.
Coexpression of GFP-VirD2 double NLS mutant and wild-type VirD2 protein in HEK 293 cells. (A) In the presence of wild-type VirD2 protein, the cytoplasmic GFP-VirD2 double NLS mutant protein (see Fig. 3D) is translocated to the nucleus and concentrated in several subnuclear regions, also shown in B. Cells were fixed and stained with propidium iodide.
VirD1 and VirD2 Are Associated in Vivo in Mammalian Cells.
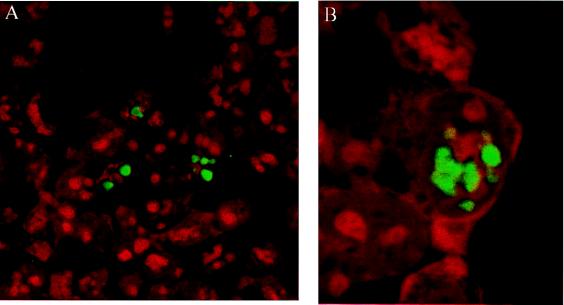
For analysis of a potential VirD1-VirD2 interaction, first the localization of the VirD1 protein was determined. The N terminus of VirD1 was fused to GFP (pGFP-D1). When overexpressed in HEK 293 or HeLa cells, the fusion protein was localized in the cytoplasm and was excluded from the nucleus (Fig. 5A). VirD1 fused to the HA epitope (pHA-D1) was cytoplasmic as well (results not shown). When GFP-D1 protein was coexpressed with wild-type VirD2 (coded by pD2), the originally cytoplasmic protein became sublocalized in the nucleus (Fig. 5B). This demonstrated that indeed VirD1 and VirD2 strongly interact. Identical results were obtained in HeLa cells comicroinjected with pGFP-D1 and pD2 (results not shown). Of interest, the observed nodular structures resembled the sublocalization of the double NLS mutant of VirD2, imported via “piggy-back” transport by wild-type VirD2 protein (see Fig. 4B). Furthermore, the same kind of structures were obtained in cells expressing the GFP-D2 fused to the N terminus of VirD1 (pGFP-D2D1) (Fig. 5C). A similar sublocalization was observed for VirE2 protein, whose NLSs have been engineered to target the protein to the nuclei of Drosophila embryos (14) (see Discussion). Because the shape of these structures resembled nucleoli and they were not stained by 4′,6-diamidino-2-phenylindole (not shown), we tested their localization by using the Rev protein of HIV (20), fused to the blue fluorescent protein as a nucleolar marker (R. Stauber, personal communication). However, the green and blue signals of the fusion proteins had a distinctly different localization (data not shown).
Figure 5.
Nuclear coimport of the GFP-VirD1 protein by the VirD2 protein in HEK 293 cells. (A) Cell expressing cytoplasmic GFP-VirD1 protein. The signal was amplified by the use of GFP-antiserum and fluorescein isothiocyanate-conjugated anti-rabbit IgG antibody. (B) When coexpressed with wild-type VirD2 protein, the GFP-VirD1 protein is translocated to the nucleus and is concentrated in distinct subnuclear regions. Indirect immunofluorescence was performed by using VirD2 antiserum and tetramethylrhodamine isothiocyanate-conjugated anti-rabbit IgG antibody. (C) When GFP-VirD2 is fused to VirD1, the resulting fusion protein is nuclear and concentrated in subnuclear regions, similar to in B. Cells in A and C were stained with propidium iodide.
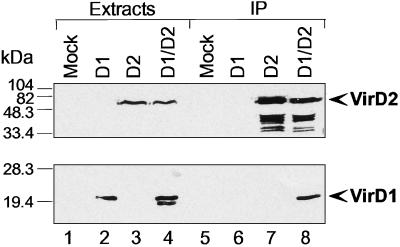
To further confirm the interaction of VirD1 and VirD2 in mammalian cells, we analyzed whether these proteins coimmunoprecipitate. VirD2 and HA-VirD1 were expressed in HEK 293 cells either alone or in combination, and their expression was analyzed by immunoblotting using specific antibodies. The anti-VirD2 antibody detected a single band of 56 kDa in the cells transfected with VirD2 and VirD2 with VirD1 (Fig. 6 Upper, lanes 3 and 4) and not in mock-transfected cells or cells expressing HA-VirD1 alone (Fig. 6 Upper, lanes 1 and 2). A protein migrating as VirD2 was detected in anti-VirD2 immunoprecipitates from corresponding cell extracts (Fig. 6 Upper, lanes 7 and 8). The 12CA5 anti-HA antibody recognized a 20-kDa band in the cells transfected with HA-VirD1 alone or in combination with VirD2 (Fig. 6 Bottom, lanes 2 and 4). However, in the extracts of the cells expressing both proteins simultaneously, a faster migrating band also was detected that could be a degradation product (Fig. 6, lane 4). The apparent molecular masses were 16 kDa for VirD1 and 56-kDa for VirD2. VirD1 is known to migrate in SDS/PAGE gels in agreement with its predicted molecular mass of 16.2 kDa whereas VirD2 is known to migrate differently from its predicted molecular mass of 47.4 kDa (26). We then analyzed anti-VirD2 immunoprecipitates by immunoblotting for the presence of HA-VirD1. The 20-kDa HA-VirD1 protein was detected in the immunoprecipitates from the cells expressing both proteins (Fig. 6, lane 8) and not from mock-transfected cells or cells transfected with HA-VirD1 or VirD2 alone (Fig. 6, lanes 5–7). These data confirm the VirD2-VirD1 association detected in vivo.
Figure 6.
Coimmunoprecipitation of VirD1 with VirD2 protein. Mock transfected HEK 293 cells or cells expressing either VirD1, VirD2, or both proteins were extracted on 10-cm dishes, and half of the lysate of each plate was used to immunoprecipitate VirD2 protein, as described in Materials and Methods. VirD2 in extracts and immunoprecipitates was detected by an anti-VirD2 antibody (Top). VirD2-specific, faster-migrating bands in lanes 7 and 8 are caused by protein degradation. HA epitope-tagged VirD1 was detected in extracts and VirD2 immunoprecipitates with an anti-HA epitope antibody. Cell extract (5%; lanes 1–4) and immunoprecipitates (lanes 5–8) were analyzed by immunoblotting. Molecular mass markers (in kDa) are shown on the left. Identical results were obtained in three independent experiments.
DISCUSSION
Interkingdom Recognition of NLS Sequences.
The monopartite NLS of the mammalian virus SV40, KKKRK has been shown in several systems to import proteins into plant nuclei: β-glucuronidase was transported to tobacco (27) and onion nuclei (28), and T7 RNA polymerase (29) was transported to the nuclei of tobacco protoplasts. Taken together, these data indicate that at least this monopartite NLS, in the context of a variety of proteins, is accepted by the plant import machinery. This conclusion is corroborated by the finding that importin α from the plant Arabidopsis thaliana binds specifically to plant NLSs as well as to the SV40 NLS coupled to human serum albumin (30).
NLSs of only a few “plant” proteins have been analyzed so far in animal systems. The results presented here show that the NLSs of the VirD2 protein, which are active in nuclear import into plant nuclei, are also functional in mammalian cells. Because it has been shown that both NLSs of the VirD2 are active in yeast (12) and that VirD2 protein is targeted to the nucleus when injected into Xenopus oocytes or Drosophila embryos (14), the VirD2 example supports the universal function of at least some nuclear localization signals in eukaryotes. Moreover, our experiments demonstrated the functionality of both the mono- and bipartite NLS of VirD2 in animal cells. Of interest, the Agrobacterium virulence protein E2 has been shown to be imported into plant nuclei because of the presence of two NLSs (31, 32). However, this protein remains in the cytoplasm of microinjected Xenopus oocytes and Drosophila embryos, unless amino acids within one of the NLSs are changed (14). Also, the yeast Matα2 NLS was shown to be nonfunctional in animal cells (33).
VirD2–VirD2 Interaction.
The Agrobacterium T-DNA transmission system shares many similarities with the conjugative transfer of the broad host range plasmid RP4 (34). Here, we will discuss only those similarities that are related to the VirD1 and VirD2 proteins. First, VirD2 shares conserved sequence motifs present in TraI and other known relaxases (35). Second, the processing of the T-DNA by VirD1 and VirD2 resembles the nicking reaction of the oriT sequence of RP4, by the TraI and TraJ proteins. In both cases, the reaction is site-specific, strand-specific, can be reproduced in vitro, and requires supercoiled DNA. Both relaxases remain covalently linked, via a phosphodiester bond between the active site tyrosine of the relaxase and the 5′ phosphate of the corresponding terminal nucleotide of the DNA-strand. Single-stranded DNA is transferred to the recipient plant cell (36, 37) and, most likely, bacterial cell (38). Termination of transfer of the RP4 conjugative plasmid seems to be achieved by a second TraI-dependent cleavage at the reconstituted nic site. However, elegant in vitro experiments have demonstrated that TraI, covalently attached to single-stranded DNA, is not able to cleave a second nic site (39). Hence, it was proposed that a second TraI molecule has to be recruited and that TraI protein acts as a dimer. The situation in A. tumefaciens is slightly different because formally there is no need for the T-DNA-VirD2 complex to cleave the left border because a second VirD1/VirD2 pair may accomplish this task. On the other hand, it also is known that binary plasmids containing a single border are functional in T-DNA transfer as well (in this case the whole plasmid is the T-DNA; ref. 40). We show here that an originally cytoplasmatically localized VirD2 double NLS mutant became nuclear in the presence of the wild-type VirD2 protein. This allows the conclusion that VirD2 can interact with VirD2. We do not know whether this interaction is necessary for virulence, but, if it is, the mechanism of termination by producing the second nick may depend on VirD2-VirD2 interaction.
The existence of such an interaction may explain results obtained in vivo, in which overexpression of the VirD2 protein, missing the C-terminal NLS, enhanced both T-strand formation inside the bacterium and plant transformation by wild-type Agrobacterium (41). If only one VirD2 molecule, i.e., the molecule that processed the T-DNA, is transported to the plant cell, the competition of a wild-type VirD2 protein and a mutant protein missing the NLS should result in decrease of plant transformation. However, if these two proteins are acting together, the NLS of the native protein may compensate for the mutant and lead to the observed enhancement.
VirD1–VirD2 Interaction.
VirD1 and VirD2 have been shown to be sufficient to process the T-DNA, when overexpressed and tested in vivo in E. coli (2), when analyzed in vitro (6), or when overexpressed in plants (4, 5). However, direct interaction of VirD1 with VirD2 has not been demonstrated so far. Of interest, in the RP4 system the relaxase TraI can act on double-stranded DNA only if TraJ had bound to DNA at oriT before (42, 38); however, also in this case, direct interaction between TraJ and TraI has not been documented (E. Lanka, personal communication). By coexpressing the cytoplasmic GFP-D1 protein with wild-type VirD2, we have provided the first evidence for a VirD1-VirD2 interaction by using two different methods: coimport of proteins monitored by direct immunofluorescence and coimmunoprecipitation analysis. Therefore the VirD1–VirD2 binding demonstrated in this report may represent an important step in understanding the functioning of relaxosomes. The VirD1–VirD2 interaction most likely plays a role in both recognition and processing of the T-DNA in Agrobacterium. Even the possibility that VirD1 protein is a member of the T-DNA complex cannot be excluded.
Subnuclear Localization of Coimported Proteins.
Although VirD2 protein distributed evenly in mammalian nuclei, coimported proteins localized to subnuclear “nodules” other than nucleoli. The same phenomena is observed for the VirD2–VirD1 fusion protein. It is intriguing that a similar sublocalization was observed for VirE2, when it is targeted to the nucleus of Drosophila embryos, because of engineered NLSs (14). Sublocalization of both viral DNA and replication proteins in a small number of nuclear sites are known in the case of adenovirus- and herpesvirus-infected cells (43, 44). Future investigations should show whether T-DNA complexes are being targeted to any of the nuclear compartments to facilitate T-DNA integration.
Coimport of Proteins to the Nucleus as a System for Analysis of Protein–Protein Interactions.
In our studies, the VirD2 nuclear protein was able to coimport the VirD1 protein or the VirD2 protein devoid of its own NLSs, each fused to the GFP reporter protein, into the mammalian nucleus. This system offers a possibility for testing in vivo interaction of proteins with nuclear or cytoplasmic localization. If both proteins to be tested are cytoplasmic, one of them could be engineered to include NLSs whereas the other could be fused to GFP. On the contrary, if both proteins are nuclear, the NLSs sequence of one of them, fused to GFP, could be deleted and tested for coimport by the nuclear protein. These tests may represent useful alternatives to immuno-coprecipitation or yeast two-hybrid analysis, at least for the study of known proteins.
Acknowledgments
We thank Andrew Matus and his group for the pβactinNGFP vector; Kurt Ballmer-Hofer for help with microinjection; Daniel Besser, Jesús Escudero, and Jan Lucht for many suggestions; and Serge Kocher for sharing pS155. We are grateful to Erich Lanka for providing us with anti-VirD2 serum and useful advice. Roland Stauber and George Pavlakis are acknowledged for their donation of the rev-bfp clone, and Brian Hemmings is acknowledged for supporting M.A. Many thanks go to Stefanie Kaech and Pawel Pelczar for critically reviewing the manuscript. This work was supported by a Swiss National Science Foundation fellowship (Swiss Priority Programme 5002-041803) to B.R., who also acknowledges support by the Friedrich Miescher Institut.
ABBREVIATIONS
- NLS
nuclear localization sequence
- GFP
green fluorescent protein
- HA
hemagglutinin
- HEK
human embryo kidney
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Sheng J, Citovsky V. Plant Cell. 1996;8:1699–1710. doi: 10.1105/tpc.8.10.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alt-Moerbe J, Rak B, Schröder J. EMBO J. 1986;5:1129–1135. doi: 10.1002/j.1460-2075.1986.tb04337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De Vos G, Zambryski P. Mol Plant–Microbe Interact. 1989;2:43–52. doi: 10.1094/mpmi-2-043. [DOI] [PubMed] [Google Scholar]
- 4.Hansen G, Chilton M-D. Proc Natl Acad Sci USA. 1996;93:14978–14983. doi: 10.1073/pnas.93.25.14978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hansen G, Shillito R D, Chilton M-D. Proc Natl Acad Sci USA. 1997;94:11726–11730. doi: 10.1073/pnas.94.21.11726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheiffele P, Pansegrau W, Lanka E. J Biol Chem. 1995;270:1269–1276. doi: 10.1074/jbc.270.3.1269. [DOI] [PubMed] [Google Scholar]
- 7.Tinland B, Hohn B. In: Genetic Engineering, Principles and Methods. Setlow J K, editor. New York: Plenum; 1995. pp. 209–229. [PubMed] [Google Scholar]
- 8.Zupan J, Zambryski P. Crit Rev Plant Sci. 1997;16:279–295. [Google Scholar]
- 9.Rossi L, Tinland B, Hohn B. In: The Rhizobiaceae. Spaink H, Hooykaas P, Kondorosi A, editors. Dordrecht, The Netherlands: Kluwer; 1998. pp. 303–320. [Google Scholar]
- 10.Makkerth J P S, Dingwall C, Laskey R A. Curr Biol. 1996;6:1025–1027. doi: 10.1016/s0960-9822(02)00648-6. [DOI] [PubMed] [Google Scholar]
- 11.Herrera-Estrella A, Van Montagu M, Wang K. Proc Natl Acad Sci USA. 1990;87:9534–9537. doi: 10.1073/pnas.87.24.9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tinland B, Koukolíková-Nicola Z, Hall M N, Hohn B. Proc Natl Acad Sci USA. 1992;89:7442–7446. doi: 10.1073/pnas.89.16.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howard E, Zupan J R, Citovsky V, Zambryski P C. Cell. 1992;68:109–118. doi: 10.1016/0092-8674(92)90210-4. [DOI] [PubMed] [Google Scholar]
- 14.Guralnick B, Thomsen G, Citovsky V. Plant Cell. 1996;8:363–373. doi: 10.1105/tpc.8.3.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shurvinton C E, Hodges L, Ream W. Proc Natl Acad Sci USA. 1992;89:11837–11841. doi: 10.1073/pnas.89.24.11837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rossi L, Hohn B, Tinland B. Mol Gen Genet. 1993;239:345–353. doi: 10.1007/BF00276932. [DOI] [PubMed] [Google Scholar]
- 17.Narasimhulu S B, Deng X, Sarria R, Gelvin S B. Plant Cell. 1997;8:873–886. doi: 10.1105/tpc.8.5.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshioka Y, Takahashi Y, Matsuoka K, Nakamura K, Koizumi J, Kojima M, Mashida Y. Plant Cell Physiol. 1996;37:782–789. [Google Scholar]
- 19.Ogawa H, Inouye S, Tsuji F I, Yasuda K, Umesono K. Proc Natl Acad Sci USA. 1995;92:11899–11903. doi: 10.1073/pnas.92.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stauber R, Gaitanaris G A, Pavlakis G N. Virology. 1995;213:439–449. doi: 10.1006/viro.1995.0016. [DOI] [PubMed] [Google Scholar]
- 21.Knauf V C, Nester E W. Plasmid. 1982;8:45–54. doi: 10.1016/0147-619x(82)90040-3. [DOI] [PubMed] [Google Scholar]
- 22.Kozak M. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludin B, Doll T, Meili R, Kaech S, Matus A. Gene. 1996;173:107–111. doi: 10.1016/0378-1119(95)00899-3. [DOI] [PubMed] [Google Scholar]
- 24.Chen C A, Okayama H. Biotechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- 25.Harlow E, Lane D. Antibodies: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. p. 418. [Google Scholar]
- 26.Yanofski M F, Porter S G, Young C, Albright L M, Gordon M P, Nester E W. Cell. 1986;47:471–477. doi: 10.1016/0092-8674(86)90604-5. [DOI] [PubMed] [Google Scholar]
- 27.van der Krol A R, Chua N-H. Plant Cell. 1991;3:667–675. doi: 10.1105/tpc.3.7.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Varagona M J, Raikhel N V. Plant J. 1994;5:207–214. doi: 10.1046/j.1365-313x.1994.05020207.x. [DOI] [PubMed] [Google Scholar]
- 29.Lassner M W, Jones A, Daubert S, Comai L. Plant Mol Biol. 1991;17:229–234. doi: 10.1007/BF00039497. [DOI] [PubMed] [Google Scholar]
- 30.Smith H M S, Hicks G R, Raikhel N V. Plant Physiol. 1997;114:411–417. doi: 10.1104/pp.114.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Citovsky V, Zupan J, Warnick D, Zambryski P. Science. 1992;256:1802–1805. doi: 10.1126/science.1615325. [DOI] [PubMed] [Google Scholar]
- 32.Citovsky V, Warnick D, Zambryski P. Proc Natl Acad Sci USA. 1994;91:3210–3214. doi: 10.1073/pnas.91.8.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chelsky D, Ralph R, Jonak G. Mol Cell Biol. 1989;9:2487–2492. doi: 10.1128/mcb.9.6.2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lessl M, Lanka E. Cell. 1994;77:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 35.Pansegrau W, Schröder W, Lanka E. J Biol Chem. 1994;269:2782–2789. [PubMed] [Google Scholar]
- 36.Tinland B, Hohn B, Puchta H. Proc Natl Acad Sci USA. 1994;91:8000–8004. doi: 10.1073/pnas.91.17.8000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yusibov V M, Steck T R, Gupta V, Gelvin S B. Proc Natl Acad Sci USA. 1994;91:2994–2998. doi: 10.1073/pnas.91.8.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pansegrau W, Lanka E. Prog Nucleic Acid Res Mol Biol. 1996;54:197–251. doi: 10.1016/s0079-6603(08)60364-5. [DOI] [PubMed] [Google Scholar]
- 39.Pansegrau W, Lanka E. J Biol Chem. 1996;271:13068–13076. doi: 10.1074/jbc.271.22.13068. [DOI] [PubMed] [Google Scholar]
- 40.Gardner R C, Knauf V C. Science. 1986;231:725–727. doi: 10.1126/science.231.4739.725. [DOI] [PubMed] [Google Scholar]
- 41.Wang K, Herrera-Estrella A, van Montagu M. J Bacteriol. 1990;8:4432–4440. doi: 10.1128/jb.172.8.4432-4440.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanka E, Wilkins B M. Annu Rev Biochem. 1995;64:141–169. doi: 10.1146/annurev.bi.64.070195.001041. [DOI] [PubMed] [Google Scholar]
- 43.Bocher J, Dawson A, Hay R T. J Virol. 1992;66:3140–3150. doi: 10.1128/jvi.66.5.3140-3150.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilcock D, Lane D P. Nature (London) 1991;349:429–431. doi: 10.1038/349429a0. [DOI] [PubMed] [Google Scholar]