Abstract
The three-dimensional structure of the human Rap30 DNA-binding domain has been solved by multinuclear NMR spectroscopy. The structure of the globular domain is strikingly similar to that of linker histone H5 and its fold places Rap30 into the “winged” helix–turn–helix family of eukaryotic transcription factors. Although the domain interacts weakly with DNA, the binding surface was identified and shown to be consistent with the structure of the HNF-3/fork head–DNA complex. The architecture of the Rap30 DNA-binding domain has important implications for the function of Rap30 in the assembly of the preinitiation complex. In analogy to the function of linker histones in chromatin formation, the fold of the Rap30 DNA-binding domain suggests that its role in transcription initiation may be that of a condensation factor for preinitiation complex assembly. Functional similarity to linker histones may explain the dependence of Rap30 binding on the bent DNA environment induced by the TATA box-binding protein. Cryptic sequence identity and functional homology between the Rap30 DNA-binding domain and region 4 of Escherichia coli σ70 may indicate that the σ factors also possess a linker histone-like activity in the formation of a prokaryotic closed complex.
Synthesis of mRNA in eukaryotes requires a complex assembly of proteins comprised of the multisubunit RNA polymerase II (Pol II) and a collection of additional factors, TFIIs, that are necessary to assemble a transcriptionally competent complex at the promoter and initiate transcription (1). This complex structure, known as the preinitiation complex (PIC), assembles in a stepwise fashion, beginning with the distortion and bending of the DNA by the TATA box-binding protein (TBP) (or as the complex TFIID) whose interaction with DNA is stabilized by TFIIA and TFIIB. The polymerase is recruited to this highly distorted promoter architecture by the factor TFIIF (2). Although eight oligomeric complexes make up the complete PIC, only TFIIF appears to play the dual role of promoting assembly of a PIC and participating in transcript elongation (3). TFIIF is a heterodimeric factor comprised of 30-kDa (Rap30) and 74-kDa (Rap74) subunits which deliver Pol II to the growing PIC as a preformed complex with the enzyme (3). Although the small subunit of TFIIF appears to be sufficient for Pol II recruitment to the PIC, genetic and biochemical analyses suggest important roles for both subunits in stabilization of the PIC through direct interactions with TFIIB and for transcription initiation (4–7).
The observation that Pol II enters the PIC as a preformed complex with TFIIF is reminiscent of the role of bacterial σ factors in bacterial core polymerase recruitment to a promoter. In Escherichia coli, σ70 is responsible for core polymerase recruitment to most bacterial housekeeping genes. σ70 is a 613-aa polypeptide with distinct functional domains which recognize the promoter sequences at two positions upstream of the transcriptional start site (−10 and −35) and form direct contacts with subunits of the core polymerase (8). Functional analysis of Rap30 localized regions of the protein responsible for Pol II binding to the middle of the molecule while an independent domain required for initiation localized to the C-terminal 86 amino acids, a region that appears to direct nonspecific binding to DNA (9, 10). This domain architecture functionally resembles the polymerase-binding and DNA-binding domains of E. coli σ70 and Bacillus subtilis σ43 despite little sequence similarity between them (8–12). Interestingly, Rap30 cosediments with E. coli core RNA polymerase in glycerol-density gradient centrifugation, an effect that can be overcome by addition of excess σ70. These data suggest a true functional homology between these two polymerase recruitment factors presumably mediated by a similar structural architecture (13).
The ability to uncouple Pol II-binding and DNA-binding functionalities of Rap30 suggested that Rap30 may be comprised of at least two structured domains. To this end, the DNA-binding domain (DBD) of human Rap30 was overproduced and its three-dimensional structure was solved by multinuclear, multidimensional NMR. The structure reveals that the DBD of human Rap30 (hRap30) is remarkably similar to linker histone H5 (14) and the hepatocyte nuclear transcription factor HNF3/fork head (15), both members of the eukaryotic “winged” helix–turn–helix (HTH) DBD family. The structure of the hRap30-DBD suggests that Rap30 may act as a PIC condensation factor by binding within the bent DNA environment induced by the TBP and helping to organize Pol II into a transcriptionally competent architecture. The functional relationship between Rap30 and bacterial σs hints that a similarly acting domain may lie within the σ factors themselves.
MATERIALS AND METHODS
Protein.
The C-terminal 86 amino acids of hRap30 (amino acids 164–249) were subcloned into pET11d (Novagen) and expressed in HMS174(DE3) in defined media containing 15N-NH4Cl and/or [13C]glucose as sole nitrogen and carbon sources, respectively. The expressed protein was purified in Tris buffer using phosphocellulose followed by chromatography on Mono-S. Chromatography on phosphocellulose revealed the presence of two forms of the domain, determined by mass spectrometry to be full length and N-terminally truncated at amino acid 169. The full-length construct was further purified on Superdex 75, concentrated to 1.2 mM, and dialyzed against 10 mM sodium phosphate, 50 mM NaCl, and 1 mM NaN3 (pH 7.2). Purity and homogeneity were determined by denaturing SDS/PAGE and matrix-associated laser desorption ionization mass spectrometry.
NMR Spectroscopy.
All NMR experiments were carried out at 24°C on either a Bruker DMX500 or DMX600 spectrometer equipped with a z-shielded gradient triple resonance probe. The sequential assignment of the 1H, 13C, and 15N chemical shifts of the hRap30-DBD was achieved by means of through-bond heteronuclear correlations along the backbone and side chains using the following three-dimensional experiments: 15N-separated HNHA, HNCACB, CBCA(CO)NH, HBHA(CO)NH, C(CO)NH, H(CCO)NH, HCCH-correlated spectroscopy, and HCCH-total correlation spectroscopy (16–18). 3JHNα, 3JCγN, 3JCγCO, and 3JNHβ coupling constants were obtained by quantitative J correlation spectroscopy (19). Nuclear Overhauser effects (NOEs) involving protons of the protein were obtained from three-dimensional 15N-separated, three-dimensional 13C-separated, and four-dimensional 13C/13C-separated NOE spectra (mixing times of 120 ms, 50 ms, and 140 ms, and 120 ms, respectively) and a three dimensional 15N-separated rotating-frame Overhauser effect spectrum recorded with a 30-ms mixing time and 6-kHz spin-locking field.
DNA-Binding Titration.
A 16-bp oligonucleotide duplex sequence was derived from the sequence of the HIV-2 promoter (5′-GATATACCCGCTGCTC-3′; C. Parada and R. G. Roeder, personal communication), synthesized by standard phosphoramidite chemistry, and purified as described (20). The DNA was exhaustively desalted by dialysis against 10 mM sodium phosphate (pH 7.2) subsequent to concentration to 4.2 mg/ml. DNA was titrated into a 0.3 mM sample of 15N-hRap30-DBD in steps up to 4 molar equivalents of DNA over protein. Continuous shift of selected resonances was monitored by 15N-1H correlation spectroscopy.
Structure Calculations.
NOEs within the protein were grouped into four distance ranges, 1.8–2.7 Å (1.8–2.9 Å for NOEs involving NH protons), 1.8–3.3 Å (1.8–3.5 Å for NOEs involving NH protons), 1.8–5.0 Å, and 1.8–6.0 Å, corresponding to strong, medium, weak, and very weak intensities. One-half angstrom was added to the upper bounds for distances involving methyl groups to account for the higher apparent intensity of NOEs involving these groups. Distances involving methyl groups, aromatic ring protons, and nonstereospecifically assigned methylene protons were represented as a (Σr−6)−1/6 sum (21). Protein backbone hydrogen-bonding restraints (rNH-O = 1.5–2.8 Å, rN-O = 2.4–3.5 Å) within areas of regular secondary structure were introduced during the final stages of refinement. φ, ψ, χ1, and χ2 torsion angle restraints were derived from rotating-frame Overhauser effect, NOE, and coupling constant data (22), and the minimum ranges used were ±30° ±30°, ±20°, and ±30°, respectively. The structures were calculated using the hybrid distance geometry dynamic-simulated annealing protocol (23) with the program XPLOR-3.843 (24) adapted to incorporate pseudopotentials for 3JHNα-coupling constants (25), secondary 13Cα and 13Cβ chemical shift restraints (26), and a conformational database potential (27). There were no hydrogen-bonding, electrostatic, or 6-12 Lennard-Jones empirical potential energy terms in the target function. The backbone amide protons for residues 242–243 were not observed experimentally and very few nonsequential NOEs were observed to their side chains. Thus, the reported models do not represent the only possible conformation for the backbone of residues 241–243. Beyond residue 243, only intraresidue and sequential NOEs were seen and, as for the N-terminal 11 amino acids, they were excluded.
RESULTS
Structure Determination.
The solution structure of the hRap30-DBD was solved by multidimensional, multinuclear NMR spectroscopy employing uniformly enriched 15N and/or 13C protein. Analysis of NMR spectra derived from protein constructions encompassing residues 90–249, 134–249, or 164–249 revealed that these truncated hRap30s contained a single, well-structured domain within the fragment 164–249 (data not shown). The balance of the protein, residues 90–163, appeared to possess no regular secondary structure in the truncated proteins prepared from bacterial expression systems. The structure described herein was therefore determined using the 164–249 construct; 1196 NOEs, 58 hydrogen bonds, 69 φ, 4 ψ, 60 χ1, and 19 χ2 restraints were defined from multinuclear nuclear Overhauser effect spectroscopy, rotating-frame Overhauser effect spectroscopy, and quantitative J correlation spectroscopy. There is some uncertainty in the orientation of the side chain of Lys236 due to the inability to resolve the chemical shifts of the side chain carbons and protons beyond Cβ. The protein domain appears to behave as a monomer in solution as evidenced from size-exclusion chromatography and estimated amide proton relaxation rates (data not shown). The coordinate precision and the extent of agreement with experimental restraints for the family of 30 structures is described in Table 1.
Table 1.
Structural statistics
| 〈SA〉 | |
|---|---|
| rms deviations from experimental distance restraints (Å)* | |
| All (1254) | 0.035 ± 0.003 |
| Sequential (|i − j| = 1) (326) | 0.033 ± 0.004 |
| Short range (1 < |i − j| ≤ 5) (314) | 0.046 ± 0.007 |
| Long range (|i − j|) > 5) (251) | 0.035 ± 0.005 |
| Intraresidue (305) | 0.020 ± 0.002 |
| H-bonds (58) | 0.042 ± 0.006 |
| rms deviations from experimental: dihedral restraints (deg) (152)* | 0.497 ± 0.07 |
| 3JHNα-coupling constants (Hz) (56)* | 0.81 ± 0.04 |
| 13Cα (ppm) (64) | 1.31 ± 0.03 |
| 13Cβ (ppm) (62) | 1.16 ± 0.04 |
| Deviations from idealized covalent geometry | |
| Bonds (Å) | 0.004 ± 0.0004 |
| Angles (degree) | 0.525 ± 0.02 |
| Impropers (degree) | 0.485 ± 0.03 |
| Coordinate precision† | |
| Backbone (residues 179–240) | 0.224 ± 0.06 |
| All nonhydrogen atoms (residues 179–240) | 0.564 ± 0.05 |
| PROCHECK statistics‡ | |
| % residues in most favorable region (1675) | 88.6% |
| % residues in allowed regions (213) | 11.4% |
| Bad contacts | 6.4 ± 1.4 |
〈SA〉 is the family of final 30 simulated annealing structures excluding residues 164–174 and residues 244–249.
None of the structures exhibited distance violations greater than 0.4 Å, dihedral angle violations greater than 5°, or 3JHNα-coupling constant violations greater than 2 Hz. No restraints between protons separated by three bonds were utilized (approximately 600 NOEs).
The precision of the coordinates is defined as the average atomic rms difference between the 30 individual simulated annealing structures and the mean coordinates SA for residues 179–240.
PROCHECK (50) was used to assess the overall quality of the structures for residues 174–243. G factors for φ/ψ, χ1/χ2, and the overall G factor were −0.15, 0.25, and 0.00, respectively, for the ensemble as calculated within PROCHECK_NMR.
The hRap30-DBD Is a Member of the Winged HTH Family of DBDs.
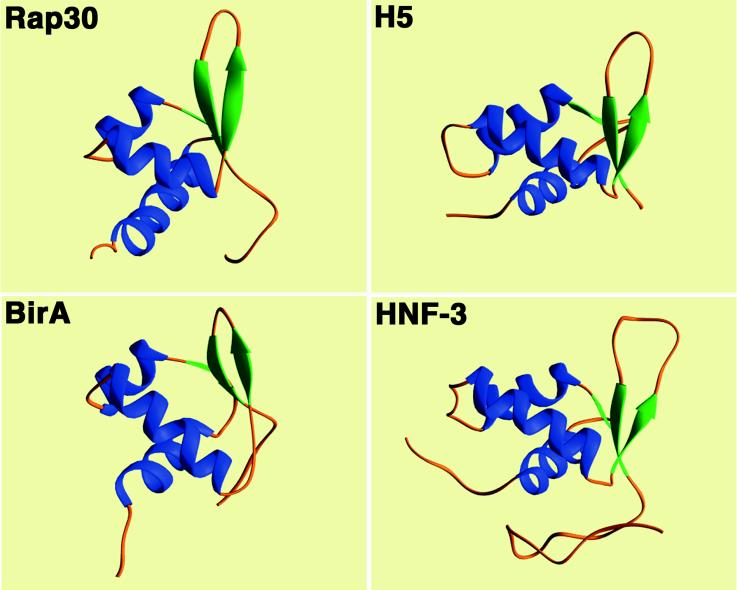
The hRap30-DBD is a compact α/β structure composed of 68 amino acids (residues 175–243) with highly disordered N (residues 164–174) and C terminii (residues 244–249). The overall fold of the domain is analogous to that of many eukaryotic transcription factors commonly referred to as the winged HTH motif (15) (Fig. 1). The N-terminal α-helix (H1, residues 179–193) is followed by a short strand (S1, residues 196–197) of anti-parallel sheet which leads into the HTH element composed of α-helix H2 (residues 199–205), a four residue loop (206–209), and α-helix H3 (residues 210–219). The wing of the domain is formed by a five-residue loop (residues 226–230) between the C-terminal strands of anti-parallel β-sheet (S2, residues 222–225 and S3, residues 231–234). This architecture closely resembles that of the hepatocyte nuclear factor HNF-3/fork head (15) and that of linker histone H5 (14) among eukaryotic factors as well as the prokaryotic domains derived from the biotin operon repressor protein BirA (28) and the LexA repressor (29) (Fig. 2). The turn of the HTH element is, in one respect, more reminiscent of the prokaryotic HTH fold in that it is composed of only four residues, although the hRap30-DBD turn lacks the conserved Gly at position 2 common to many prokaryotic HTH motifs (30). This contrasts with the five to eight residues seen in HNF-3/fork head, histone H5, the protooncogene products c-myb (31) and ETS1 (32–34,) and the POU-specific domain (35). The helix crossing angle of ≈70° between H2 and H3 follows the paradigm for this DNA-binding motif.
Figure 1.
The three-dimensional structure of the hRap30-DBD. Stereo superposition of the family of 30 simulated annealing structures of the hRap30-DBD. The protein backbone (residues 175–243) is shown in blue with selected amino acid side chains in green. The side chains shown are His-182, Val-183, Leu-184, Leu-187, Phe-188, Ala-190, Phe-191, Tyr-197, Leu-199, Leu-202, Val-203, Val-211, Tyr-212, Leu-213, Leu-217, Ile-220, Val-222, Val-225, Thr-232, Trp-233, Leu-235, and Tyr-239.
Figure 2.
Comparison of domain folds for eukaryotic and prokaryotic “winged” HTH motif proteins. The solution structure of the hRap30-DBD is compared with the x-ray crystal structures of the linker histone H5 (14), the HNF-3/fork head-DBD (15), and the prokaryotic biotin operon repressor protein BirA (28). Cα superpositions were 1.9 Å (45 residues), 2.1 Å (42 residues), and 1.7 Å (38 residues) for H5, HNF-3, and BirA, respectively.
The three-stranded anti-parallel sheet is highly twisted, a common feature among proteins of this class (Fig. 2). The wing formed by residues in the loop of the β-hairpin in hRap30 displays a characteristic kink with a positive φ angle near 70° for the third residue of the loop (His-229). This feature appears to be an important component in presenting this region to the phosphodiester backbone of the DNA in HNF-3/fork head (15). The DNA-binding mechanism has not been experimentally established for H5; however, for such a common motif it would be expected that the general features observed in HNF-3 would also be present in H5 and therefore presumably also present in hRap30 (see below). The C terminus of the hRap30-DBD contains a pseudohelical turn (residues 235–239) which brings Tyr-239 into van der Waals contact with Ile220; numerous NOEs are observed between the side chains of these residues. The pseudohelical turn also promotes contacts to form among His-241, Tyr-242, and residues near the amino end of helix H1, acting as a sort of cap to close the hydrophobic core (Fig. 1). Beyond residue 243, essentially no nonsequential interresidue NOEs were observed to the rest of the domain and these residues have been excluded from presentation. The same can be said for residues 164–174 for which only intraresidue or sequential NOEs were observed.
DNA-Binding Surface.
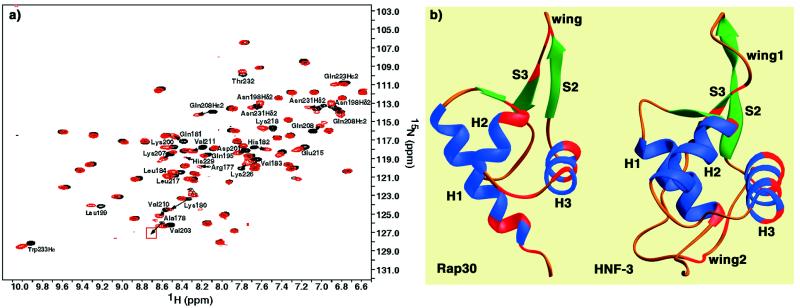
The DNA-binding surface of the hRap30-DBD was examined by titration of an oligonucleotide duplex whose sequence was derived from a strong class II promoter in HIV-2 (Fig. 3). The specific region chosen was determined by comparison to the adenovirus major late promoter to which hRap30 has been shown to crosslink at position −19 (36). Fig. 3 displays the result of the DNA titration. A continual shift of selected resonances was observed between 0.2 and 4 molar equivalents of added DNA. Although most residues were not affected by the DNA, significant changes in chemical shift were observed for main chain and side chain residues at the head of helix H2 (Asn-198-Lys-200), within the HTH-turn (Lys-207 and Gln-208), the exposed surface of helix H3 (Val-210, Val-211, Glu-215, and Lys-218), and residues in the wing (Lys-226, His-229, and Asn-231). The crosspeak for His229 was already very weak in the unliganded domain and almost completely disappears on addition of DNA. The most significant changes occur at the amino end of helix H1, specifically residues Arg-177, Ala-178, and Lys-180-His-182, suggestive of a local change in the position of the helix that may be due to direct interaction with the DNA.
Figure 3.
Identification of the DNA-binding surface of the hRap30-DBD. (a) The beginning (black) and end point (red) are shown of a titration of HIV-2 promoter DNA (16 bp) as monitored by 15N-1H correlation spectroscopy. The end point represents 4 molar equivalents of added DNA. Peaks are labeled for those selected backbone (NH) and/or side chain (Asn or Gln) resonances which displayed significant changes in chemical shift in the presence of DNA. Arrows indicate the direction of movement along the entire titration for these residues. The red box indicates the position of Ala178 at the end point which falls below the plotted contour level in the figure. (b) Comparison of the hRap30-DBD and HNF-3/fork head DNA-binding surfaces. Helices and sheets are shown in blue and green, respectively. Residues shifted by the presence of DNA in hRap30 or that were observed contacting DNA in the cocrystal structure of HNF-3 (15) are shown in red. The surfaces of interaction for the two protein domains appear to be qualitatively identical as exhibited by the red surfaces in each molecule.
DISCUSSION
Functional dissection of hRap30 has shown that the heterodimerization surface for Rap74 and the DBD are located on opposite poles of the polypeptide, the N and C terminii, respectively (7, 9–10). The physical separation of these domains mirrors their apparent functional independence. Internal deletions within the C-terminal DBD do not disrupt heterodimerization but abolish transcription initiation, presumably due to a loss in the ability of hRap30 to interact with DNA or bind Pol II (10). This can be readily understood from the structure of the hRap30-DBD. The derivative Δ181–195 would obliterate the hydrophobic core of the domain, removing Val-183, Leu-184, Leu-187, Phe-188, Ala-190, and Phe-191, thereby destabilizing the entire structure of the domain. Δ196–210 and Δ226–240 would be expected to have two-fold effects, both removing part of the hydrophobic core (e.g., Leu-199, Leu-202, and Val-203 in Δ196–210) as well as removing some of the anticipated participants in DNA binding, namely, the helix H2 and HTH-turn (Δ196–210) or the β-hairpin (S2-wing-S3) (Δ226–240), respectively. It is more difficult to assess the defect imparted by Δ166–180 because this derivative appears to delete an unstructured albeit highly basic segment at the N terminus of the domain.
The deletions Δ136–150 and Δ136–165 map within the Pol II-binding/elongation region and are defective in transcription initiation (10). These regions are essentially unstructured in domain constructions extending from residue 90 to the C terminus (data not shown), suggesting that if the region 90–173 is structured in the context of the PIC, it would be induced to form either by Pol II binding or TFIIF oligomerization. Efficient reconstitution of transcription and elongation functions of TFIIF is dependent on coexpression of the two proteins (5). Heterodimerization of independently purified subunits requires partial denaturation and renaturation for stable association (37). Although the purified subunits are each capable of binding Pol II in vitro, simple mixing of the subunits does not reconstitute elongation activity of TFIIF (5). These observations suggest that there is a conformational change in TFIIF that is dependent on both subunits. Since the elongation and Pol II-binding activities of Rap30 overlap in the unstructured 90–173 region (10), it is plausible that an induced conformation is necessary for reconstitution of fully functional TFIIF. The possibility that the N-terminally deleted constructs simply failed to fold properly cannot be excluded; however, the requirement for partial denaturation/renaturation for stable heterodimerization of independently expressed subunits argues against this. Denaturation/renaturation of the N-terminally deleted constructs does not introduce additional structured regions into the hRap30-DBD; NMR spectra of the refolded protein are identical to the native protein (data not shown).
Implications for Preinitiation Complex Assembly.
The emerging theme of the PIC is one in which elements of nucleosomal architecture imposed by histones may be approximately preserved by histone-like factors in the general transcription machinery (38). First observed for the N-terminal portions of two Drosophila TBP-associated factors, TAFII42 and TAFII62 (39), the three-dimensional structure of the hRap30-DBD represents the second example of a component of the eukaryotic preinitiation complex which possesses a histone-like fold. The similarity in the DBD fold of Rap30 to the globular domain of linker histones suggests a functional similarity with respect to the effects of each protein on their nucleoprotein targets.
Biochemical and genetic analyses of linker histones have established at least two functional surfaces within the globular domain (40–42). The probable primary DNA-binding surface of the linker histone is essentially analogous to that observed in the x-ray structure of the HNF-3/DNA complex (15) and as defined in this study for the hRap30-DBD (Fig. 3). A second functional surface is comprised of basic residues in the loop between H1 and H2 (Lys-40 and Arg-42) and a residue in the middle of H2 (Lys-52) which are associated with chromatin compaction (40–42). Mutagenesis of this second surface precludes chromatosome formation in globular H5 (40) but not in a full-length linker histone variant H1° (41). The importance of these residues to linker histone function was reinforced by gain-of-function mutations introduced into analogous regions of HNF-3 which enabled the formation of chromatosomes at a low level (42). These data have led to the proposal that the primary nucleosomal binding surface for linker histones is within the major groove of DNA and that the second surface of contact is possibly responsible for binding adjacent regions of the nucleoprotein complex within or between nucleosome core particles (43).
The orientation of linker histones on DNA has important implications for the role of Rap30 in PIC formation. Both linker histone and Rap30 bind DNA without sequence specificity, but prefer nonlinear DNA conformations. Crosslinking of Rap30 to DNA is TBP dependent and therefore presumably dependent on the bent DNA environment induced by TBP (36, 44). Rap30 crosslinks in the adjacent turn of helix upstream and downstream of the TATA box (44, 45). The observation that TFIIF may be a heterotetramer in solution (46) would suggest that one monomer of Rap30 is crosslinked in each major groove upstream and downstream of TBP, presumably via the DBD. Electron micrographs of PIC/DNA complexes indicate an ≈50 bp shortening of the DNA in the presence of IIF (44, 45). It has been postulated that additional wrapping or compaction of promoter DNA occurs upon PIC formation on the basis of these electron micrographic images (44, 45). Thus, in analogy to the properties of linker histones, Rap30 may to bind to a nonlinear DNA environment adjacent to the TATA box in the major groove and participate in the condensation of the PIC. The dramatic reduction in the copper phenanthroline footprint of IIB, IID, and Pol II on addition of a IIE/IIF extract supports a model in which either Rap30 or IIF is responsible for condensing the PIC, thereby accounting for the change in footprint size (47). The question remains whether the portion of Rap30 which may be responsible for this effect is equivalent to the second functional region in H5 associated with nucleosome compaction. In Rap30, the equivalent segment between helix-1 and sheet-1 is only two amino acids in length and is not basic. An alternative possibility within Rap30 could be the unstructured 90–173 segment which might form an interaction surface either within the IIF heterotetramer or upon binding within the PIC. Further study will be necessary to identify the regions of Rap30 or IIF associated with DNA wrapping or condensation.
Relationship to Prokaryotic σ Factors.
The ability of hRap30 to bind to Pol II and its role in recruiting Pol II to the PIC in eukaryotic transcription reflects the properties of the prokaryotic σ factors (8). It is surprising to find that Rap30 could cosediment with the E. coli core polymerase depleted of σ70, an effect which could be suppressed by competition with excess σ70 (13). Despite these observations, there is no statistically significant sequence identity between the two proteins (48, 49). The hRap30-DBD has been proposed to be most similar to region 4.2 of σ70 and region 4.2 is thought to contain a HTH motif (11, 12). It would be intriguing to find a similar structural topology in region 4.2 of σ70, although no direct proof presently exists. In contrast to the hRap30-DBD, isolated fragments of σ70 region 4 are expressed in inclusion bodies and are insoluble above a few micromolar. Moreover, the hRap30-DBD fails to bind a factor which specifically targets region 4 of σ70, namely, the anti-σ protein AsiA (C. Groft and M. Werner, unpublished results). Thus, if it can be shown that there is structural homology between these protein domains, their functional surfaces are expected to be quite distinct in composition, consistent with the lack of significant amino acid identity between them.
Acknowledgments
We are grateful to Camilo Parada, Bob Roeder, and Jim Kadonaga for supplying cDNAs of human and Drosophila Rap30; Nick Loizos, Seth Darst, Debbie Hinton, and Alicia Dombroski for σ factor derivatives and AsiA; Brian Chait, Bob Roeder, and Stephen Burley for useful discussions, Bob Roeder for critical reading of the manuscript; and Jeff Bonnano, Andy LiWang, Dan Garrett, Frank Delaglio, and Marius Clore for technical support. M.H.W. is a scholar of the Sidney Kimmel Cancer Foundation.
ABBREVIATIONS
- Pol II
polymerase II
- PIC
preinitiation complex
- TBP
TATA box-binding protein
- DBD
DNA-binding domain
- hRAP30
human Rap30
- NOE
nuclear Overhauser effect
- NOESY
nuclear Overhauser effect spectroscopy
- ROESY
rotating-frame Overhauser effect
- HTH
helix–turn–helix
Footnotes
Data deposition: The coordinates for the minimized average structure and the ensemble of 30 structures have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID codes 1bby and 2bby).
References
- 1.Roeder R G. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 2.Zawel L, Reinberg D. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 3.Reines D, Conaway J W, Conaway R C. Trends Biochem Sci. 1996;21:351–355. [PMC free article] [PubMed] [Google Scholar]
- 4.Zawel L, Reinberg D. Prog Nucleic Acids Res Mol Biol. 1993;44:67–108. doi: 10.1016/s0079-6603(08)60217-2. [DOI] [PubMed] [Google Scholar]
- 5.Tan S, Aso T, Conaway R C, Conaway J W. J Biol Chem. 1994;269:25684–25691. [PubMed] [Google Scholar]
- 6.Sun F-W, Hempsey M. Proc Natl Acad Sci USA. 1995;92:3127–3131. doi: 10.1073/pnas.92.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fang S M, Burton Z F. J Biol Chem. 1996;271:11703–11709. doi: 10.1074/jbc.271.20.11703. [DOI] [PubMed] [Google Scholar]
- 8.Gross C A, Lonetto M, Losick R. In: Transcriptional Regulation. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. pp. 129–176. [Google Scholar]
- 9.Tan S, Garrett K P, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1994;91:9808–9812. doi: 10.1073/pnas.91.21.9808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tan S, Conaway R C, Conaway J W. Proc Natl Acad Sci USA. 1995;92:6042–6046. doi: 10.1073/pnas.92.13.6042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardella T, Moyle T, Susskind M M. J Mol Biol. 1989;206:579–590. doi: 10.1016/0022-2836(89)90567-6. [DOI] [PubMed] [Google Scholar]
- 12.Siegele D A, Hu J C, Walter W A, Gross C A. J Mol Biol. 1989;206:591–603. doi: 10.1016/0022-2836(89)90568-8. [DOI] [PubMed] [Google Scholar]
- 13.McCracken S, Greenblatt J. Science. 1991;253:900–902. doi: 10.1126/science.1652156. [DOI] [PubMed] [Google Scholar]
- 14.Ramakrishnan V, Finch J T, Graziano V, Lee P L, Sweet R M. Nature (London) 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 15.Clark K L, Halay E D, Lai E, Burley S K. Nature (London) 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 16.Bax A, Grzesiek S. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- 17.Clore G M, Gronenborn A M. Science. 1991;252:1390–1399. doi: 10.1126/science.2047852. [DOI] [PubMed] [Google Scholar]
- 18.Clore G M, Gronenborn A M. Protein Sci. 1994;3:372–390. doi: 10.1002/pro.5560030302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bax A, Vuister G W, Grzesiek S, Delaglio F, Wang A C, Tschudin R, Zhu G. Methods Enzymol. 1994;239:79–106. doi: 10.1016/s0076-6879(94)39004-5. [DOI] [PubMed] [Google Scholar]
- 20.Werner M H, Bianchi M E, Gronenborn A M, Clore G M. Biochemistry. 1995;34:11998–12004. doi: 10.1021/bi00037a042. [DOI] [PubMed] [Google Scholar]
- 21.Nilges M A. Proteins Struct Funct Genet. 1993;17:297–309. doi: 10.1002/prot.340170307. [DOI] [PubMed] [Google Scholar]
- 22.Nilges M A, Clore G M, Gronenborn A M. Biopolymers. 1990;29:813–822. doi: 10.1002/bip.360290415. [DOI] [PubMed] [Google Scholar]
- 23.Nilges M A, Clore G M, Gronenborn A M. FEBS Lett. 1988;229:317–324. doi: 10.1016/0014-5793(88)81148-7. [DOI] [PubMed] [Google Scholar]
- 24.Brünger A T. X-PLOR Version 3.1: A System for X-ray Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1993. [Google Scholar]
- 25.Garrett D S, Kuszewski J, Hancock T J, Lodi P J, Vuister G W, Gronenborn A M, Clore G M. J Magn Reson Ser B. 1994;104:99–103. doi: 10.1006/jmrb.1994.1061. [DOI] [PubMed] [Google Scholar]
- 26.Kuszewski J, Qin J, Gronenborn A M, Clore G M. J Magn Reson Ser B. 1995;106:92–96. doi: 10.1006/jmrb.1995.1017. [DOI] [PubMed] [Google Scholar]
- 27.Kuszewski J, Gronenborn A M, Clore G M. J Magn Reson. 1997;125:171–177. doi: 10.1006/jmre.1997.1116. [DOI] [PubMed] [Google Scholar]
- 28.Wilson K P, Shewchuk L M, Brenan R G, Otsuka A J, Matthews B W. Proc Natl Acad Sci USA. 1992;89:9257–9261. doi: 10.1073/pnas.89.19.9257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fogh R H, Ottleben G, Ruterjans H, Schnarr M, Boelens R, Kaptein R. EMBO J. 1994;13:3936–3944. doi: 10.1002/j.1460-2075.1994.tb06709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pabo C O, Sauer R T. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 31.Ogata K, Hojo H, Aimoto S, Nakai T, Nakamura H, Sarai A, Ishii S, Nishimura Y. Proc Natl Acad Sci USA. 1992;89:6428–6432. doi: 10.1073/pnas.89.14.6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kodandapani R, Pio F, Ni C Z, Piccialli G, Klemsz M, McKercher S, Maki R A, Ely K R. Nature (London) 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 33.Werner M H, Clore G M, Fisher C L, Fisher R J, Trinh L, Shiloach J, Gronenborn A M. J Biomol NMR. 1997;10:317–328. doi: 10.1023/a:1018399711996. [DOI] [PubMed] [Google Scholar]
- 34.Batchelor A, Piper D E, dela Brousse F C, McKnight S L, Wolberger C. Science. 1998;279:1037–1041. doi: 10.1126/science.279.5353.1037. [DOI] [PubMed] [Google Scholar]
- 35.Assa-Munt N, Mortishire-Smith R J, Aurora R, Herr W, Wright P E. Cell. 1993;73:193–205. doi: 10.1016/0092-8674(93)90171-l. [DOI] [PubMed] [Google Scholar]
- 36.Robert F, Forget D, Li J, Greenblatt J, Coulombe B. J Biol Chem. 1996;271:8517–8520. doi: 10.1074/jbc.271.15.8517. [DOI] [PubMed] [Google Scholar]
- 37.Wang B Q, Kostrub C F, Finkelstein A, Burton Z F. Protein Expression Purif. 1993;4:207–214. doi: 10.1006/prep.1993.1027. [DOI] [PubMed] [Google Scholar]
- 38.Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V. Proc Natl Acad Sci USA. 1996;93:9384–9388. doi: 10.1073/pnas.93.18.9384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie X, Kokubo T, Cohen S L, Hoffmann A, Chait B T, Roeder R G, Nakatani Y, Burley S K. Nature (London) 1996;380:316–322. doi: 10.1038/380316a0. [DOI] [PubMed] [Google Scholar]
- 40.Goytisolo F A, Gerchman S E, Yu X, Rees C, Graziano V, Ramakrishnan V, Thomas J O. EMBO J. 1996;15:3421–3429. [PMC free article] [PubMed] [Google Scholar]
- 41.Hayes J J, Kaplan R, Ura K, Pruss D, Wolffe A. J Biol Chem. 1996;271:25817–25822. doi: 10.1074/jbc.271.42.25817. [DOI] [PubMed] [Google Scholar]
- 42.Cirillo L A, McPherson C E, Bossard P, Stevens K, Cherian S, Shim E Y, Clark K L, Burley S K, Zaret K S. EMBO J. 1998;17:244–254. doi: 10.1093/emboj/17.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruss D, Bartholomew B, Persinger J, Hayes J, Arents G, Moudrianakis E N, Wolffe A P. Science. 1996;274:614–617. doi: 10.1126/science.274.5287.614. [DOI] [PubMed] [Google Scholar]
- 44.Forget D, Robet F, Grondin G, Burton Z F, Greenblatt J, Coulombe B. Proc Natl Acad Sci USA. 1997;94:7150–7155. doi: 10.1073/pnas.94.14.7150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim T-Y, Lagrane T, Wang Y-H, Griffith J D, Reinberg D, Ebright R H. Proc Natl Acad Sci USA. 1997;94:12268–12273. doi: 10.1073/pnas.94.23.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Flores O, Ha I, Reinberg D. J Biol Chem. 1990;265:5629–5634. [PubMed] [Google Scholar]
- 47.Buratowski S, Sopta M, Greenblatt J, Sharp P A. Proc Natl Acad Sci USA. 1991;88:7509–7513. doi: 10.1073/pnas.88.17.7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sopta M, Burton A F, Greenblatt J. Nature (London) 1989;341:410–414. doi: 10.1038/341410a0. [DOI] [PubMed] [Google Scholar]
- 49.Garrett K P, Serizawa H, Hanley J P, Bradsher J N, Tsuboi A, Arai N, Yokota T, Arai K-I, Conaway R C, Conaway J W. J Biol Chem. 1992;25:23942–23949. [PubMed] [Google Scholar]
- 50.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]