Abstract
Small heat shock proteins (sHSPs) belong to a family of 12- to 43-kDa proteins that are ubiquitous and are conserved in amino acid sequence among all organisms. A sHSP homologue of Methanococcus jannaschii, a hyperthermophilic Archaeon, forms a homogeneous multimer comprised of 24 monomers with a molecular mass of 400 kDa in contrast to other sHSPs that show heterogeneous oligomeric complexes. Electron microscopy analysis revealed a spherically shaped oligomeric structure ≈15–20 nm in diameter. The protein confers thermal protection of other proteins in vitro as found in other sHSPs. Escherichia coli cell extracts containing the protein were protected from heat-denatured precipitation when heated up to 100°C, whereas extracts from cells not expressing the protein were heat-sensitive at 60°C. Similar results were obtained when purified sHSP protein was added to an E. coli cell lysate. The protein also prevented the aggregation of two purified proteins: single-chain monellin (SCM) at 80°C and citrate synthase at 40°C.
Exposure to high temperature or other stress conditions can be lethal to cells, and thus, organisms have developed stress-induced responses to protect themselves. This is manifested by the heat-induced synthesis of highly conserved proteins called heat shock proteins (HSPs), many of which have chaperone activity (1–4). There are four classes of high molecular weight HSPs and one family of small heat shock proteins (sHSPs). The sHSPs are found in all three kingdoms: Archaea, Bacteria, and Eukarya (5, 6) and range in size from 12 to 43 kDa. They share a conserved sequence with the α-crystallin proteins of the vertebrate eye lens (7, 8), called the α-crystallin or sHSP domain. This domain is preceded by an N-terminal region that varies in size and sequence and is followed by a C-terminal sequence that is not conserved.
Small HSPs are known to form large multimeric complexes that can vary in size. The recombinant sHSP from Caenorhabditis elegans, HSP16–2, forms a heterogeneous mixture of particles comprised of ≈14 and 24 subunits, respectively (9). The human recombinant αB-crystallin recently was shown by cryo-electron microscopy to have an asymmetric, variable quaternary structure of spherical multimers 8–18 nm in diameter (10). On the other hand, the Mycobacterium tuberculosis Hsp16.3 forms an oligomeric complex comprised of a trimer of trimers that gives rise to a triangular structure (11).
The sHSPs have been reported to have a wide range of cellular functions including endowment of thermotolerance to cells expressing sHSPs in vivo (12, 13) and molecular chaperone activity in vitro (14–19). They also are expressed at various developmental stages in plants (20). It has been suggested that sHSPs bind to their substrate proteins under denaturing temperatures and form a stable complex with folding intermediates of the substrates (21, 22). The amount of sHSPs required to prevent substrate denaturation varies depending on the sHSP and the substrate. For example, a 1:0.5 molar ratio of pea sHSP18.1 to malate dehydrogenase prevents the latter from heat denaturation (22), whereas 14–15 C. elegans HSP16–2 molecules are needed to protect one citrate synthase (CS) monomer (9).
In an effort to understand the nature of the oligomerization and the chaperone activity of a sHSP, we have expressed the gene Mj 0285, coding for a 16.5-kDa sHSP homolog from Methanococcus jannaschii, a hyperthermophile (23), and biochemically characterized its gene product, Mj HSP16.5. The α-crystallin domain of Mj HSP 16.5 has 20.7% sequence identity with human αA-crystallin and 31.4% identity with rice HSP16.9 (14, 24). We show that Mj HSP16.5 forms a unique spherical oligomer of 24 subunits and has the ability to protect most proteins in E. coli cell extracts and other purified proteins from thermal aggregation at a very high temperature.
MATERIALS AND METHODS
Materials.
Porcine heart CS (43.5 kDa), egg white lysozyme (14.4 kDa), and cytochrome C (12.5 kDa) were obtained from Sigma. SCM (11.1 kDa) was provided by LG Chemicals (Seoul, Korea). Purified Mj 577 protein (18.4 kDa), a heat-stable protein from M. jannaschii, was provided by Tom Zarembinski of the University of California, Berkeley, CA.
Strain and Plasmid.
The Mj 0285 gene from M. jannaschii (6) was cloned into the pET21a vector (Novagen) and transformed into E. coli BL21(DE3) {F−dcm, ompT (lon) hsdSB (rB− mB−), gal [DE3]} carrying the plasmid pSJS1240 (25) that contains the genes coding for the rare E. coli tRNA codons for arginine (AGA) and isoleucine (ATA), which are necessary for the translation of this hyperthermophilic gene.
Protein Purification.
Protein purification was performed following the procedure described by Kim et al. (23).
Gel Filtration.
Analytical gel filtration chromatography was carried out on a Tosohaas (Montgomeryville, PA) TSK G4000SW column (7.5 × 30 cm) connected to a Perseptive Biosystems Biocad instrument (Cambridge, MA). The column was equilibrated with buffer A (25 mM Hepes, pH 7.5/0.1 M NaCl) at a flow rate of 0.5 ml/min. Gel filtration standard proteins from Sigma [carbonic anhydrase (29 kDa), albumin (66 kDa), apoferritin (443 kDa), and thyroglobulin (699 kDa)] were used to calibrate the column. The void volume was determined by the use of dextran blue (2,000 kDa). Proteins were detected by absorbance at 280 nm.
Electron Microscopy.
Mj HSP16.5 (1 mM) in buffer A was negatively stained with 2% uranyl acetate on formvar carbon-coated films and imaged at ×50,000 in a JEOL 100 CS transmission electron microscope (EM) (JEOL) operated at 80 kV.
Thermal Stability of Soluble Extracts Expressing Mj HSP16.5.
BL21(DE3)/pSJS1240 transformed with pET21a/Mj 0285 were grown at 37°C in Luria–Bertani medium containing 50 μg/ml ampicillin and 30 μg/ml spectinomycin to an OD600 of 1.0. Cells were induced with 0.5 mM isopropyl-1-thio-β-d-galacto-pyranoside for 2 h. The cell pellet was harvested at 4°C by centrifugation at 5,000 × g for 6 min, washed with buffer B (25 mM Tris⋅HCl, pH 7.5/10% glycerol/2 mM DTT/1 mM EDTA), sonicated three times at 80 W for 30 s, and centrifuged for 20 min at 20,000 × g in a Beckman Type 60 Ti rotor. The protein concentration of the cell extract was diluted to 4 mg/ml. E. coli host BL21(DE3)/pSJS1240 cell extract was prepared in the same way except that the cells were grown in the presence of spectinomycin. The cell extracts were covered with mineral oil and heated at various temperatures (60–110°C) for 20 min. After being allowed to cool to room temperature, the mineral oil was removed and the samples were centrifuged at 10,000 × g for 5 min. The soluble supernatants (10 μl) were analyzed on a 15% SDS/PAGE gel following the method of Laemmli (26). Protein concentrations were measured by using the Bradford assay (27).
Thermal Protection of E. coli Proteins by Purified Mj HSP16.5.
A cell extract of the background E. coli host strain BL21(DE3)/pSJS1240 was prepared in buffer A following the same procedure as described above. The assay was performed by incubating 200 μg of cell extract with 100 or 200 μg of purified Mj HSP16.5 protein or 200 μg of cytochrome C in a final volume of 100 μl at 80°C for 20 min. The samples were centrifuged at room temperature at 10,000 × g for 5 min. The pellet was resuspended in 100 μl of buffer, and 10 μl of supernatant and pellet samples was analyzed on a 15% SDS/PAGE gel.
Thermal Protection of SCM by Purified Mj HSP16.5.
SCM was incubated at a concentration of 90 μM in the presence of Mj HSP16.5, lysozyme, or Mj 577 protein at equimolar ratios in buffer A in a final volume of 70 μl. The samples were covered with mineral oil and heated at 80°C for 20 min. After being allowed to cool to room temperature, the samples were centrifuged at 10,000 × g for 5 min. The supernatants were removed, and the pellets were resuspended in 70 μl of the above buffer. Ten microliters of supernatant and pellet samples was analyzed by electrophoresis on a 15% SDS/PAGE gel.
Thermal Aggregation Measurements.
The aggregation of CS on thermal denaturation was determined by measuring the absorption caused by increased turbidity at 360 nm in a Unikon 933 double beam UV/Vis spectrophotometer at 40°C. All experiments were performed at a CS concentration of 5.8 μM in 50 mM potassium phosphate (pH 7). The CS and Mj HSP16.5 concentration was determined by absorbance at 280 nm by using an extinction coefficient of 1.55 × 10−5 M−1 cm−1 (28) and 3.42 × 10−5 M−1 cm−1, respectively.
RESULTS
Multimer Formation by Mj HSP16.5 as Detected by Size Exclusion Chromatography (SEC) and Electron Microscopy.
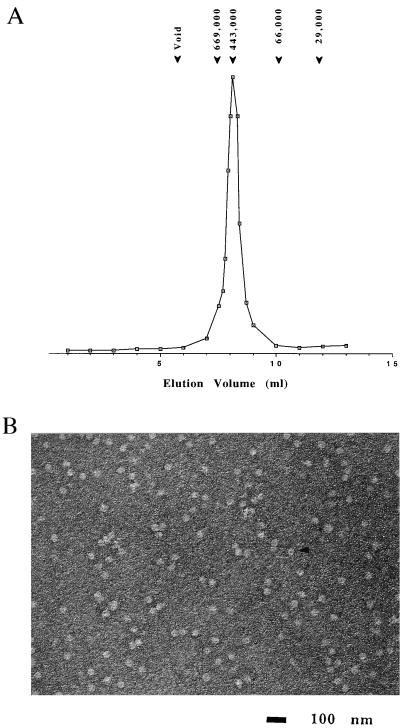
Analysis by SEC was performed on a calibrated Tosohaas G4000SW column. Mj HSP16.5 eluted as a single peak with an estimated molecular mass of ≈440 kDa (Fig. 1A). Sedimentation equilibrium analysis indicated a molecular mass of ≈400 kDa, which corresponds to 24 monomers (unpublished results). The crystal structure of this sHSP homologue at 2.9 Å (29) also supports the observation that Mj HSP16.5 is a 24 mer.
Figure 1.
SEC and electron microscopy (EM) of Mj HSP16.5. (A) SEC was performed by using a Tosohaas TSK G4000SW column as described in Materials and Methods. Purified Mj HSP16.5 elutes as a single peak of molecular mass ≈440 kDa. The column was standardized with the following markers as indicated above the figure: carbonic anhydrase (29 kDa); albumin (66 kDa); apoferritin (443 kDa); thyroglobulin (699 kDa). (B) Negative stain EM image of Mj HSP16.5 (1 μM) appears as small particles ≈15–20 nm in diameter. The arrow shows a particle displaying a hole. (Bar = 100 nm.)
Negative stain EM of Mj HSP16.5 at room temperature showed spherical particles of a relatively uniform size of ≈15–20 nm in diameter from a variety of directions (Fig. 1B). An opening can be seen in some of the particles. The hole in the center of the particle also was observed in its three-dimensional crystal structure (29). Taken together, these results suggest that Mj HSP16.5 forms a spherical, homogeneous complex, different from other sHSPs that form oligomers of variable sizes and shapes (9, 10).
Protection from Heat Denaturation by Mj HSP16.5.
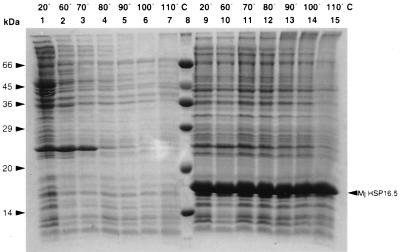
Soluble proteins after heat treatment are considered as being protected from heat denaturation. Fig. 2 shows that proteins in an E. coli cell extract expressing Mj HSP16.5 remained soluble to temperatures as high as 100°C (Fig. 2, lane 14), whereas those in the background host cell extract started to precipitate at a temperature of 60°C (Fig. 2, lane 2). Bradford protein assays of cell extract of the background strain heated to 80°C for 20 min showed that only 22% of the proteins of the cell remained soluble, whereas the extracts from cells expressing Mj HSP16.5 retained >75% of the protein of the cell in a soluble state at 80°C. This indicates either that Mj HSP16.5 expressed in E. coli can protect E. coli proteins or that the process of overexpressing Mj HSP16.5 triggers the expression of unknown E. coli proteins that are capable of protecting other E. coli proteins from heat denaturation.
Figure 2.
Thermal stability of an E. coli crude extract expressing Mj HSP16.5. Extract of soluble E. coli proteins from induced BL21(DE3)/pSJS1240 and BL21(DE3)/pSJS1240 cells expressing Mj HSP16.5 were prepared as described in Materials and Methods. Aliquots of cell extract (4 mg/ml) were heated for 20 min at various temperatures, as indicated. After being allowed to cool to room temperature, samples were centrifuged at 10,000 × g for 5 min. The supernatants (10 μl) were analyzed by SDS/PAGE. Lanes: 1–7, BL21(DE3)/pSJS1240 cell extract heated at 20°, 60°, 70°, 80°, 90°, 100°, and 110°C, respectively; 8, molecular mass markers in kilodaltons; 9–15, transformed BL21(DE3)/pSJS1240 cell extract expressing Mj HSP16.5 heated at 20°, 60°, 70°, 80°, 90°, 100°, and 110°C, respectively.
Purified Mj HSP16.5 Can Protect E. coli Cell Extract from Thermal Aggregation.
To show that thermal protection was caused solely by Mj HSP16.5 and not by induction of other E. coli proteins in response to Mj HSP16.5 overexpression, Mj HSP16.5 was purified to 95% homogeneity (23) and added to E. coli BL21 (DE3)/pSJS1240 cell extracts. The assay was performed at 80°C because at this temperature the background cell extract was very sensitive to precipitation (Fig. 2). When cell extracts (2 mg/ml) were incubated at 80°C for 20 min, cooled, and centrifuged, only 22% of the proteins of the cell remained soluble (Fig. 3, lane 4). When the cell extracts were incubated in the presence of purified Mj HSP16.5 at a 2:1 or 1:1 (wt/wt) ratio at 80°C, 72% and 94%, respectively, of the E. coli proteins remained in the soluble fraction (Fig. 3, lanes 7 and 10). As a control, the cell extract was incubated with cytochrome C at a 1:1 (wt/wt) ratio. Cytochrome C was chosen because it is a small protein in a size range comparable to Mj HSP16.5 and is resistant to precipitation at 80°C. Cytochrome C was not able to protect the cell extract from heat denaturation (Fig. 3, lane 12). This finding shows that Mj HSP16.5 (24 mer) itself is active in preventing the thermal aggregation of E. coli proteins upon heating at 80°C.
Figure 3.
Thermal protection of E. coli cell extract by purified Mj HSP16.5. Cell extract (CE) from E. coli BL21(DE3)/pSJS1240 (2 mg/ml) was incubated with Mj HSP16.5 or cytochrome C at 2:1 or 1:1 (wt/wt) ratio at 80°C for 20 min. Samples were centrifuged, and the pellets were resuspended in the original volume. Ten microliters of each sample were run on a 15% SDS/PAGE gel. Lanes: 1, molecular mass markers in kilodaltons; 2, cell extract (CE), unheated (U); 3–4, CE, heated (H) 80°C, 20 min: insoluble pellet (I), supernatant (S); 5, CE:Mj HSP16.5 (2:1 wt/wt ratio), U; 6–7, CE:Mj HSP16.5 (2:1 wt/wt ratio), H 80°C, 20 min: I, S; 8, CE:Mj HSP16.5 (1:1 wt/wt ratio), U; 9–10, CE:Mj HSP16.5 (1:1 wt/wt ratio), H 80°C, 20 min: I, S; 11, CE:cytochrome C (1:1 wt/wt ratio), U; 12–13, CE:cytochrome C (1:1 wt/wt ratio), H 80°C, 20 min: I, S; 14, 1 μg Mj HSP16.5; 15, 5 μg cytochrome C.
Single Chain Monellin Is Protected from Denaturation by Purified Mj HSP16.5 at High Temperature.
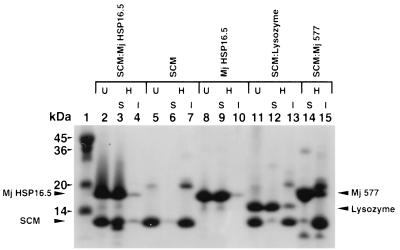
To exclude the possibility that E. coli cell extracts contain one or more components that, in conjunction with purified Mj HSP16.5, protect E. coli cell extracts from heat denaturation, we have tested pure proteins as a substrate for Mj HSP16.5. Monellin is a two-polypeptide chain protein that is responsible for the sweetness of the African berries of Dioscoreophyllum cumminsii (30, 31). A fusion of the two-polypeptide chains resulted in a SCM, a 94-residue polypeptide. SCM is more stable than the two-chain monellin to heat denaturation when heated up to 60°C but is denatured at 80°C (32). Thus, SCM is a good substrate for in vitro studies of the role of Mj HSP16.5 in preventing thermal aggregation at 80°C. As shown in Fig. 4, the incubation of SCM and Mj HSP16.5 at a 1:1 molar monomer ratio at 80°C for 20 min resulted in both proteins remaining soluble (Fig. 4, lane 3).
Figure 4.
Thermal protection of SCM by purified Mj HSP16.5. SCM (90 μM) was incubated at 80°C for 20 min in the presence of Mj HSP16.5, lysozyme, or Mj 577 protein. Samples were centrifuged, and the pellets were resuspended in the original volume. Ten microliters of each sample was run on a 15% SDS/PAGE gel. Lanes: 1, molecular mass markers in kilodaltons; 2, SCM:Mj HSP16.5 (1:1 molar ratio), unheated (U); 3–4, SCM:Mj HSP16.5 (1:1 molar ratio), heated (H): supernatant (S), insoluble pellet (I); 5, SCM U; 6–7, SCM, H: S, I; 8, Mj HSP16.5, U; 9–10, Mj HSP16.5, H: S, I; 11, SCM:lysozyme (1:1 molar ratio), U; 12–13, SCM:lysozyme (1:1 molar ratio), H: S, I; 14–15, SCM:Mj 577 protein (1:1 molar ratio) H: S, I.
As controls, the incubation at 80°C of SCM alone resulted in the protein precipitating (Fig. 4, lane 6), whereas the incubation of Mj HSP16.5 alone showed it to remain in the soluble fraction (Fig. 4, lane 9). SCM was incubated with lysozyme (14.4 kDa), another protein of comparable size to Mj HSP16.5 and known to be heat-tolerant at 80°C. Fig. 4, lanes 12–13, shows that, although lysozyme was heat-resistant to 80°C for 20 min, it did not protect SCM from thermal aggregation. Another control was conducted by using the Mj 577 protein, a M. jannaschii hypothetical protein that is heat-stable to 80°C. SCM was incubated with the Mj 577 protein at a 1:1 molar ratio at 80°C for 20 min. Fig. 4, lanes 14 and 15, shows that the Mj 577 protein could not protect SCM from precipitation upon heating.
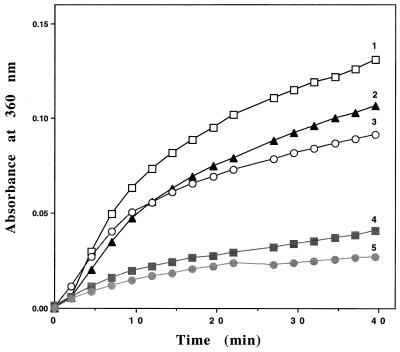
Prevention of Thermal Aggregation of Citrate Synthase by Mj HSP16.5 at an Intermediate Temperature.
Pig heart CS, a dimer of identical 43.5-kDa subunits, was used as the substrate for folding studies because its behavior in this type of study is well characterized (33). The results in Fig. 5 indicate that Mj HSP16.5 effectively inhibits the thermal aggregation of CS at 40°C. This temperature was chosen because CS aggregates at ≈40°C and cannot be incubated at 80°C because it precipitates at that temperature. There was no suppression of CS aggregation when lysozyme was added under the same assay conditions (results not included). It is interesting to note that, in this study, a molar ratio of 1:40 of CS:Mj HSP16.5 protein is needed to prevent thermal aggregation at 40°C, whereas our previous result with SCM shows that a molar ratio of 1:1 of SCM:Mj HSP16.5 is sufficient to inhibit thermal aggregation at 80°C.
Figure 5.
Thermal aggregation of CS was prevented by the addition of Mj HSP16.5. CS dimer at a 5.8-μM concentration was incubated in a spectrophotometer cell thermostatted at 40°C in 50 mM potassium phosphate (pH 7), with various concentrations of Mj HSP16.5 (1: 0, 2: 29.3 μM; 3: 58.6 μM; 4: 177 μM; 5: 234 μM). The aggregation of CS was monitored by measuring apparent light scattering (A360).
DISCUSSION
In contrast to some sHSPs that form heterogeneous oligomeric complexes, we show that a sHSP from M. jannaschii, a 16.5-kDa protein, forms a unique oligomeric structure of homogeneous size. Electron microscopy of Mj HSP16.5 revealed a spherical structure of ≈15–20 nm in diameter with possible or potential openings. Our results from size exclusion chromatography and sedimentation velocity measurements are consistent with the crystal structure (29) that shows the complex to be a hollow sphere comprised of 24 subunits with many windows.
In an E. coli cell extract expressing Mj HSP16.5, 75% of the proteins were protected from heat aggregation at 80°C. Furthermore, purified Mj HSP16.5 had the same effect on total E. coli cell extracts. We also have shown that Mj HSP16.5 can interact with a purified protein such as SCM and prevent aggregation of SCM at 80°C, whereas lysozyme and another heat stable M. jannaschii protein Mj 577 do not. The above results prove that Mj HSP16.5 alone provides thermal protection to aggregation prone proteins without any auxillary proteins. This confirms that Mj HSP16.5 has the same chaperone activity that has been shown for other sHSPs from bacteria and eukaryotes. Mj HSP16.5 exhibits broad substrate specificity as shown by protection of the E. coli cell lysate, in a manner similar to that of murine Hsp25 (21).
SCM is an appropriate substrate for the hyperthermophilic protein Mj HSP16.5 because it is stable at 60°C but showed 85% precipitation at 80°C. This thermal aggregation experiment showed that, at a monomer molar ratio of 1:1, Mj HSP16.5 can protect SCM. Thermal protection by Mj HSP16.5 was tested for another purified protein, CS, which aggregates at a much lower temperature than SCM. Light scattering experiments were performed with CS at 40°C. Under those conditions, a monomer molar ratio of 1:40 of CS:Mj HSP16.5 was necessary to prevent the thermal aggregation of CS. A requirement for a much higher ratio could be due to Mj HSP16.5 needing a higher temperature to be fully active. This difference in activity dependence on temperature has been found with hyperthermophilic enzymes (34). Lee et al. (22) have shown that pea HSP18.1 has a large binding capacity for heat-denatured malate dehydrogenase. It was shown that each subunit of HSP18.1 can bind up to one malate dehydrogenase monomer, as measured by light scattering at 40°C. This is similar to the ratio that we have found for the binding capacity of Mj HSP16.5 to SCM at 80°C. However, the molar ratio can vary depending on the sHSPs and substrates studied (18).
There is the possibility that the sphere formed by 24 subunits of Mj HSP16.5 traps its protein substrates inside or on the surface of the sphere. From the binding studies discussed above, the molar ratio of SCM:Mj HSP16.5 is 1:1. The volume of the cavity as revealed by the crystal structure (29) of this sHSP would not be large enough to allow 24 SCMs to be accommodated inside the sphere without changing its size. Previous biochemical and EM studies of pea HSP18.1 have suggested that the substrate coats the surface of the HSP particles (22), but presently, the exact mechanism by which Mj HSP16.5 interacts with and prevents denatured proteins from aggregating is not known.
Acknowledgments
We are grateful to Reena Zalpuri for help in obtaining the electron micrographs, Dr. Tom Zarembinski for providing the Mj 577 protein, Steve Santoso and Dimitry Lerner for their technical assistance, Lan Huang for performing the sedimentation velocity measurements, and Dr. David Boisvert for helpful discussions. This work was supported by the Director, Office of Energy Research, Office of Biological and Environmental Research, of the U.S. Department of Energy under Contract DE-AC03-76SF0098 (R.K. and S.-H.K.).
ABBREVIATIONS
- HSP
heat shock proteins
- sHSP
small HSP
- SCM
single-chain monellin
- CS
citrate synthase
- SEC
size exclusion chromatography
References
- 1.Gething M J, Sambrook J. Nature (London) 1992;355:33–45. doi: 10.1038/355033a0. [DOI] [PubMed] [Google Scholar]
- 2.Hendrick J P, Hartl F-U. Annu Rev Biochem. 1993;62:349–384. doi: 10.1146/annurev.bi.62.070193.002025. [DOI] [PubMed] [Google Scholar]
- 3.Parsell D A, Lindquist S. In: The Biology of Heat Shock Proteins and Molecular Chaperones. Morimoto R I, Tissieres A, Georgopoulos C, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press; 1994. pp. 457–494. [Google Scholar]
- 4.Buchner J. FASEB J. 1996;10:10–19. [PubMed] [Google Scholar]
- 5.Caspers G-J, Leunissen J A M, de Jong W W. J Mol Evol. 1995;40:238–248. doi: 10.1007/BF00163229. [DOI] [PubMed] [Google Scholar]
- 6.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, Fitzgerald L M, Clayton R A, Gocayne J D, et al. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 7.Plesofsky-Vig N, Brambl R. Proc Natl Acad Sci USA. 1995;92:5032–5036. doi: 10.1073/pnas.92.11.5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Jong W W, Leunissan J A M, Voorter C E M. Mol Biol Evol. 1993;10:103–126. doi: 10.1093/oxfordjournals.molbev.a039992. [DOI] [PubMed] [Google Scholar]
- 9.Leroux M R, Melki R, Gordon B, Batelier G, Candido E P M. J Biol Chem. 1997;272:24646–26656. doi: 10.1074/jbc.272.39.24646. [DOI] [PubMed] [Google Scholar]
- 10.Haley D A, Horwitz J, Stewart P L. J Mol Biol. 1998;277:27–35. doi: 10.1006/jmbi.1997.1611. [DOI] [PubMed] [Google Scholar]
- 11.Chang Z, Primm T P, Jakana J, Lee I H, Serysheva I, Chiu W, Gilbert H F, Quiocho F A. J Biol Chem. 1996;271:7218–7223. [PubMed] [Google Scholar]
- 12.Schirmer E C, Lindquist S, Vierling E. Plant Cell. 1994;6:1899–1909. doi: 10.1105/tpc.6.12.1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van den IJssel P R, Overkamp P, Knauf U, Gaestel M, de Jong W W. FEBS Lett. 1994;355:54–56. doi: 10.1016/0014-5793(94)01175-3. [DOI] [PubMed] [Google Scholar]
- 14.Horwitz J. Proc Natl Acad Sci USA. 1992;89:10449–10453. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacob U, Gaestel M, Engel K, Buchner J. J Biol Chem. 1993;268:1517–1520. [PubMed] [Google Scholar]
- 16.Merck K B, Groenen P J T A, Voorter C E M, de Haard-Hoekman W A, Horwitz J, Boemendal H, de Jong W W. J Biol Chem. 1993;268:1046–1052. [PubMed] [Google Scholar]
- 17.Lee G J, Pokala N, Vierling E. J Biol Chem. 1995;270:10432–10438. doi: 10.1074/jbc.270.18.10432. [DOI] [PubMed] [Google Scholar]
- 18.Chang Z, Primm T P, Jakana J, Lee I H, Serysheva I, Chiu W, Gilbert H F, Quiocho F A. J Biol Chem. 1996;271:7218–7223. [PubMed] [Google Scholar]
- 19.Jaenicke R, Creighton T E. Curr Biol. 1993;3:234–235. doi: 10.1016/0960-9822(93)90342-l. [DOI] [PubMed] [Google Scholar]
- 20.Waters E R, Lee J L, Vierling E. J Exp Bot. 1996;47:325–338. [Google Scholar]
- 21.Ehrnsperger M, Graber S, Gaestel M, Buchner J. EMBO J. 1997;16:221–229. doi: 10.1093/emboj/16.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee G J, Roseman A M, Saibil H R, Vierling E. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K K, Yokota H, Santoso S, Lerner D, Kim R, Kim S-H. J Struct Biol. 1998;121:76–80. doi: 10.1006/jsbi.1998.3969. [DOI] [PubMed] [Google Scholar]
- 24.Tseng T S, Yeh K W, Yeh C H, Chang F C, Chen Y M, Lin C Y. Plant Mol Biol. 1992;18:963–965. doi: 10.1007/BF00019209. [DOI] [PubMed] [Google Scholar]
- 25.Kim R, Sandler S J, Goldman S, Yokota H, Clark A J, Kim S-H. Biotech Lett. 1998;20:207–210. [Google Scholar]
- 26.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Singh M, Brooks G C, Srere P A. J Biol Chem. 1970;245:4636–4640. [PubMed] [Google Scholar]
- 29.Kim, K. K., Kim, R. & Kim, S.-H. Nature (London), in press.
- 30.Morris J A, Cagan R H. Biochim Biophys Acta. 1972;261:114–122. doi: 10.1016/0304-4165(72)90320-0. [DOI] [PubMed] [Google Scholar]
- 31.van der Wel H. FEBS Lett. 1972;21:88–90. doi: 10.1016/0014-5793(72)80170-4. [DOI] [PubMed] [Google Scholar]
- 32.Kim S-H, Kang C-H, Kim R, Cho J M, Lee Y-B, Lee T-K. Protein Eng. 1989;2:571–575. doi: 10.1093/protein/2.8.571. [DOI] [PubMed] [Google Scholar]
- 33.Zhi W, Landry S J, Gierasach L M, Srere P A. Protein Sci. 1992;1:522–529. doi: 10.1002/pro.5560010407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Danson M J, Hough D W, Russell R J M, Taylor G L, Pearl L. Protein Eng. 1996;9:629–630. doi: 10.1093/protein/9.8.629. [DOI] [PubMed] [Google Scholar]