Abstract
The gp41 subunit of the envelope protein complex from human and simian immunodeficiency viruses (HIV and SIV) mediates membrane fusion during viral entry. The crystal structure of the HIV-1 gp41 ectodomain core in its proposed fusion-active state is a six-helix bundle. Here we have reconstituted the core of the SIV gp41 ectodomain with two synthetic peptides called SIV N36 and SIV C34, which form a highly helical trimer of heterodimers. The 2.2 Å resolution crystal structure of this SIV N36/C34 complex is very similar to the analogous structure in HIV-1 gp41. In both structures, three N36 helices form a central trimeric coiled coil. Three C34 helices pack in an antiparallel orientation into highly conserved, hydrophobic grooves along the surface of this coiled coil. The conserved nature of the N36-C34 interface suggests that the HIV-1 and SIV peptides are functionally interchangeable. Indeed, a heterotypic complex between HIV-1 N36 and SIV C34 peptides is highly helical and stable. Moreover, as with HIV-1 C34, the SIV C34 peptide is a potent inhibitor of HIV-1 infection. These results identify conserved packing interactions between the N and C helices of gp41 and have implications for the development of C peptide analogs with broad inhibitory activity.
Human and simian immunodeficiency viruses (HIV and SIV) use envelope glycoproteins to enter cells. The envelope glycoprotein complex consists of the surface subunit gp120 and the transmembrane subunit gp41, which are produced through proteolytic cleavage of the precursor gp160. gp120 binds to CD4 and one of several coreceptors that are members of the chemokine receptor family. Subsequently, gp41 undergoes conformational changes that facilitate fusion of viral and host cell membranes (reviewed in ref. 1). Structural studies of these envelope proteins will be essential in elucidating the mechanism of membrane fusion in these viruses.
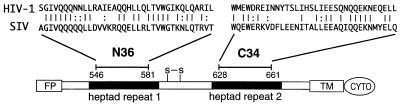
The gp41 ectodomain contains several notable features (Fig. 1). It has a fusion peptide at the amino terminus that is thought to insert directly into the target membrane during the membrane fusion process. Carboxyl-terminal to the fusion peptide are two regions containing 4,3 hydrophobic (heptad) repeats, sequence motifs suggestive of coiled-coil structures. We have used protein-dissection methods to identify stable, protease-resistant cores in the ectodomains of gp41 from HIV-1 (2, 3) and SIV (4). In both cases, the core consists of two peptides, termed N and C peptides, that correspond to the two regions with hydrophobic heptad repeats. These biophysical studies show that the N and C peptides form a stable, helical trimer of heterodimers (2, 4).
Figure 1.
A schematic view of gp41, showing the locations of the N36 and C34 peptides from SIV and HIV-1. Identical residues between the SIV and HIV-1 peptides are indicated with a bar, and similar residues are indicated by dots. To facilitate comparisons between the HIV-1 and SIV structures, the peptide residues are numbered according to their positions in HIV-1 gp160 (HXB2 strain; GenBank accession no. K03455). For the SIV peptides (SIV Mac239 strain; GenBank accession no. M33262), N36 actually corresponds to positions 559–594, and C34 corresponds to 637–670. The 4,3 hydrophobic repeats are indicated with black shading. FP, fusion peptide; S—S, disulfide bond; TM, transmembrane region; CYTO, cytoplasmic domain.
Crystallographic analysis of the HIV-1 gp41 core confirmed that the N/C complex is a 6-helix bundle in which three N helices form an interior, trimeric coiled coil (5–7). Three C helices pack in an antiparallel orientation into hydrophobic grooves on the surface of the N peptide coiled coil. This structure likely represents the fusion-active conformation of HIV-1 gp41 and resembles the fusion-active conformation of the transmembrane fusion proteins of influenza (8) and Moloney murine leukemia virus (9). Interestingly, synthetic C peptides are potent inhibitors of infection for both laboratory-adapted strains and primary isolates of HIV-1 (10, 11). These C peptides likely inhibit infection, in a dominant-negative manner, by binding to a transiently exposed, coiled-coil trimer formed by the N peptide region of intact gp41 (reviewed in ref. 1).
A high-resolution structure of the related SIV gp41 core would contribute to our understanding of HIV-2, the second subtype of HIV. The envelope proteins of HIV-2 are serologically distinct from those of HIV-1, but are closely related to those of SIV. Sequence analysis indicates that SIV and HIV-2 are closely related viruses (12, 13). HIV-2 infection in humans can lead to the development of AIDS, although the virus appears to be less pathogenic than HIV-1. A detailed structure of the SIV gp41 core also would allow a direct comparison of its helical interactions with those in HIV-1 gp41. Such a comparison could have implications for the design and development of drugs based on C peptide analogs.
In our current study, we have reconstituted the core of the SIV gp41 ectodomain by using two synthetic peptides called SIV N36 and SIV C34. Each of these peptides in isolation is poorly structured, but they form a highly helical complex when mixed together. Crystallographic analysis of the SIV N36/C34 complex at 2.2 Å resolution reveals a trimeric structure that is remarkably similar to the analogous complex from HIV-1 gp41. This high degree of structural conservation motivated us to study the biophysical properties of a heterotypic complex between HIV-1 N36 and SIV C34. We find that such a hybrid is highly helical and stable. Moreover, the SIV C34 peptide is a potent inhibitor of HIV-1 infection. These results demonstrate that divergent immunodeficiency viruses contain envelope proteins that are highly conserved structurally and functionally, and they provide a structural explanation for the broad inhibitory activity of the C peptides.
MATERIALS AND METHODS
Peptides and CD Spectroscopy.
SIV N36 and SIV C34 peptides (sequences shown in Fig. 1) were synthesized by using fluorenylmethoxycarbonyl peptide chemistry. Both peptides were acetylated at the amino termini and amidated at the carboxyl termini. After cleavage from the resin, the peptides were desalted on a Sephadex G-25 column (Pharmacia) and lyophilized. The N36 peptide was then dissolved in water and purified by reverse-phase HPLC (Waters) with a Vydac C18 preparative column (water-acetonitrile gradient in the presence of 0.1% trifluoroacetic acid). The C34 peptide, because of its lower solubility at low pH, was purified by using a Jupiter C18 preparative column at pH 6.8 (water-acetonitrile gradient in the presence of 50 mM triethylamine-acetic acid buffer). The peptide identities were checked by MALDI mass spectrometry (Voyager Elite, PerSeptive Biosystems). CD spectroscopy was performed in PBS (50 mM sodium phosphate, pH 7.0/150 mM NaCl) on an Aviv Model 62DS CD spectrometer (Aviv Associates, Lakewood, NJ) as described previously (2). Peptide concentration was determined by absorbance at 280 nm in 6 M guanidine hydrochloride (14).
Crystallization and Structure Determination.
Stocks of purified N36 and C34 peptide were dissolved in water and 10 mM sodium phosphate, pH 7.5, respectively. Equimolar amounts of N36 and C34 were mixed, and the final protein concentration was adjusted to 10 mg/ml. Initial crystallization conditions were found by using the vapor diffusion method with sparse matrix crystallization kits (Crystal Screen I and II, Hampton Research, Riverside, CA) and then optimized. Several crystal forms were obtained, but the best diffracting crystals were grown by equilibrating 4-μl drops (protein solution mixed 1:1 with reservoir solution) against a reservoir solution containing 18–19% PEG8000/0.1 M sodium cacodylate (adjusted to pH 6.5 with HCl)/0.2 M magnesium acetate. Crystals belong to space group P212121 (a = 55.74 Å, b = 57.59 Å, and c = 66.75 Å; α = β = γ = 90°). The appearance of one trimer in the asymmetric unit results in a solvent content of 0.44 (15). The crystals demonstrate high mosaicity and diffract anisotropically to a maximum resolution of 1.9 Å, but to only 2.2 Å in the orthogonal directions. Isomorphous crystals of similar size also were grown by using 30% PEG4000/0.1 M Tris⋅HCl, pH 8.5/0.2 M MgCl2, but their diffraction did not exceed 2.3 Å.
For data collection, crystals were harvested and flash frozen from a drop containing reservoir solution plus 10% 2-methyl-2,4-pentanediol. Diffraction data were collected at 100 K by using an X-stream cryosystem and an R-AXIS IV detector mounted on a Rigaku RU300 rotating-anode x-ray generator (Molecular Structure, The Woodlands, TX). Diffraction intensities were integrated by using the denzo and scalepack software (16) and reduced to structure factors by using the program truncate from the CCP4 program suite (17). The structure of SIV N36/C34 was solved by molecular replacement by using amore (18), with an SIV N36/C34 model built from the x-ray structure of HIV-1 N36/C34 (5). Solvent flattening, histogram matching, and 3-fold noncrystallographic averaging with the program dm (19) were used to remove model bias. This produced an electron density map of good quality, which clearly revealed differences from the initial model (Fig. 3A). Noncrystallographic symmetry restraints were not used in the refinement. Density interpretation and model building were done with the program o (20). Bulk solvent and anisotropic overall B-factor corrections, as implemented in the programs x-plor (21) and refmac (17), were crucial for crystallographic refinement of SIV N36/C34 (Table 1).
Figure 3.
Stereoviews of representative electron density maps. (A) Initial 2Fo-Fc map after density modification and phase improvement with dm (19) superimposed onto the initial molecular replacement solution model. A shift of the N36 helix (left side of figure) and other clear differences between the trial model and the observed density are seen (white arrows). (B) Final 2Fo-Fc map superimposed onto the refined model. Water molecules are indicated with small, red balls. Both maps are contoured at 1.5 σ. Figures were generated with o (20).
Table 1.
Data collection and refinement statistics
| Data collection | |
| Highest resolution, Å | 2.20 |
| Observed reflections | 52,353 |
| Unique reflections | 11,157 |
| Completeness, % | 96.9 |
| Rmerge* | 0.058 |
| Refinement | |
| Resolution, Å | 20.0–2.20 |
| Protein nonhydrogen atoms | 1,809 |
| Water molecules | 184 |
| Rcryst† | 0.211 |
| Rfree† | 0.288 |
| Average B-factor, Å2 | 36.2 |
| rmsd from ideal geometry | |
| Bond length, Å | 0.009 |
| Bond angles, ° | 1.1 |
| Torsion angles, ° | 20.8 |
Rmerge = ΣΣj|Ij(hkl) − 〈I(hkl)〉 |/Σ| 〈I(hkl)〉|, where Ij is the intensity measurement for reflection j and 〈I〉 is the mean intensity over j reflections.
Rcryst (Rfree) = Σ∥Fobs(hkl)| − |Fcalc(hkl)∥/Σ|Fobs(hkl)|, where Fobs and Fcalc are observed and calculated structure factors, respectively. Statistics are given for F > 2σ. Ten percent of the reflections were excluded from refinement and used to calculate Rfree.
Viral Fusion Assay.
The inhibitory activity of HIV-1 and SIV C34 peptides was determined by using a recombinant luciferase-encoding HIV-1 (22). This replication-incompetent virus was produced by cotransfection of 293T cells with the plasmid NL43LucR–E– (22) and the HXB2 gp160 expression vector pCMVHXB2 gp160 (D.C.C. and P.S.K., unpublished data). Viral supernatants were used to infect HOST4LES cells (N. Landau, National Institutes of Health AIDS Reagent Program) in the presence of varying concentrations of peptide to obtain a dose-response curve. Each infection was done in triplicate. Cell lysates were prepared 48 hr postinfection, and luciferase activity was measured in a Wallac AutoLumat LB953 luminometer (EG & G, Salem, MA).
RESULTS
Stability of the SIV N36/C34 Complex in Solution.
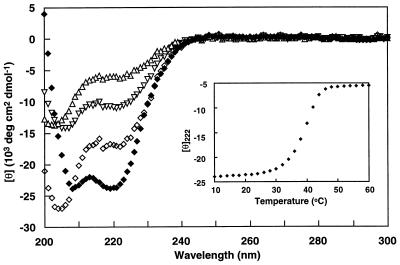
CD spectra for SIV N36 and SIV C34 show that neither peptide in isolation has significant helical structure. However, when SIV N36 and C34 are mixed together in equimolar amounts, there is a large increase in helicity (Fig. 2). In this assay, the SIV N36/C34 complex is estimated to be 70% helical. Upon thermal denaturation, this complex unfolds cooperatively and reversibly with a transition midpoint of 40°C (Fig. 2). The corresponding HIV-1 N36/C34 complex has an apparent Tm of 66°C (Fig. 6A) (3).
Figure 2.
CD spectra of SIV N36 and C34 peptides. CD spectra of SIV N36 (▿), SIV C34 (▵), and the SIV N36 C34 complex (♦) in PBS. The predicted spectrum for noninteracting peptides (⋄) is shown for comparison. Inset shows the thermal dependence of the CD signal at 222 nm for the SIV N36/C34 complex.
Figure 6.
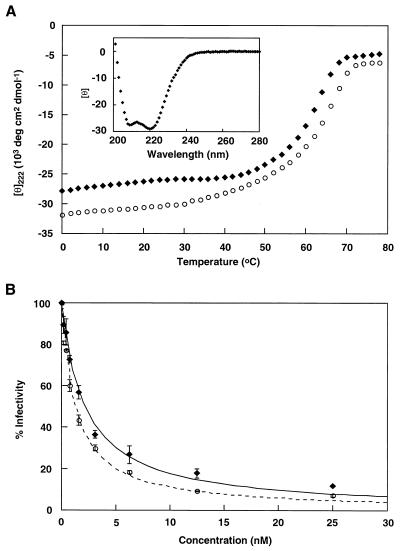
HIV/SIV heterotypic complexes. (A) Thermal dependence of the CD signal at 222 nm for the HIV N36/SIV C34 heterotypic complex (♦) and the HIV N36/HIV C34 complex (○). Inset shows the CD spectrum for the HIV N36/SIV C34 heterotypic complex. (B) Inhibition of HIV-1 (HXB2 strain) infection by SIV C34 (♦ and solid line) and HIV C34 peptide (○ and dashed line). Error bars indicate the SE from triplicate experiments.
Crystal Structure of the SIV gp41 Core.
Crystals of the SIV N36/C34 complex are obtained by using the hanging-drop vapor-diffusion method. Crystal density calculations (15) indicate that the SIV N36/C34 crystals contain one N36/C34 trimer in the asymmetric unit; each monomer is related by an approximate noncrystallographic 3-fold axis. In contrast, the crystal forms of the HIV-1 gp41 core contain trimers centered on a crystallographic 3-fold axis (5–7). Using the molecular replacement method, various trimeric SIV N36/C34 models based on the HIV-1 ectodomain structure yielded the same rotation–translation solution. This solution was consistent with the orientation of the noncrystallographic 3-fold axis deduced from a self-rotation function. Phase improvement with dm (19) increased the quality of the initial electron density map and served to reduce model bias, as indicated by the appearance of clear 2Fo-Fc electron density not dictated by the trial model (Fig. 3A). The final 2Fo-Fc electron density map is of good quality (Fig. 3B) and reveals the positions of all the amino acid residues except for a few disordered side chains at the helix termini and on the surface.
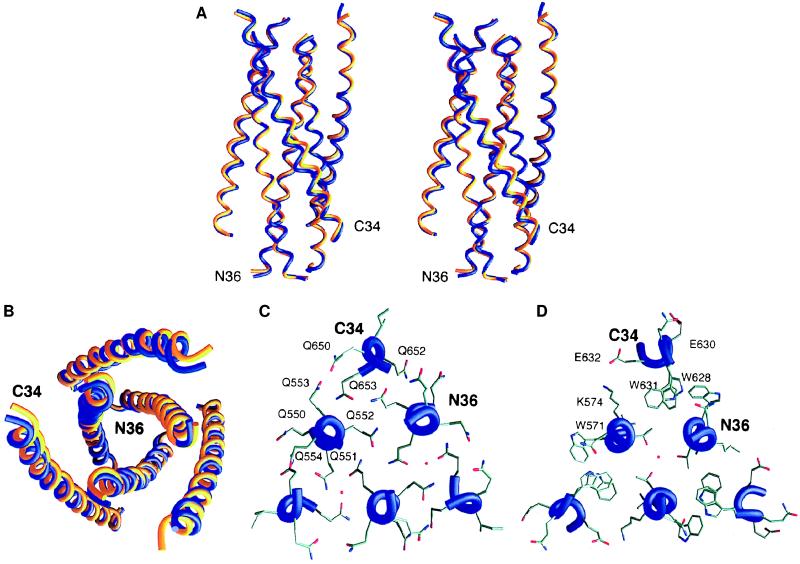
The overall topology of the SIV gp41 core is the same as that of the HIV gp41 core. Three SIV N36 helices form a central, parallel coiled coil with a left-handed superhelical pitch, whereas three C34 helices pack, in an antiparallel orientation, into grooves along the surface of the N36 coiled coil (Fig. 4 A and B). The interface between each C34 helix and the N36 core is predominantly hydrophobic. The root mean square deviation (rmsd) between all Cα atoms of the N36 coiled-coil core in SIV and HIV is 0.54 Å. If Cα atoms of the entire structure are used for superposition, the rmsd increases to 0.92 Å because of the pronounced shift and bending of the C34 helix in the SIV structure (Fig. 4A). The shift is most pronounced near Gln-640, in the middle of the C34 helix, where the rmsd is 2.0–2.5 Å.
Figure 4.
Structure of SIV N36/C34 complex and comparison with that of HIV-1. (A) Stereoview from the side. The amino termini of N36 and the carboxyl termini of C34 are at the top of the figure. Helices in the N36 coiled-coil trimer from SIV (blue) and HIV-1 (yellow) were used for the superposition. The inner N36 coiled-coil trimers superimpose well; however, a pronounced shift in the central region of the outer C34 helices is visible. (B) View from the top, along the noncrystallographic 3-fold axis. The same color coding is used. (C) A cross-section of the SIV ectodomain composed almost exclusively of glutamine residues, six of which are conserved in the HIV ectodomain sequence and two that are conservatively replaced by asparagines. Glutamine side chains are involved in a web of hydrogen bonds that also involve some carbonyl oxygens of the main chain. (D) A cross-section of the SIV N36/C34 structure near the conserved, deep hydrophobic cavity involving three tryptophan residues. Figures were generated with grasp (28).
Conserved Helical Interactions.
Two notable features of the N36-C34 interface are conserved between the SIV and HIV-1 gp41 structures. First, although most of the N36-C34 interface involves hydrophobic interactions, one layer of interactions is polar in nature and includes conserved clusters of glutamines (Fig. 4C). The SIV N36 sequence contains five glutamines in a row (Fig. 1); it remains to be determined whether there is any relationship between the glutamine clusters seen here (Fig. 4C) and those found in polyglutamine repeats implicated in neurological diseases (23). Second, each of the grooves on the surface of the SIV N36 coiled coil contains a deep cavity lined by hydrophobic side chains from two N36 helices. The side chains of two tryptophans and a valine (isoleucine in the case of HIV-1) from the C34 helices pack into this cavity (Figs. 4D and 5B). A conserved salt bridge is formed on a side of the cavity between Lys-574 from N36 and Glu-632 from C34 (Fig. 4D).
Figure 5.
The conserved hydrophobic cavity on the surface of the N36 coiled coil. (A) Structural overlay of the published HIV-1 gp41 ectodomain structures. The three structures [Chan et al. (5), yellow; Weissenhorn et al. (6), red; and Tan et al. (7), green] deviate substantially in this region. (B) Interactions in the N36 hydrophobic cavity of SIV (blue) are most similar to the Chan et al. (5) HIV-1 structure (yellow). The same superposition as in Fig. 4 A and B was used. The SIV N36 coiled coil is represented as a molecular surface, and the C34 helices are shown as ribbons with selected side chains. The molecular surface is colored white for residues that are identical in SIV and HIV, and green for residues that are not identical. Figures were generated with grasp (28).
The analogous deep cavity in the N36 coiled coil of HIV gp41 has been suggested to be a potential drug target (5). Three reasons for this suggestion are: (i) earlier genetic studies indicate that two of the N36 residues that form the cavity (Leu-568 and Trp-571) are critical for membrane-fusion activity (24), (ii) the N36 residues that form the cavity are highly conserved in different HIV-1 isolates, and even between HIV-1 and SIV, so that drug-escape mutants are expected to be less frequent, and (iii) the combined molecular mass of the C34 residues that insert into the N36 cavity is ≈600 Da, which is within the size range for the development of orally bioavailable, small-molecule drugs.
For effective drug design, it is crucial to obtain an accurate, high-resolution view of this cavity and the residues that project into it. However, the three existing HIV-1 gp41 crystal structures (5–7) show different rotamers for two key residues that are involved in forming the cavity: Trp-571 of N36 and Trp-628 of C34 (Fig. 5A). Our previous crystallographic analysis of HIV-1 gp41 showed unambiguous experimental electron density in this region (see figure 2 in ref. 5). These side chain discrepancies may be secondary to differences in crystal packing contacts, the use of different crystal forms, chain termini effects, or an inherent flexibility in these side chains. It is also possible that in one study (7) Trp-628 may be distorted because of an artificial linker that immediately precedes it, whereas in a second study (6) Trp-628 may be poorly ordered, as suggested by very high individual atomic B factors.
The SIV structure described here is highly consistent (Fig. 5B) with the HIV-1 gp41 structure that we described previously (5). Because this HIV-1 structure was used to solve the SIV gp41 structure by molecular replacement, we were concerned that these similarities might arise from model bias. However, electron density for key residues that form the cavity (e.g., Leu-568, Val-570, Trp-571, Gly-572, Thr-573, Asn-575, Leu-576, and Gln-577 of N36 and Trp-628, Trp-631, and Val-635 of C34) was unambiguous after density modification (19) (Fig. 3A). Moreover, each of these residues was confirmed with simulated annealing omit maps calculated with x-plor (21). Chain termini effects are unlikely to explain the similar side chain conformations observed with Trp-628, because preliminary analysis of a larger SIV gp41 construct (V.N.M. and P.S.K., unpublished observations) in a distinct crystal form indicates the same side chain conformations.
Heterotypic Complexes of the gp41 Core.
The remarkable conservation of the N–C interface suggests that the N and C peptides of SIV and HIV-1 might be functionally interchangeable, at least in the formation of this six-helix structure. Simple modeling indicates that the SIV C34 helix can fit easily into the hydrophobic groove on the surface of the HIV-1 N36 core. Indeed, CD measurements show that a heterotypic complex consisting of HIV-1 N36 and SIV C34 is highly helical (Fig. 6A Inset). Remarkably, the apparent Tm of this heterotypic complex is 62°C, close to that of the HIV-1 gp41 core (Tm = 66°C; Fig. 6A), and significantly greater than that of the SIV gp41 core (Tm = 40°C; Fig. 2).
The high stability of this heterotypic complex suggests that SIV C34, like HIV-1 C34, might inhibit HIV-1 infection. Indeed, SIV C34 potently inhibited infection by HIV-1 (HXB2 strain) with an IC50 of 2.2 ± 0.2 nM (mean ± SE), compared with an IC50 of 1.3 ± 0.07 nM for HIV-1 C34 (Fig. 6B).
DISCUSSION
Although the similarity of the HIV-1 and SIV gp41 core structures was anticipated (4), several features of SIV and HIV-1 envelope proteins are different. First, the SIV envelope proteins are serologically related to those of HIV-2 but are distinct from those of HIV-1 (12). These serological relations are well supported by phylogenetic sequence analysis of these lentiviruses. Second, the gp120/gp41 complex of SIV appears to be more tightly associated than that of HIV-1, at least when compared with some laboratory-adapted strains of HIV-1, including the HXB2 strain (25, 26). Finally, the gp120 subunit of SIV has been reported to bind CD4 with lower affinity than HIV-1 gp120 (26).
A comparison of the HIV-1 and SIV gp41 cores reveals a high degree of structural similarity. The overall structure of both complexes consists of a six-helix bundle, in which three N helices form an interior, parallel coiled coil. Three C34 helices wrap around the outside of this coiled coil to form a highly conserved interface. These structural similarities indicate that the fusion mechanisms of HIV-1 and SIV are closely related.
The high degree of similarity at the interfaces between the N and C peptide helices in the HIV-1 and SIV gp41 cores provides a structural explanation for the broad inhibitory activity of the C peptides. C peptides (derived from the laboratory-adapted strain HXB2) potently inhibit a wide range of HIV-1 isolates, including primary isolates that are typically resistant to neutralization by soluble CD4 and neutralizing antibodies (10, 11). At higher concentrations, the C peptide DP178 also is capable of inhibiting HIV-2 strains (11, 27). We show that SIV C34 is a potent inhibitor of HIV-1 infection. Our results reinforce the notion that future development of C peptide analogs should target conserved structural features of the N36 coiled coil.
Acknowledgments
We thank Michael Burgess, James Pang, and Roberta Moro for peptide synthesis support, and Dr. Benjamin Chen for reagents and helpful advice on viral infection assays. D.C.C. is supported by a postdoctoral fellowship from the Jane Coffin Childs Memorial Fund for Medical Research. This research was funded by the National Institutes of Health (PO1 GM56552) and utilized the W. M. Keck Foundation X-Ray Crystallography Facility at the Whitehead Institute.
ABBREVIATIONS
- SIV
simian immunodeficiency virus
- HIV-1 and HIV-2
HIV types 1 and 2
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, Biology Department, Brookhaven National Laboratory, Upton, NY 11973 (PDB ID code 2 siv) and are available immediately at the website http://www.wi.mit.edu/kim/home.html.
References
- 1.Chan D C, Kim P S. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 2.Lu M, Blacklow S C, Kim P S. Nat Struct Biol. 1995;2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 3.Lu M, Kim P S. J Biomol Struct Dyn. 1997;15:465–471. doi: 10.1080/07391102.1997.10508958. [DOI] [PubMed] [Google Scholar]
- 4.Blacklow S C, Lu M, Kim P S. Biochemistry. 1995;34:14955–14962. doi: 10.1021/bi00046a001. [DOI] [PubMed] [Google Scholar]
- 5.Chan D C, Fass D, Berger J M, Kim P S. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 6.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Nature (London) 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 7.Tan K, Liu J, Wang J, Shen S, Lu M. Proc Natl Acad Sci USA. 1997;94:12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bullough P A, Hughson F M, Skehel J J, Wiley D C. Nature (London) 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 9.Fass D, Harrison S C, Kim P S. Nat Struct Biol. 1996;3:465–469. doi: 10.1038/nsb0596-465. [DOI] [PubMed] [Google Scholar]
- 10.Jiang S, Lin K, Strick N, Neurath A R. Nature (London) 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 11.Wild C T, Shugars D C, Greenwell T K, McDanal C B, Matthews T J. Proc Natl Acad Sci USA. 1994;91:9770–9774. doi: 10.1073/pnas.91.21.9770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy J A. HIV and the Pathogenesis of AIDS. Washington, DC: Am. Soc. Microbiol.; 1998. [Google Scholar]
- 13.Luciw P A. In: Fields Virology. Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Strauss S E, editors. Philadelphia: Lippincott; 1996. pp. 1881–1952. [Google Scholar]
- 14.Edelhoch H. Biochemistry. 1967;6:1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- 15.Matthews B W. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 16.Otwinowski Z. In: Data Collection and Processing. Sawer L, Isaacs N, Bailey S, editors. Daresbury Laboratory, Warrington, England: Science and Engineering Research Council; 1993. pp. 55–62. [Google Scholar]
- 17.CCP4. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 18.Navaza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 19.Cowtan K D. Joint CCP 4 and ESF-EACBM Newslett Prot Crystallogr. 1994;31:43–38. [Google Scholar]
- 20.Jones T A, Zou J W, Cowan S W, Kjelgaard M. Acta Crystallogr D. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 21.Brünger A T. x-plor, A System for Crystallography and NMR. New Haven, CT: Yale Univ. Press; 1992. , Version 3.1. [Google Scholar]
- 22.Chen B K, Saksela K, Andino R, Baltimore D. J Virol. 1994;68:654–660. doi: 10.1128/jvi.68.2.654-660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ross C A. Neuron. 1997;19:1147–1150. doi: 10.1016/s0896-6273(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 24.Cao J, Bergeron L, Helseth E, Thali M, Repke H, Sodroski J. J Virol. 1993;67:2747–2755. doi: 10.1128/jvi.67.5.2747-2755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sattentau Q J, Moore J P. Philos Trans R Soc B. 1993;342:59–66. doi: 10.1098/rstb.1993.0136. [DOI] [PubMed] [Google Scholar]
- 26.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Furuta R A, Wild C T, Weng Y, Weiss C D. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 28.Nichols A, Sharp K, Honig B. Proteins Struct Funct Genet. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]