Abstract
The cell wall protects fungi against lysis and determines their cell shape. Alpha-glucan is a major carbohydrate component of the fungal cell wall, but its function is unknown and its synthase has remained elusive. Here, we describe a fission yeast gene, ags1+, which encodes a putative alpha-glucan synthase. In contrast to the structure of other carbohydrate polymer synthases, the predicted Ags1 protein consists of two probable catalytic domains for alpha-glucan assembly, namely an intracellular domain for alpha-glucan synthesis and an extracellular domain speculated to cross-link or remodel alpha-glucan. In addition, the predicted Ags1 protein contains a multipass transmembrane domain that might contribute to transport of alpha-glucan across the membrane. Loss of Ags1p function in a temperature-sensitive mutant results in cell lysis, whereas mutant cells grown at the semipermissive temperature contain decreased levels of cell wall alpha-glucan and fail to maintain rod shapes, causing rounding of the cells. These findings demonstrate that alpha-glucan is essential for fission yeast morphogenesis.
Cell morphogenesis involves a number of discrete molecular mechanisms, such as establishment of cell polarity and structural maintenance of cell shape. Fission yeast, Schizosaccharomyces pombe, has been used as a model eukaryote to study systematically polarity of cell growth (1). Fission yeast cells restrict cell growth to their ends and form a cylindrical rod-like cell shape. Mutants have been isolated that no longer restrict growth toward the two ends, but instead grew from three ends; these tea mutants display a T shape (2). In addition, mutants have been identified that have lost their ability to grow in a polarized fashion; those orb mutants were rounded (3).
To display a rod shape, fission yeast cells not only need to polarize cell growth, they also must maintain cell shape. In fission yeast, the cell wall is responsible for maintenance of cell shape. The cell wall is an extracellular matrix with a layered organization, consisting of an outer layer of glycoproteins and an inner layer of carbohydrate polymers. This carbohydrate layer is composed mainly of β(1 → 3)-glucan (55% of cell wall carbohydrate) and α(1 → 3)-glucan (28%), but it also contains some β(1 → 6)-glucan (6%) and chitin (0.5%) (4–6). This carbohydrate layer, in turn, is composed of two layers, an outer layer of amorphous β(1 → 3)-glucan and a rigid inner layer, which appears fibrillar (7). Although this inner layer appears to have a continuous fibrillar structure, it is composed of two types of microfibrils that are interwoven, namely fibrils consisting predominantly of β(1 → 3)-glucan and fibrils consisting predominantly of α(1 → 3)-glucan. Importantly, this fibrillar layer is responsible for maintaining cell shape (7).
Although β(1 → 3)-glucan has been studied in great detail, little is known about α-glucan. β(1 → 3)-glucan generally is accepted to be a principal support of cell shape. For example, in fission yeast, inhibition of β(1 → 3)-glucan synthesis by the antifungal drug aculeacin A resulted in cell lysis (8), whereas partial inhibition resulted in cell walls with a looser, more flexible structure, causing rounding of the cells under the intrinsic turgor pressure (9). In contrast, the function of α-glucan is not clear, and it has remained unknown whether α-glucan contributes to maintenance of cell shape. In S. pombe, α-glucan consists of straight-chain polymers, approximately 200 glucose moieties in length, linked by α(1 → 3)-glucosidic bonds [and some 7% of α(1 → 4)-glucosidic bonds] (4). Although genes for β(1 → 3)-glucan synthases have been cloned, genes for α(1 → 3)-glucan synthases have not been reported.
Here, we describe a fission yeast mutant that no longer is able to maintain its rod shape. Carbohydrate biochemical analyses show that its cell wall contains reduced levels of alpha-glucan, and amino acid sequence similarities indicate that the mutated gene encodes a catalytic subunit for alpha-glucan synthesis. Interestingly, this gene, denoted ags1+, for alpha-glucan synthase I, encodes an integral membrane protein that appears to contain multiple functional domains, providing a unique model to study the molecular mechanisms of fungal cell wall assembly.
MATERIALS AND METHODS
General Methods.
YEA medium contained 5 g/liter of yeast extract, 30 g/liter of glucose, and 250 mg/liter of adenine sulfate, and YEAS medium was YEA medium with 1.2 M sorbitol. Glucanase sensitivity of whole cells was evaluated as described (10), except that five times lower concentrations of Zymolyase-20T (Seikagaku, Tokyo) were used. To determine genetic distances between ade6, ags1, and hta1-htb1, crosses of strains FH001 (h− ags1–1ts ade6-M216 ura4-D18) and FH040 (h+ hta1-htb1∷ura4+ ura4-D18) were subjected to tetrad analysis.
Mutant Isolation.
FYC15 cells (h+ ade6-M216; American Type Culture Collection) were incubated in 90 mM ethylmethanesulfonate (Sigma) for 3 hr. Cells were washed three times, allowed to recover in YEA medium for 15 hr at 32°C, spread on YEA plates, and grown for two generations at 32°C. Microcolonies were screened visually for cell shape mutants, and rounded mutants were isolated with a micromanipulator. One cell shape mutant, ags1–1ts, showed a temperature-sensitive (ts) lethal phenotype and did not grow at 37°C.
Electron Microscopy.
Cells grown in YEAS medium at 19°C were harvested, taken up in fresh YEAS medium prewarmed at 37°C, and grown to midexponential phase in YEAS medium at 37°C for 10 hr. Then, freshly prepared paraformaldehyde and electron microscopy-grade glutaraldehyde (Fluka) were added to the shaking cell culture to final concentrations of 3% (wt/vol) and 0.2% (wt/vol), respectively. After an incubation at 37°C of 30 min, cells were harvested and postfixed at 20°C in 100 mM Pipes, pH 6.9, 1 mM EGTA, 1 mM magnesium sulfate, 1.2 M sorbitol (PEMS) containing 3% paraformaldehyde and 0.2% glutaraldehyde for 90 min, followed by a 60-min incubation in PEMS containing 1% (wt/vol) potassium permanganate (Sigma). After three wash steps in water, cells were embedded in epon resin and sectioned with a diamond knife. Thin sections were stained with lead citrate (11), and micrographs were obtained by using a JEOL 1010 electron microscope operating at 60 kV.
Measurements of Cell Wall Alpha-Glucan Content.
Cell walls were isolated as described (12). In short, cells were grown in 250 ml of YEAS medium to a final OD595 of 2, washed twice in breaking buffer [1 mM phenylmethylsulfonyl fluoride (PMSF)/10 mM Tris⋅HCl, pH 7.6], and resuspended in breaking buffer at a density of 0.5 g (wet weight) per ml. Cells were subjected to mechanical breakage by addition of glass beads (0.45 mm diameter) to just below the surface of the suspension and by shaking using a Flask Shaker (Griffin and George, Loughborough, U.K.) at half maximum speed at 4°C for 20 min, until more than 95% of the cells had lysed. Ten milliliters of ice-cold 1 M NaCl were added, glass beads were allowed to settle, and the supernatant was recovered. Glass beads were washed three times with 10 ml of 1 M NaCl, and supernatants were collected and centrifuged at 1,600 × g at 4°C for 10 min. The pellet was washed three times with ice-cold 1 M NaCl and once with 1 mM PMSF. Then, the pellet was resuspended at a concentration of 0.5 g (wet weight) per 5 ml in extraction buffer [40 mM 2-mercaptoethanol/2% (wt/vol) SDS/100 mM EDTA/50 mM Tris⋅HCl, pH 7.6], and boiled for 5 min to remove cytosolic contaminants. The suspension was centrifuged at 1,600 × g at 4°C for 10 min, and the pellet was resuspended in the same amount of extraction buffer and boiled again for 5 min. After three wash steps with 1 mM PMSF, the cell wall containing pellet was resuspended to a concentration of 0.5 g (wet weight) per ml of 1 mM PMSF, aliquoted, and stored at −70°C.
For β-glucanase digestions, 10 mg (wet weight) of cell walls were taken up in 100 μl of 40 mM 2-mercaptoethanol, 0.02% (wt/vol) sodium azide in 50 mM buffer; the buffer was citrate-phosphate, pH 5.6, for digestion with Zymolyase-100T, and Tris⋅HCl, pH 7.6, for digestion with Quantazyme. Then, 3 μl of 10 mg/ml Zymolyase-100T (Seikagaku) or 3 μl of 20 units/μl Quantazyme ylg (Quantum Biotechnologies, Montreal) were added. Samples were digested at 37°C for 16 hr. One-hundred microliters of 4% (wt/vol) SDS then were added, and samples were incubated at 100°C for 5 min to remove solubilized cell wall components and centrifuged at 16,000 × g for 5 min. The carbohydrate contents of the β-glucanase resistant cell wall pellets were measured by using phenol-sulfuric acid (13). We confirmed that digestions were complete by redigesting a treated cell wall pellet with fresh glucanase and by observing no further decrease in its carbohydrate content.
Cloning and Sequence Analysis of ags1.
FH021 cells were transformed with genomic fragments by using a lithium acetate method (14) and grown on YEA plates. After 1 day at 20°C, cells were selected for growth at 37°C to test the ability of genomic fragments to rescue the ags1–1 ts-lethal phenotype. First, cosmid DNA was used from two ordered cosmid libraries, namely cosmids c23E4, c25F2, c17A7, c8C8, c16E6, and c11E10 (15), and cosmids 338, 1818, 622, and 61 (16). Second, three genomic fragments of approximately 8 kb were cloned into pYC12 vector (17) and tested, namely a XhoI–XhoI fragment derived from cosmid 338, and, derived from cosmid c17A7, a BamHI—NheI and an EcoRI—SalI fragment. Third, an overlapping series of PCR amplification products was used, spanning the ags1+ ORF from the start codon to codon 1798. These PCR products were generated with cloned Pfu DNA polymerase (Stratagene) by using cosmid c17A7 DNA as a template, and they were used directly for transformation without previous subcloning into vectors. Transformation with 1 μg of cosmid c17A7 DNA resulted routinely in 150–300 colonies able to grow at 37°C, transformations with positive PCR products resulted in the range of 1 to 400 colonies per μg of DNA, whereas mock transformations did not give rise to any colonies at 37°C.
The 8-kb EcoRI–SalI fragment that rescued the ags1–1 ts-lethal phenotype and part of an adjacent 9.4-kb SalI–SalI fragment also derived from cosmid c17A7 were sequenced from both strands by primer walking using the dideoxy chain termination method with T7 sequenase version 2.0 and 7-deaza-dGTP (Amersham) and [α-35S]dATP (Amersham). The 3′-end of the ags1+ gene was identified by PCR amplifying and cloning its 3′-untranslated region from total RNA of wild-type strain 972 by using the 3′-AmpliFinder RACE kit (CLONTECH), and by DNA sequencing an isolated clone, denoted pAGS1–3′UTR#3. To identify the ags1–1 mutation, PCR amplification products were generated by using cloned Pfu DNA polymerase, primers RS45 (5′-ACATTCATAGCATCCGAA-3′) and RS48 (5′-AGACGAAACAACAGATAA-3′), and, as templates, genomic DNA of strains FH021 or FH023. Both PCR products were sequenced directly, without previous subcloning, from both strands. To confirm the ags1–1 mutation, these PCR amplification products also were digested with the restriction enzyme GsuI (Eurogentec, Brussels). Those derived from genomic DNA of strains FH021 and FH022 were sensitive to GsuI digestion (generating fragments of approximately 360 and 220 bp), whereas those derived from strains FH023 and FH024 were resistant to GsuI digestion (showing the undigested PCR amplification product of approximately 580 bp). DNA and protein sequences were analyzed by using the Wisconsin Package, version 8.1 (Genetics Computer Group), including the algorithms bestfit to calculate percentages of sequence identity and similarity and pileup to generate multiple sequence alignments. blast was used to identify amino acid sequences with significant similarity (18). genefinder (P. Green, personal communication) with integral prediction tables configured using a model set of S. pombe genes is part of PomBase, an AceDB database.
RESULTS AND DISCUSSION
Morphological and Cell Wall Defects of the ags1–1ts Mutant.
We mutagenized rod-shaped cells of strain FYC15 with ethylmethanesulfonate and visually screened for mutants with a rounded-shape phenotype. Here, we characterize a ts cell shape mutant, ags1–1ts. Tetrad analyses showed that both the ts and the cell shape phenotype were caused by a single genetic locus and both phenotypes were recessive, indicating a loss-of-function mutation. The ags1–1ts mutant was back-crossed to wild-type strains 972 and 975 to cross out secondary mutations, and the strains derived from a single tetrad of the eighth backcross, FH021 (h− ags1–1ts), FH022 (h+ ags1–1ts), FH023 (h−), and FH024 (h+), were used for detailed analyses.
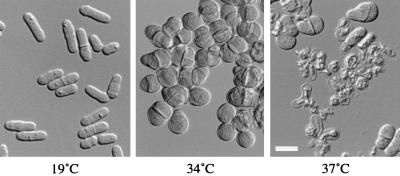
ags1–1ts cells display a remarkable temperature-dependent shape phenotype (Fig. 1). At a permissive temperature of 19°C, cells show a cylindrical rod shape, like that of wild-type cells (Fig. 1, Left). Also, they grow with a generation time similar to that of wild-type cells (generation times of strains FH021 and FH023 are 4.5 ± 0.3 hr and 4.7 ± 0.5 hr, respectively; mean ± SD). In contrast, ags1–1ts cells grown at a semipermissive temperature of 34°C are rounded and pear-shaped (Fig. 1, Center). When ags1–1ts cells are shifted from 19°C to a restrictive temperature of 37°C, cell lysis can be observed after 5 hr of growth, and by 12 hr, most cells have lysed (Fig. 1, Right). This cell lysis phenotype could be suppressed partially by adding an osmotic stabilizer, sorbitol, allowing an exacerbation of the ags1–1ts cell shape phenotype. After 15 hr of growth at 37°C in YEAS medium, all cells were rounded. The observed change in morphology indicates that the cell wall structure may be affected, whereas the cell lysis phenotype indicates that the cell wall lost its ability to maintain cell integrity.
Figure 1.
Temperature-dependent morphology of ags1–1ts cells. ags1–1ts cells (strain FH021) were grown at 19°C (Left) or 34°C (Center) in YEA medium for 2 days, or shifted from 19°C to 37°C in YEA medium for 9 hr (Right). Wild-type cells grown under these conditions are rod shaped. (Bar = 10 μm.)
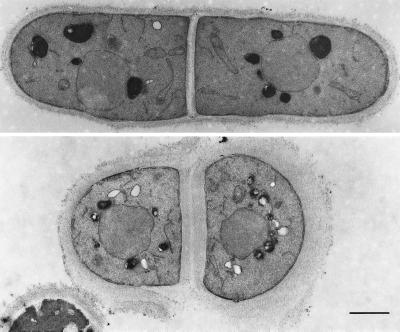
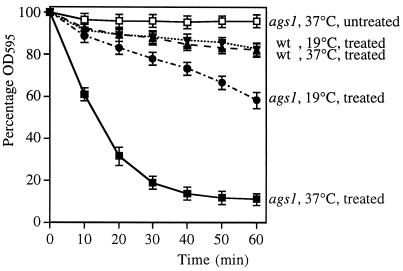
To determine whether the cell wall was affected by the ags1–1ts mutation, we analyzed its ultrastructure. This analysis showed that ags1–1ts cell walls had become looser and 2–4 times thicker than those of wild-type controls, demonstrating that cell wall structure had changed dramatically (Fig. 2). We confirmed this conclusion by showing that ags1–1ts cells grown at 37°C for only 4 hr were hypersensitive to digestion by β(1 → 3)-glucanase (Fig. 3). These data demonstrate that the composition and/or the organization of the cell wall has changed.
Figure 2.
Defective cell wall ultrastructure of ags1–1ts cells. Electron micrographs of thin sections from a wild-type cell (Upper) and an ags1–1ts cell (Lower). Cells of ags1–1ts strain FH021 and wild-type strain FH023 were grown at 37°C for 10 hr, followed by fixation. (Bar = 1 μm.)
Figure 3.
β-glucanase hypersensitivity of ags1–1ts cells. ags1–1ts cells (strain FH021) and wild-type cells (strain FH023) were grown first at 19°C and then at 37°C for 4 hr (1–2 generation times). Cells then were incubated in 20 μg/ml of Zymolyase-20T at 28°C under continuous shaking. Cell lysis was monitored by measurement of OD595 (mean ± SD; n = 3). (■) ags1–1ts cells, 4 hr at 37°C, treated with Zymolyase; (□) ags1–1ts cells, 4 hr at 37°C, untreated (all untreated controls showed overlapping curves; data not shown); (•) ags1–1ts cells, grown at 19°C, treated; (▾) wild-type cells, grown at 19°C, treated; (▴) wild-type cells, 4 hr at 37°C, treated.
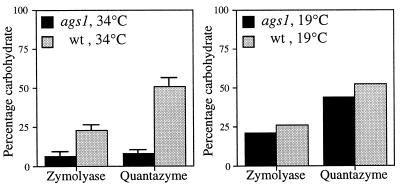
Next, we determined the levels of α-glucan and β-glucan in the ags1–1ts cell wall. To ensure that most cells expressed the rounded cell shape phenotype while remaining physically intact, ags1–1ts cells (and wild-type cells) were grown at 34°C in YEAS for 2 days. Cell walls then were isolated and digested with Zymolyase-100T, a purified β-glucanase preparation that extracts most β-glucan. The remaining carbohydrate fraction was measured and considered to be α-glucan. The validity of this procedure was confirmed by using x-ray diffraction analyses. The Zymolyase-100T resistant cell wall fraction from wild-type strain FH023 showed solely diffraction lines characteristic for α(1 → 3)-glucan (19) and no diffraction lines characteristic for β(1 → 3)-glucan, whereas both patterns were clearly visible in untreated cell walls (F. H. and J. H. Sietsma, unpublished observations). Cell walls from wild-type strain FH023 grown in YEAS medium at either 19°C or 34°C contained 23 ± 3% α-glucan (as a percentage of total carbohydrate; mean ± SD; n = 3) (Fig. 4), which accords well with the published value of 28% of total cell wall dry weight (4). Importantly, cell walls isolated from ags1–1ts strain FH021 grown at 34°C contained only 7 ± 3% α-glucan (mean ± SD; n = 3), a 3-fold reduction in α-glucan levels (Fig. 4, Left). This dramatic reduction was not observed when ags1–1ts cells were grown at 19°C, because under these conditions the cell wall contained almost wild-type levels of α-glucan (Fig. 4, Right). Thus, in the ags1–1ts mutant, the decrease in cell wall α-glucan coincides with a change in cell shape. The temperature-dependent reduction in β-glucanase-resistant carbohydrate was confirmed by using a cloned endo-β-glucanase, Quantazyme; however, this recombinant enzyme was able to extract only a portion of the total β-glucan from wild-type cell walls (Fig. 4). From these carbohydrate biochemical results we conclude that ags1–1ts cells have a defect in α(1 → 3)-glucan biosynthesis.
Figure 4.
Reduction of α-glucan in the ags1–1ts cell wall. Cell walls isolated from ags1–1ts strain FH021 and wild-type strain FH023 grown at 34°C (Left) or 19°C (Right) were digested with Zymolyase-100T, and the residual carbohydrate was measured and regarded as α-glucan. Cell walls also were digested with Quantazyme.
Three Domains in the Predicted Ags1 Protein.
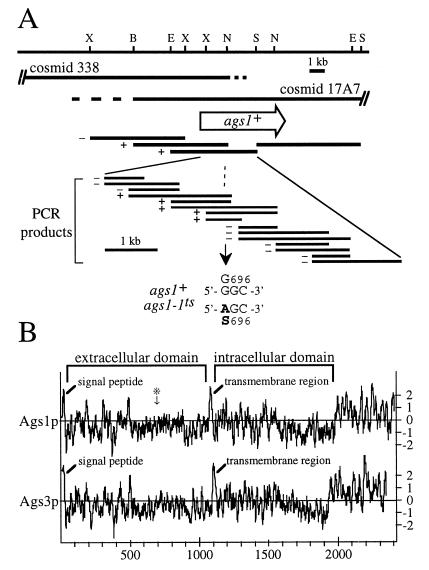
To determine whether the defect in α(1 → 3)-glucan biosynthesis was caused by a mutation in a regulatory or catalytic component, we cloned the ags1+ gene. In a positional cloning approach, we mapped the ags1 locus between ade6 and hta1-htb1 on chromosome III. The genetic distance between ade6 and ags1 was 5.0 centiMorgans (166 parental ditype, 12 tetratype, 1 nonparental ditype), localizing ags1 to approximately 33 kb to the telomeric side of ade6. Cosmids 17A7 (15) and 338 (16), covering this genomic region, rescued both the ts and the cell shape phenotypes (Fig. 5A). Using DNA cloning and sequencing analyses, we identified the ags1+ gene as a contiguous ORF of 7,230 bp, which appears not to contain hidden introns based on a prediction by a S. pombe version of the genefinder algorithm. This ORF encodes a protein, Ags1p, of 2,410 amino acids with a calculated molecular mass of 272 kDa. We verified that this ORF was indeed expressed, by using a rapid amplification of cDNA ends–PCR (RACE-PCR) with one primer localized near the ags1+ termination codon and another primer localized to the start of the poly(A) tail. Cloning and sequence analysis of this cDNA indicated the presence of a 3′-untranslated region of 357 bp after the termination codon.
Figure 5.
Characterization of the ags1+ locus and gene product. (A) Restriction map of the ags1+ gene locus and mutation site of the ags1–1ts allele. Both cosmid c17A7 and 338 rescue the ts and cell shape phenotypes of ags1–1 cells. The extent of overlap of the two cosmids has not been mapped precisely (indicated by dashed lines). The open arrow indicates the location and orientation of the ags1+ ORF. The ability of genomic fragments and PCR amplification products to rescue the ts phenotype is indicated to the left of each fragment by + or −. DNA sequence analyses of PCR amplification products identified the ags1–1ts mutation site as codon 696, which was changed from a Gly to a Ser codon. B, BamHI; E, EcoRI; N, NheI; S, SalI; X, XhoI.(B) Predicted topologies of the ags1+ and ags3+ gene products. Hydropathy plots (20) were generated by using a window size of 13. ∗ indicates the ags1–1ts mutation site. Like Ags1p, the Ags3 protein contains an amino-terminal signal peptide predicted to be cleavable (between residues 22 and 23), a single membrane-spanning region at residues 1091–1107, and a multipass transmembrane domain. Only partial sequence is known for Ags2p.
A hydropathy profile (20) of the predicted Ags1 protein reveals three major hydrophobic regions (Fig. 5B). The first was located at the amino terminus. An algorithm identifying signal peptides for entry into the secretory pathway (21) predicted the Ags1p amino terminus to be a cleavable signal peptide and identified the peptide bond between Ala-26 and Ala-27 as the most likely cleavage site for signal peptidase. The second hydrophobic region (residues 1069–1091) is located approximately in the middle of the protein and is predicted to form a transmembrane region (22). Because transmembrane regions function as stop-transfer peptides during protein translocation, we predict the polypeptide preceding this middle hydrophobic region to be localized to the extracytoplasmic space and the polypeptide after this region to be localized to the cytoplasm. The third hydrophobic region, located at the carboxy-terminus of Ags1p (residues 1987–2410), may span the membrane multiple times. The precise number of membrane passes was difficult to predict, because one algorithm (22) indicated nine potential transmembrane regions, whereas another (23) predicted 13. In summary, the predicted Ags1 protein appears to contain three domains, namely an extracellular domain (residues 27–1068), an intracellular domain (residues 1092–1986), and a multipass transmembrane domain (residues 1987–2410). Because it does not contain known retention or targeting signals, the Ags1 protein is predicted to traverse the secretory pathway from the endoplasmic reticulum to the plasma membrane.
Sequence Similarity to Alpha-Glucan Synthases and Transglycosylases.
A blast search of the GenBank/EMBL/DDBJ database (December 8, 1997) revealed that the S. pombe genome contains two homologs of ags1+, both identified as hypothetical genes by the systematic genome sequencing project. Cosmid 23D3 derived from chromosome I contained only part of a homologous ORF, denoted 23D3.15, whose 3′-sequence is not yet known. Cosmid 1750 derived from chromosome II contained a full-length ORF (nucleotides 18838–25896), denoted 1750.11, encoding a polypeptide of 2,352 amino acids with a calculated molecular mass of 266 kDa. Because the deduced amino acid sequence of 23D3.15 showed a higher sequence identity to Ags1p than that of 1750.11 (51% versus 42%, comparing corresponding polypeptides), we propose to designate ORF 23D3.15 as ags2+ and ORF 1750.11 as ags3+. The Ags3 protein shows a predicted polypeptide topology remarkably similar to that of Ags1p (Fig. 5B). The database search also revealed that, at present, no homologs have been identified in filamentous fungi. Importantly, no ags homolog exists in the genome of the budding yeast Saccharomyces cerevisiae, which has been sequenced completely. The absence of ags-like genes in S. cerevisiae is consistent with the absence of α(1 → 3)-glucan from its cell walls.
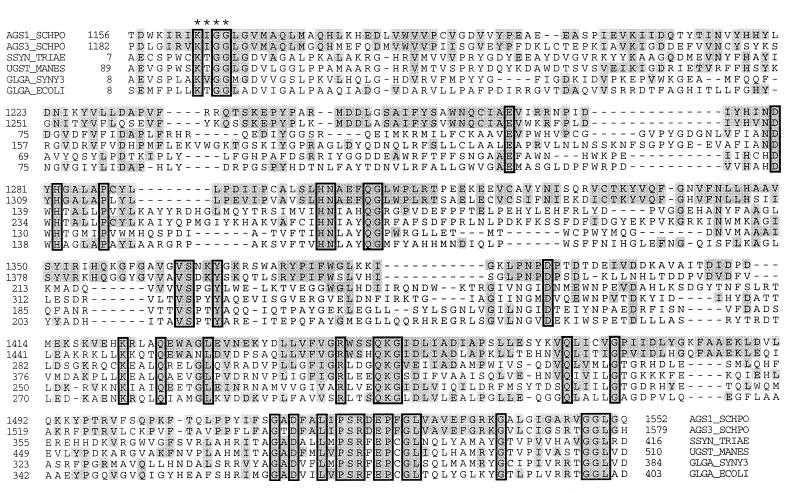
To identify a potential function of the putative Ags1p intracellular domain, we analyzed blast results related to this domain. Starch synthases of several plants and the glycogen synthases of the cyanobacterium Synechocystis sp. and of Escherichia coli showed amino acid sequence similarities to the Ags1p intracellular domain (Fig. 6). Pair-wise alignments of the starch synthases of wheat and cassava and the glgA glycogen synthases of Synechocystis and E. coli to Ags1p residues 1156–1552 showed sequence identities of 29%, 28%, 24%, and 24% and sequence similarities of 51%, 50%, 48%, and 48%, respectively. A lysine, which participates in the catalytic reaction of E. coli glgA glycogen synthase (24), is conserved at the corresponding position in Ags1p (residue 1422) and Ags3p (residue 1449) (Fig. 6). Also a sequence motif, Lys/Arg-X-Gly-Gly (X represents any residue), that is essential in E. coli glgA glycogen synthase for efficient binding of its substrate ADP-glucose (25), is conserved at the corresponding positions in Ags1p and Ags3p (Fig. 6). Starch and glycogen synthases catalyze biosynthetic reactions in which the substrate UDP-glucose or ADP-glucose is polymerized into linear α(1 → 4)-glucan. However, cell walls of S. pombe contain few α(1 → 4)-glucosidic linkages, whereas they do contain substantial amounts of α(1 → 3)-glucan. In addition, we showed that the levels of α(1 → 3)-glucan in ags1–1ts cell walls were reduced dramatically. Therefore, we propose that Ags1p synthesizes α(1 → 3)-glucan.
Figure 6.
Amino acid sequence similarities to the predicted intracellular domain of Ags1p. The sequences of Ags1p (AGS1_SCHPO) and Ags3p (AGS3_SCHPO) were aligned with starch synthases of Triticum aestivum (wheat; SSYN_TRIAE; U66377) and Manihot esculenta (cassava; UGST_MANES; Q43784), and with glycogen synthases of Synechocystis sp. (gene product of glgA; GLGA_SYNY3; P74521) and E. coli (gene product of glgA; GLGA_ECOLI; P08323). The Lys/Arg-X-Gly-Gly motif is indicated by ∗. Boxes indicate sequence identities among the proteins shown, shading indicates sequence similarities to Ags1p, and dashes indicate gaps in the pileup alignment.
To identify a potential function of the putative Ags1p extracellular domain, we analyzed blast results related to this domain. Interestingly, we again identified sequence similarities to enzymes involved with α-glucan metabolism, namely to bacterial α-amylases. Four enzymes showing the highest amino acid sequence similarities to this Ags1p domain (residues 73–586) were cyclomaltodextrinase of Thermoanaerobacter ethanolicus, neopullulanase of Bacillus stearothermophilus, and α-amylases of Streptomyces lividans and Paenibacillus polymyxa (with sequence identities of 23–26% and sequence similarities of 46–52%). These bacterial enzymes are classified in the same family of glycosyl hydrolases, namely family 13, also known as the α-amylase family (26). Based on crystal structures, this family contains a characteristic catalytic (β/α)8-barrel domain, with two invariant aspartate residues and one invariant glutamate residue constituting a catalytic triad (27). These three catalytic residues and three of five additional highly conserved residues are present in all three Ags proteins (data not shown). Enzymes of this family catalyze reactions such as hydrolysis of α(1 → 4)- or α(1 → 6)-glucosidic linkages and transglycosylation to form α(1 → 4)- or α(1 → 6)-glucosidic linkages. We speculate that the Ags1p extracellular domain might function as a transglycosylase, perhaps remodeling α(1 → 3)-glucan or cross-linking it via α(1 → 4)-glucosidic linkages to the existing cell wall matrix.
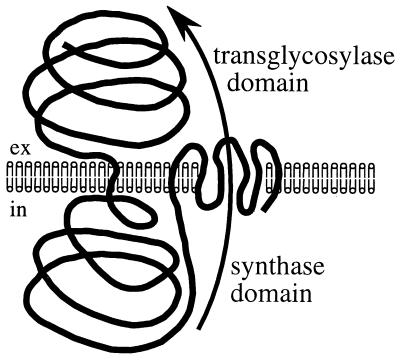
These data are consistent with the following speculative, but testable, model for Ags1p function (Fig. 7). The intracellular synthase domain may produce α(1 → 3)-glucan homopolymers at the plasma membrane. These would need be transported across the membrane, perhaps by the Ags1p multipass transmembrane domain, which might form a pore-like structure. Once transported, the Ags1p extracellular domain might either cross-link α-glucan homopolymers to other cell wall carbohydrates or remodel them. Accordingly, we speculate that Ags1p might function as a complete α-glucan synthesis-transport-modeling machine.
Figure 7.
Speculative model for Ags1p function (see text for details).
The ags1–1ts Mutation Site Is in the Extracellular Domain.
We mapped the mutation site of the ags1–1ts allele to determine which Ags1p domain was affected. Overlapping PCR amplification products generated from wild-type DNA were tested for their ability to rescue the ags1–1ts phenotype (Fig. 5A). We presume that this rescue activity is caused by replacement of the mutation site with wild-type sequences through homologous recombination. In these rescue experiments, we narrowed down the mutation site to a region of 580 bp. Sequence analyses of PCR amplification products of mutant and wild-type genomic DNA revealed that ags1–1ts contains a mutation in codon 696, changing this codon from GGC to AGC, and creating a recognition site of the restriction enzyme GsuI. The presence of this GsuI site was confirmed in PCR amplification products from genomic DNA of ags1–1ts strains FH021 and FH022, whereas it was absent from those of wild-type strains FH023 and FH024 (data not shown). The mutation causes an amino acid change from Gly to Ser in the predicted extracellular domain (see ∗ in Fig. 5B). Although this amino acid falls outside the transglycosylase similarity domain (residues 73–586), it is part of a tetrapeptide, Gly-Cys-(Ile/Leu)-Pro, that is conserved among the three predicted Ags proteins. At present, we do not know the fate of the mutant Ags1 protein at the restrictive temperature. We speculate that the G696S mutation either might lead to a defect in its putative cross-linking or remodeling function at the plasma membrane or might cause misfolding, triggering retention of inactive Ags1 protein in the endoplasmic reticulum by the quality-control mechanism.
CONCLUSIONS
In this report, we identify in S. pombe a candidate gene, denoted ags1+, for an α(1 → 3)-glucan synthase. Remarkably, this α(1 → 3)-glucan synthase appears to be composed of multiple functional domains. This multidomain structure is unique among known cell wall carbohydrate synthases and may allow detailed studies into three discrete steps involved in deposition of α(1 → 3)-glucan into the cell wall, namely synthesis, transport across the membrane, and transglycosylation.
We propose that, like other fungal cell wall carbohydrate polymers, α(1 → 3)-glucan plays a critical role in maintaining cell shape and integrity. Not only fission yeast, but also many dimorphic and filamentous fungi contain substantial amounts of α-glucan in their cell wall (on the order of 30%); these fungi include the pathogens Aspergillus fumigatus, Histoplasma capsulatum, Paracoccidioides brasiliensis, and Blastomyces dermatitidis. Importantly, in the latter three organisms, mutant strains have been isolated in which decreased amounts of cell wall α(1 → 3)-glucan correlated with a decreased virulence for mice (28–30). Together, these data reveal α(1 → 3)-glucan as a therapeutic target. Specifically, development of drugs that inhibit α(1 → 3)-glucan synthase or transglycosylase activities could aid in the fight against fungal infection.
Acknowledgments
We thank Ravi Dhar, Henri Levin, Jürg Kohli, and Jagmohan Singh for S. pombe strains. Cosmid clones were made available by the Reference Library, Imperial Cancer Research Fund (London), and by Richard McCombie. We also would like to thank Mike Lyne for the genefinder analysis and the anonymous reviewers for comments on the manuscript. F.H. was a visiting fellow of the National Institute for Child Health and Human Development and currently is supported by a fellowship of the Royal Netherlands Academy of Arts and Sciences.
ABBREVIATIONS
- ags1
alpha-glucan synthase I
- ts
temperature sensitive
- YEA
yeast extract/glucose/adenine sulfate
- YEAS
YEA plus sorbitol
Footnotes
References
- 1.Snell, V. & Nurse, P. (1993) Dev. Suppl. 289–299. [PubMed]
- 2.Mata J, Nurse P. Cell. 1997;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 3.Verde F, Mata J, Nurse P. J Cell Biol. 1995;131:1529–1538. doi: 10.1083/jcb.131.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bush D A, Horisberger M, Horman I, Wursch P. J Gen Microbiol. 1974;81:199–206. doi: 10.1099/00221287-81-1-199. [DOI] [PubMed] [Google Scholar]
- 5.Manners D J, Meyer M T. Carbohydr Res. 1977;57:189–203. [Google Scholar]
- 6.Sietsma J H, Wessels J G H. J Gen Microbiol. 1990;136:2261–2265. doi: 10.1099/00221287-136-11-2261. [DOI] [PubMed] [Google Scholar]
- 7.Kopecká M, Fleet G H, Phaff H J. J Struct Biol. 1995;114:140–152. doi: 10.1006/jsbi.1995.1013. [DOI] [PubMed] [Google Scholar]
- 8.Miyata M, Kitamura J, Miyata H. Arch Microbiol. 1980;127:11–16. doi: 10.1007/BF00414349. [DOI] [PubMed] [Google Scholar]
- 9.Miyata M, Kanbe T, Tanaka K. J Gen Microbiol. 1985;131:611–621. [Google Scholar]
- 10.Levin D E, Bishop J M. Proc Natl Acad Sci USA. 1990;87:8272–8276. doi: 10.1073/pnas.87.21.8272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds E S. J Cell Biol. 1963;17:208–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schreuder M P, Brekelmans S, Van den Ende H, Klis F M. Yeast. 1993;9:399–409. doi: 10.1002/yea.320090410. [DOI] [PubMed] [Google Scholar]
- 13.Dubois M, Gilles K A, Hamilton J K, Rebers P A, Smith F. Anal Chem. 1956;28:350–356. [Google Scholar]
- 14.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoheisel J D, Maier E, Mott R, McCarthy L, Grigoriev A V, Schalkwyk L C, Nizetic D, Francis F, Lehrach H. Cell. 1993;73:109–120. doi: 10.1016/0092-8674(93)90164-l. [DOI] [PubMed] [Google Scholar]
- 16.Mizukami T, Chang W I, Garkavtsev I, Kaplan N, Lombardi D, Matsumoto T, Niwa O, Kounosu A, Yanagida M, Marr T G, Beach D. Cell. 1993;73:121–132. doi: 10.1016/0092-8674(93)90165-m. [DOI] [PubMed] [Google Scholar]
- 17.Chikashige Y, Kinoshita N, Nakaseko Y, Matsumoto T, Murakami S, Niwa O, Yanagida M. Cell. 1989;57:739–751. doi: 10.1016/0092-8674(89)90789-7. [DOI] [PubMed] [Google Scholar]
- 18.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 19.Kreger D R. Biochim Biophys Acta. 1954;13:1–9. doi: 10.1016/0006-3002(54)90264-4. [DOI] [PubMed] [Google Scholar]
- 20.Kyte J, Doolittle R F. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 21.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 22.Klein P, Kanehisa M, DeLisi C. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 23.Rost B, Casadio R, Fariselli P, Sander C. Protein Sci. 1995;4:521–533. doi: 10.1002/pro.5560040318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Furukawa K, Tagaya M, Tanizawa K, Fukui T. J Biol Chem. 1994;269:868–871. [PubMed] [Google Scholar]
- 25.Furukawa K, Tagaya M, Tanizawa K, Fukui T. J Biol Chem. 1993;268:23837–23842. [PubMed] [Google Scholar]
- 26.Henrissat B, Bairoch A. Biochem J. 1996;316:695–696. doi: 10.1042/bj3160695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svensson B. Plant Mol Biol. 1994;25:141–157. doi: 10.1007/BF00023233. [DOI] [PubMed] [Google Scholar]
- 28.Klimpel K R, Goldman W E. Infect Immun. 1988;56:2997–3000. doi: 10.1128/iai.56.11.2997-3000.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.San-Blas G, San-Blas F, Serrano L E. Infect Immun. 1977;15:343–346. doi: 10.1128/iai.15.2.343-346.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hogan L H, Klein B S. Infect Immun. 1994;62:3543–3546. doi: 10.1128/iai.62.8.3543-3546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]