Abstract
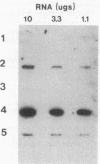
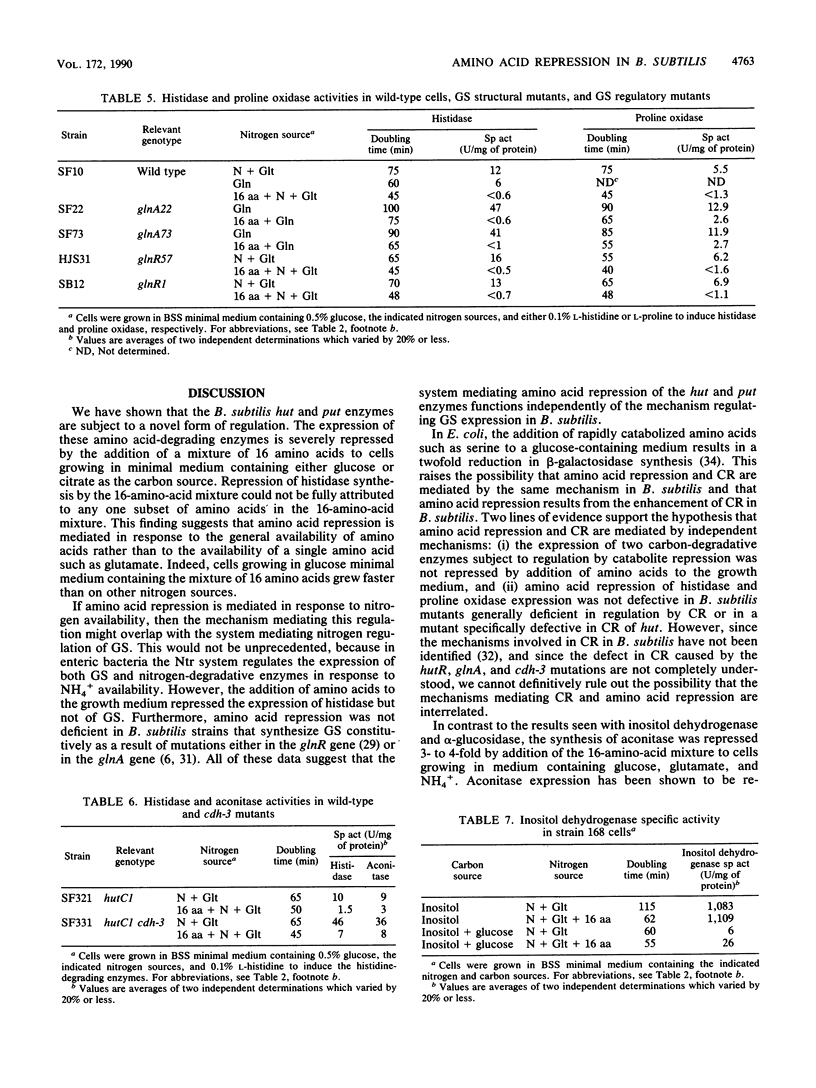
The first enzymes of the histidine (hut) and proline degradative pathways, histidase and proline oxidase, could not be induced in Bacillus subtilis cells growing in glucose minimal medium containing a mixture of 16 amino acids. Addition of the 16-amino-acid mixture to induced wild-type cells growing in citrate minimal medium repressed histidase synthesis 25- to 250-fold and proline oxidase synthesis 16-fold. A strain containing a transcriptional fusion of the hut promoter to the beta-galactosidase gene was isolated from a library of Tn917-lacZ transpositions. Examination of histidase and beta-galactosidase expression in extracts of a hut-lacZ fusion strain grown in various media showed that induction, catabolite repression, and amino acid repression of the hut operon were mediated at the level of transcription. This result was confirmed by measurement of the steady-state level of hut RNA in cells grown in various media. Since amino acid repression was not defective in B. subtilis mutants deficient in nitrogen regulation of glutamine synthetase and catabolite repression, amino acid repression appears to be mediated by a system that functions independently of these regulatory systems.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bender R. A., Snyder P. M., Bueno R., Quinto M., Magasanik B. Nitrogen regulation system of Klebsiella aerogenes: the nac gene. J Bacteriol. 1983 Oct;156(1):444–446. doi: 10.1128/jb.156.1.444-446.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan S. A., Chun K. T., Edson B. A., Price C. W. Early-blocked sporulation mutations alter expression of enzymes under carbon control in Bacillus subtilis. Mol Gen Genet. 1988 May;212(2):271–280. doi: 10.1007/BF00334696. [DOI] [PubMed] [Google Scholar]
- Brehm S. P., Staal S. P., Hoch J. A. Phenotypes of pleiotropic-negative sporulation mutants of Bacillus subtilis. J Bacteriol. 1973 Sep;115(3):1063–1070. doi: 10.1128/jb.115.3.1063-1070.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chasin L. A., Magasanik B. Induction and repression of the histidine-degrading enzymes of Bacillus subtilis. J Biol Chem. 1968 Oct 10;243(19):5165–5178. [PubMed] [Google Scholar]
- Cox D. P., Hanson R. S. Catabolite repression of aconitate hydratase in Bacillus subtilis. Biochim Biophys Acta. 1968 Apr 16;158(1):36–44. doi: 10.1016/0304-4165(68)90069-x. [DOI] [PubMed] [Google Scholar]
- Dean D. R., Hoch J. A., Aronson A. I. Alteration of the Bacillus subtilis glutamine synthetase results in overproduction of the enzyme. J Bacteriol. 1977 Sep;131(3):981–987. doi: 10.1128/jb.131.3.981-987.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S. H., Magasanik B. 2-Ketoglutarate and the regulation of aconitase and histidase formation in Bacillus subtilis. J Bacteriol. 1984 Apr;158(1):379–382. doi: 10.1128/jb.158.1.379-382.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S. H., Magasanik B. Isolation of Bacillus subtilis mutants pleiotropically insensitive to glucose catabolite repression. J Bacteriol. 1984 Mar;157(3):942–944. doi: 10.1128/jb.157.3.942-944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S. H., Sonenshein A. L. Bacillus subtilis glutamine synthetase mutants pleiotropically altered in glucose catabolite repression. J Bacteriol. 1984 Feb;157(2):612–621. doi: 10.1128/jb.157.2.612-621.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher S. H., Wray L. V., Jr Regulation of glutamine synthetase in Streptomyces coelicolor. J Bacteriol. 1989 May;171(5):2378–2383. doi: 10.1128/jb.171.5.2378-2383.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden K. J., Bernlohr R. W. Nitrogen catabolite repression of the L-asparaginase of Bacillus licheniformis. J Bacteriol. 1985 Nov;164(2):938–940. doi: 10.1128/jb.164.2.938-940.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HARTWELL L. H., MAGASANIK B. THE MOLECULAR BASIS OF HISTIDASE INDUCTION IN BACILLUS SUBTILIS. J Mol Biol. 1963 Oct;7:401–420. doi: 10.1016/s0022-2836(63)80033-9. [DOI] [PubMed] [Google Scholar]
- Hanson R. S., Cox D. P. Effect of different nutritional conditions on the synthesis of tricarboxylic acid cycle enzymes. J Bacteriol. 1967 Jun;93(6):1777–1787. doi: 10.1128/jb.93.6.1777-1787.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood C. R., Baumberg S. Arginine hydroxamate-resistant mutants of Bacillus subtilis with altered control of arginine metabolism. J Gen Microbiol. 1977 May;100(1):177–188. doi: 10.1099/00221287-100-1-177. [DOI] [PubMed] [Google Scholar]
- Kimhi Y., Magasanik B. Genetic basis of histidine degradation in Bacillus subtilis. J Biol Chem. 1970 Jul 25;245(14):3545–3548. [PubMed] [Google Scholar]
- LEVIN A. P., MAGASANIK B. Enzyme synthesis in guanine-starved cells. J Biol Chem. 1961 Jun;236:1810–1815. [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Oda M., Sugishita A., Furukawa K. Cloning and nucleotide sequences of histidase and regulatory genes in the Bacillus subtilis hut operon and positive regulation of the operon. J Bacteriol. 1988 Jul;170(7):3199–3205. doi: 10.1128/jb.170.7.3199-3205.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F. L., Coote J. G. Glutamine synthetase and glutamate synthase activities during growth and sporulation in Bacillus subtilis. J Gen Microbiol. 1979 Jun;112(2):373–377. doi: 10.1099/00221287-112-2-373. [DOI] [PubMed] [Google Scholar]
- Perkins J. B., Youngman P. J. A physical and functional analysis of Tn917, a Streptococcus transposon in the Tn3 family that functions in Bacillus. Plasmid. 1984 Sep;12(2):119–138. doi: 10.1016/0147-619x(84)90058-1. [DOI] [PubMed] [Google Scholar]
- Rebello J. L., Strauss N. Regulation of synthesis of glutamine synthase in Bacillus subtilis. J Bacteriol. 1969 May;98(2):683–688. doi: 10.1128/jb.98.2.683-688.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenkrantz M. S., Dingman D. W., Sonenshein A. L. Bacillus subtilis citB gene is regulated synergistically by glucose and glutamine. J Bacteriol. 1985 Oct;164(1):155–164. doi: 10.1128/jb.164.1.155-164.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier H. J., Smith T. M., Bernlohr R. W. Regulation of nitrogen catabolic enzymes in Bacillus spp. J Bacteriol. 1982 Aug;151(2):971–975. doi: 10.1128/jb.151.2.971-975.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier H. J., Sonenshein A. L. Altered regulation of the glnA gene in glutamine synthetase mutants of Bacillus subtilis. J Bacteriol. 1986 Jul;167(1):35–43. doi: 10.1128/jb.167.1.35-43.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenshein A. L., Cami B., Brevet J., Cote R. Isolation and characterization of rifampin-resistant and streptolydigin-resistant mutants of Bacillus subtilis with altered sporulation properties. J Bacteriol. 1974 Oct;120(1):253–265. doi: 10.1128/jb.120.1.253-265.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B. L., Kodaira R., Neidhardt F. C. Regulation of lac operon expression: reappraisal of the theory of catabolite repression. J Bacteriol. 1978 Dec;136(3):947–954. doi: 10.1128/jb.136.3.947-954.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]