Abstract
Noninvasive cellular import of synthetic peptides can be accomplished by incorporating a hydrophobic, membrane-permeable sequence (MPS). Herein, we describe a facile method that expedites synthesis of biologically active, cell-permeable peptides by site-specific ligation of two free peptide modules: one bearing a functional sequence and the second bearing a MPS. A nonpeptide thiazolidino linkage between the two modules is produced by ligation of the COOH-terminal aldehyde on the MPS and the NH2-terminal 1,2-amino thiol moiety on the functional sequence. This thiazolidine ligation approach is performed with stoichiometric amounts of fully unprotected MPS and functional peptide in an aqueous buffered solution, eliminating the need for additional chemical manipulation and purification prior to use in bioassays. Two different MPSs were interchangeably combined with two different functional sequences to generate two sets of hybrid peptides. One set of hybrid peptides, carrying the cytoplasmic cell adhesion regulatory domain of the human integrin β3, inhibited adhesion of human erythroleukemia cells to fibrinogen-coated surfaces. A second set of hybrid peptides, carrying the nuclear localization sequence of the transcription factor NF-κB, inhibited nuclear import of transcription factors NF-κB, activator protein 1, and nuclear factor of activated T cells in agonist-stimulated Jurkat T lymphocytes. In each assay, these nonamide bond hybrids were found to be functionally comparable to peptides prepared by the conventional method. Cumulatively, this new ligation approach provides an easy and rapid method for engineering of functional, cell-permeable peptides and demonstrates the potential for synthesis of cell-permeable peptide libraries designed to block intracellular protein–protein interactions.
The average nucleated cell contains approximately 10,000 proteins that participate in signal transduction, gene transcription, cell–cell communication, and intracellular protein trafficking. These fundamental processes in the life of a cell depend on intracellular protein–protein and protein–DNA interactions. Structure-function analysis of intracellular proteins is hampered by the inability of sequence-specific antibodies or synthetic peptides to penetrate the plasma membrane. Therefore, invasive methods of microinjection or application of membrane-disrupting pore-forming reagents are currently used to introduce antibodies, synthetic peptides, or other non-cell membrane-permeable molecules into cells (1). Alternatively, transfection experiments are used to introduce DNA encoding truncated or mutated intracellular proteins (2, 3). Although such approaches yield significant information, their inherent limitations impede structure-function analysis of existing and newly sequenced intracellular proteins.
To circumvent these limitations and allow easy, noninvasive delivery of functional peptides to intact cells in bulk concentrations, new methods have been developed (see ref. 4 for review). One such technique utilizes the core hydrophobic (H) region of the signal sequence as a “carrier” for cellular import of relevant intracellular protein motifs (5, 6). Originally, the hydrophobic membrane-permeable peptide (MPS) was added to each individual functional peptide by stepwise synthesis, thus creating cell-permeable peptides with hydrophobic and functional sequences linked by a normal peptide bond (5, 6). We have used this approach to prepare biologically active cell-permeable peptides that inhibit adhesion of human erythroleukemia (HEL) cells to fibrinogen-coated surfaces and prevent the inducible nuclear import of transcription factors in human monocytic, endothelial, and T lymphocyte cell lines (ref. 4 and T.R.T., A.D.C., J. Donahue, Y.-Z. Lin, and J.H., unpublished results). Although conventional solid-phase peptide synthesis of cell-permeable peptides is effective, a modular approach to their synthesis would be more practical, allowing the rapid preparation of cell-permeable peptides from pre-prepared hydrophobic and functional peptide modules.
In this paper, we report the development of a modular approach to synthesis of cell-permeable peptides. We have previously described a general and mild method for site-specific biotinylation of peptides and glycoproteins that is based on the chemoselective ligation of an aldehyde on one reactant with a 1,2-amino thiol moiety on a second reactant (7, 8). Ligation of the aldehyde with the 1,2-amino thiol is rapid and site specific, forming a stable thiazolidine ring. The reaction can be carried out using fully unprotected peptide or protein segments in aqueous buffered solutions. Using this approach, independently prepared hydrophobic and functional peptide modules were linked by single-step ligation through a nonamide thiazolidino linkage. Since purified peptide modules were ligated in aqueous solution, the resulting functional cell-permeable peptides required no further purification prior to use in biological assays. The linkage formed by this approach is a nonpeptide bond; therefore, it was important to determine whether these hybrid peptides retained the same biological activity as their counterparts synthesized by the conventional method. Peptides were tested in HEL cells for their ability to inhibit adhesion to fibrinogen-coated surfaces and in Jurkat T lymphocytes for their ability to inhibit agonist-stimulated nuclear import of transcription factors. Results of the biological assays indicate that hybrid peptides prepared by our new approach were functionally equipotent with those synthesized by the conventional method using an amide linkage.
MATERIALS AND METHODS
Peptide Synthesis.
Solid-phase peptide synthesis (9) was performed on a CS-Bio Peptide Synthesizer (fluorenylmethyloxycarbonyl chemistry) or an ABI 430A Peptide Synthesizer (tert-butyoxycarbonyl chemistry) using a 4-fold excess of amino acid and benzotriazol-1-yl-oxy-tris-dimethylamino-phosphonium hexafluorophosphate, and a 6-fold excess of N,N-diisopropylethylamine. Peptides from Fmoc synthesis were cleaved from the resin with trifluoroacetic acid/thioanisole/triisopropylsilane/methanol (90:5:2.5:2.5, vol/vol/vol/vol) at 20°C for 4 h and those from Boc synthesis with HF/anisole (9:1, vol/vol) at 4°C for 1 h. Aldehyde precursor peptides were oxidized by NaIO4 to aldehydes prior to RP-HPLC purification whereas NH2-terminal cysteinyl-peptides were directly purified by reverse-phase (RP)-HPLC. RP-HPLC for all peptides was performed on a Vydac column (250 × 4.6 mm) with a 1-min isocratic gradient of 30% buffer B and a 30-min linear gradient of 30–100% buffer B at a flow rate of 1 ml/min (buffer A: 0.045% trifluoroacetic acid in H2O; buffer B: 0.039% trifluoroacetic acid in 60% CH3CN in H2O).
Synthesis of C-Terminal Peptides Aldehyde.
Preparation of 3-(9-fluorenylmethyloxycarbonyl)-amino-1,2-propanediol 1.9-Fluorenylmethyl chloroformate (5.2 g, 20 mmol) was added at 4°C to a solution of 3-amino-1,2-propanediol (1.8 g, 20 mmol)/triethylamine (3.1 ml, 22 mM) in H2O/1,4-dioxane (1:3, vol/vol, 100 ml). After 18 h, the solvent was evaporated and the residue was dissolved in ethyl acetate and crystallized to give 5.6 g (89% yield) of compound 1: melting point, 133–134°C; fast atom bombardment mass spectrometry [M + H]+, 314.4 (calc), 314.1 (found).
Preparation of the C-terminal diol trityl resin 2.
1 (1.25 g, 4 mmol) was added to 2-chlorotritylchloride resin (1 g, 1 mmol)/4-dimethylaminopyridine (0.5 g, 2 mmol) in dry N,N-dimethylacetamide (10 ml). After 24 h at 22°C, the resin was washed sequentially with dimethylformamide, methanol, and dichloromethane. Based on detection of the 9-fluorenylmethyl chromophore at 290 nm, the coupling yield was 80% (0.8 mmol/g resin). Free hydroxy groups were blocked with t-butyl-2,2,2-trichloroacetimidate.
Preparation of peptide aldehyde precursor, AAVALLPAVLLALLA-NHCH2CH(OH)CH2OH 3.
The hydrophobic peptide, containing a 1,2-propanediol moiety, was synthesized on resin 2 using Fmoc chemistry in 85% yield: fast atom bombardment mass spectrometry [M + H]+, 1491.9 (calc), 1491.1 (found).
Preparation of peptide glycoaldehyde, AAVALLPAVLLALLA-NHCH2CHO 4.
3 (30 mg, 0.02 mmol) was dissolved in 33% acetic acid (6 ml) and subsequently oxidized for 15 min with 20 mM NaIO4 (600 μl, in water). The desired product was obtained in a yield of 85% after preparative RP-HPLC (30–100% linear gradient of buffer B). The purity of the product was checked by analytical HPLC (tR: 31.5 min) and fast atom bombardment mass spectrometry [M + H]+: 1459.9 (calc), 1459.9 (found).
Synthesis of COOH-Terminal Nɛ-Glyoxylyl-Lysyl Peptides.
Preparation of Boc-Lys(Fmoc)-4-methylbenzhydrylamine resin 5. Boc-Lys(Fmoc)-OH (2.8 g, 6 mmol), benzotriazol-1-yl-oxy-tris-dimethylamino-phosphonium hexafluorophosphate (2.7 g, 6 mmol), and N,N-diisopropylethylamine (2.3 ml, 13.2 mmol) were added to 4-methylbenzhydrylamine resin (5 g, 3 mmol) in dimethylformamide (40 ml). The reaction was complete after 2 h as confirmed by ninhydrin test.
Synthesis of peptide aldehyde precursors.
AAVALLPAVLLALLAK(S)-NH2 6 and VTVLALGALAGVGVGK(S)-NH2 7 were synthesized using Boc chemistry and Boc-Ser(Bzl) was coupled to the Nɛ amino side chain of Lys after removal of the Fmoc group. After HF cleavage, crude peptides 6 and 7 were obtained in 85 and 80% yields, respectively. Purity of the products was confirmed by analytical HPLC (tR for 6, 26.73 min; tR for 7, 15.59 min) and matrix-assisted laser desorption ionization mass spectrometry (MALDI-MS): [M + H]+: 6, 1634.0 (calc), 1634 (found); 7, 1511.8 (calc), 1512 ± 1 (found).
Oxidation to peptide-Lys(COCHO)-NH2, 8 and 9.
6 (50 mg, 0.03 mmol) was dissolved in buffer B (20 ml) and subsequently oxidized with 20 mM NaIO4 (2 ml, in water) to yield 8. The reaction was complete after 3 h as monitored by analytical HPLC (tR for 8, 27.75 min). 8 was purified by preparative HPLC (60 min, 30–100% linear gradient of buffer A) in a yield of 90%. MALDI-MS [M + H]+: 8, 1602.0 (calc), 1602 ± 1 (found). 7 (55 mg, 0.036 mmol) was dissolved in 6 M guanidinium hydrochloride (GnHCl; 10 ml, in water) and subsequently oxidized with 20 mM NaIO4 (2 ml) for 1 h to yield 9. The product was dialyzed against water using a Spectra/Por CE dialysis membrane (molecular weight cut off: 500) and found to be 85% pure by analytical HPLC (tR for 9: 16.01). MALDI-MS [M + Na-H]+: 9, 1502.8 (calc), 1503 ± 1 (found).
Synthesis of Functional Peptides.
CYQRKRQKLMP 10, CYKEATSTFTAITYRGT 11 and CYKEATPTFTAITYRGT 12 were prepared by solid-phase peptide synthesis using Boc chemistry and purified by preparative HPLC (0–50% linear gradient of buffer B). MALDI-MS [M + H]+: 10, 1549.9 (calc), 1549 ± 1 (found); 11, 1956.1 (calc), 1959 ± 1 (found); 12, 1966.2 (calc), 1966 ± 1 (found).
Peptide Ligation via Thiazolidine Formation.
General procedure. 4 or 8 (5 μmol) was dissolved in dimethylformamide (0.5 ml) and 10, 11, or 12 (6 μmol) was added in 0.2 M sodium acetate buffer, pH 5.4 (0.8 ml). After ligation for 8 h, the product was isolated by preparative HPLC (30–100% buffer B). Products were analyzed by RP-HPLC and characterized by MALDI-MS. 13, (4 + 10): tR: 24.97 min, MALDI-MS [M + H]+: 2991 (calc), 2991 ± 1 (found); 14, (8 + 10): tR: 25.89 min, MALDI-MS [M + H]+ 3133 (calc), 3132 ± 1 (found); 15, (4 + 11): tR: 26.54 min, MALDI-MS [M + H]+: 3398 (calc), 3398 ± 1 (found); 16, (8 + 11): tR: 25.64 min, MALDI-MS [M + H]+: 3540 (calc), 3540 ± 1 (found); 17, (4 + 12): tR: 26.51 min, MALDI-MS [M + H]+: 3408 (calc), 3408 ± 1 (found).
Ligation of peptide aldehyde 9.
9 (1 μmol) was dissolved in 6 M GnHCl solution (5 ml, pH 5) and added to 11 (2 μmol) to yield 18. After the ligation was complete (17 h), tris(2-carboxyethyl)phosphine was added, and the mixture was diluted with 50 ml of water and dialyzed against water using a Spectra/Por CE dialysis membrane (molecular weight cutoff: 2000). 18 was obtained in 79% yield. Analytical HPLC: tR: 16.99 min. MALDI-MS [M + H]+: 18, 3417 (calc), 3419 ± 1 (found).
Antibodies and Cell Lines.
Polyclonal antibodies against the integrin β3-1 peptide lacking the membrane-permeable sequence were raised in rabbits immunized with this peptide conjugated to keyhole limpet hemocyanin. The antibodies were monospecific for the β3-1 peptide, as measured by ELISA (6). The HEL cell line (10, 11) was obtained from Thalia Papayannopoulou (University of Washington, Seattle, WA). The human Jurkat T cell line was obtained from Dean Ballard (Vanderbilt University, Nashville, TN). Both cell lines were maintained in RPMI 1640 (Cellgro) supplemented with 50 units/ml penicillin, 50 μg/ml streptomycin, and 10% heat-inactivated fetal bovine serum.
HEL Cell Adhesion Assay.
The cell adhesion assay was conducted as reported previously (6). Briefly, microtiter plates were coated with purified human fibrinogen (1.25 μg/ml) (12) overnight at 4°C, washed, and nonspecific sites blocked with 1% BSA. To measure the effect of peptides on cell adhesion, HEL cells (105 cells/well) were incubated with peptides at the indicated concentration for 30 min at room temperature in RPMI 1640/10% serum, centrifuged, and resuspended in RPMI 1640/10% serum. Cells were stimulated with phorbo 12-myristate 13-acetate (PMA; 10 nM) and plated on fibrinogen-coated microtiter plates. After incubation at 37°C for 120 min, plates were washed and adherent cells were quantitated with the cellular acid phosphatase assay (13). Percentage of inhibition of cell adhesion was determined after subtracting the background value obtained in ELISA. Trypan blue exclusion and fluorescein diacetate tests (14) were used to confirm that the cell-permeable peptides were not cytotoxic within the concentrations used (≤200 μM).
Cell-Permeable Peptide Import Detection.
Import of cell-permeable peptides was analyzed by confocal laser scanning microscopy of peptide-treated HEL cells cytocentrifuged onto glass slides (5). Adherent cells were fixed with 3.5% paraformaldehyde, permeabilized with 0.25% Triton X-100, and reacted with the anti-β3-1 peptide antiserum for 1 h at room temperature. Intracellular peptide–antibody complexes were detected with rhodamine-conjugated anti-rabbit IgG (Kirkgaard & Perry Laboratories) and visualized using a Leitz confocal laser scanning microscope with a ×100 oil immersion lens as previously described (5, 6).
Transcription Factor Nuclear Import Assay.
Peptides were dissolved in 0.5 ml of Hanks’ buffered salt solution without Ca2+ or Mg2+ (Cellgro), added to 4.5 ml of media containing 5 × 106 Jurkat T cells, and incubated for 30 min. Cells were then stimulated with PMA (5 nM; Sigma) and ionomycin (2 μM; Calbiochem) for 30 min, and cytoplasmic and nuclear extracts were prepared according to a modified method of Schreiber et al. (15). Briefly, cells were washed with ice-cold PBS and then lysed by incubation in 200 μl of buffer α [10 mM Hepes (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.4% Nonidet P-40, 1 mM DTT, 0.5 mM phenylmethylsulfonyl fluoride, 1 μg/ml each of leupeptin, aprotinin, pepstatin, chymostatin, and antipain] on ice for 5 min. Nuclei were pelleted by centrifugation, the supernatant was saved as the cytoplasmic extract, and the nuclear pellet was washed in 1 ml of buffer α. Nuclei were resuspended in 100 μl of buffer β [20 mM Hepes (pH 7.9), 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM phenymethylsulfonyl fluoride, and 1 μg/ml each of leupeptin, aprotinin, pepstatin, chymostatin, and antipain]. Samples were vortexed for 15 min at 4°C and centrifuged, and the supernatant was saved as the nuclear extract. The protein concentrations of the nuclear extracts were determined using the Pierce BCA protein assay and equalized among samples using buffer β.
Electrophoretic Mobility Shift Assay.
The following oligonucleotides were used (transcription factor recognition sites underlined): NF-κB, 5′-AGCTTAGAGGGGACTTTCCGAGAGGA-3′ (16); activator protein 1 (AP-1), 5′-GATCCATGACTCAGAGGAAAACA-3′ (17); nuclear factor of activated T cells (NFAT)/AP-1, 5′-GATCCAGAAAGGAGGAAAAACTGTTTCATACAG-3′ (18); and nuclear factor-Y box binding (NF-Y), 5′-GATCTGAGAATTTTCTGATTGGTTCTGGCGAGTTTGG-3′ (19). Double-stranded oligonucleotides (40 pmol) were labeled with 50 μCi of [α-32P]dATP (3000 Ci/mmol, DuPont-New England Nuclear) and 5 units of Escherichia coli DNA polymerase I (Klenow fragment) at room temperature for 30 min. Labeled oligonucleotides were separated from unincorporated [α-32P]dATP by chromatography on a Sephadex G50 spin column (Sigma). DNA-binding reactions were done in a final volume of 20 μl. NF-κB- and AP-1-binding reactions contained 5 μl of nuclear extract, 1 μg of poly(dI-dC), 1 μg of salmon sperm DNA, 2 μl of 10× electrophoretic mobility shift assay buffer [200 mM Hepes (pH 7.9), 50% glycerol, and 10 mM EDTA], and 100,000 cpm of 32P-labeled oligonucleotide. NFAT-binding reactions contained 5 μl of nuclear extract, 2 μg of poly(dI-dC), and 100,000 cpm of 32P-labeled oligonucleotide. NF-Y-binding reactions contained 5 μl of nuclear extract, 2 μg of poly(dI-dC), and 50,000 cpm of 32P-labeled oligonucleotide. All binding reactions were incubated at room temperature for 20 min and terminated by the addition of 2 μl of 10× electrophoretic mobility shift assay gel-loading dye (0.25% bromophenol blue, 0.25% xylene cyanol, and 50% glycerol). Samples were run in 4% polyacrylamide/0.5× TBE (45 mM Tris-borate and 1 mM EDTA) gels pre-run in 0.5× TBE for 60 min at 100 V. Gels were dried onto chromatography paper (Whatman 3Chr) and exposed to Kodak Biomax MR autoradiography film.
RESULTS
Synthetic Strategy to Generate an Aldehyde on the Hydrophobic MPS.
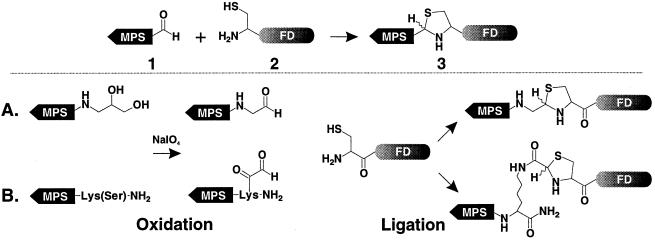
Two hydrophobic sequences, AAVALLPAVLLALLA and VTVLALGALAGVGVG, derived from the signal sequences of Kaposi’s fibroblast growth factor and human integrin β3, respectively, were used as MPSs (5, 6). Two general approaches were developed to prepare the COOH-terminal peptide aldehydes. The first involved synthesis of peptides containing a COOH-terminal aminopropanediol moiety (type A linkage, Fig. 1) and the second involved synthesis of peptides containing a 1,2-amino alcohol moiety (type B linkage, Fig. 1). The aldehyde functionality was generated on each peptide by periodate oxidation of the 1,2-cis-diol or 1,2-amino alcohol moiety (20, 21).
Figure 1.
General scheme for peptide ligation via thiazolidine ring formation using unprotected peptides. 1. A membrane-permeable peptide aldehyde. 2. NH2-terminal cysteinyl functional domain. 3. Ligation product linked by thiazolidine ring. (A) Thiazolidine ring formation between COOH-terminal peptide glycoaldehyde and NH2-terminal cysteinyl peptide. (B) Thiazolidine ring formation between lysinyl side chain aldehyde and NH2-terminal cysteinyl peptide.
Type A aldehyde precursor peptides were synthesized on a new functionalized resin generated by derivatizing chlorotrityl resin with 3-(9-fluorenylmethyloxycarbonyl)-amino-1,2-propanediol 1. The resulting resin 2 was used for synthesis of the Kaposi fibroblast growth factor hydrophobic sequence 3. Periodate oxidation of 3 generated the desired COOH-terminal peptide aldehyde 4. The type B aldehyde precursor peptides were synthesized on Boc-Lys(Fmoc)-4-methylbenzhydrylamine resin 5 using Boc chemistry. After assembly of the hydrophobic peptide sequences, Fmoc was removed and Boc-Ser(Bzl) was attached to the ɛ amino group of the Lys side chain to generate 6 and 7, respectively. The 1,2-amino alcohol moiety of Ser was converted to an aldehyde by periodate oxidation to give 8 and 9. Depending on the solubility of each hydrophobic aldehyde precursor peptide, different periodate oxidation conditions were used to optimize the yield. For example, 3 was oxidized in 33% acetic acid for 15 min whereas 7 was oxidized in 6 M GnHCl (pH 5) for 10 min. Alternatively, oxidation of 7 was also performed in 60% aqueous acetonitrile (pH 2) for 3 h.
Ligation.
Preparation of functional cell-permeable peptides by the modular approach described here involves ligation of a MPS prepared as a hydrophobic peptide aldehyde and a functional sequence with a NH2-terminal cysteine. Preparation of the MPS and functional peptide modules has been described above. The functional peptide sequences used include a wild-type (CYKEATSTFTNITYRGT) and loss-of-function mutant (CYKEATPTFTNITYRGT) of the human integrin β3 cytoplasmic tail and the NF-κB p50 nuclear localization signal (NLS) sequence (CYVQRKRQKLMP). Peptide ligation through thiazolidine formation required adjustment of conditions to optimize solubility of both hydrophobic and functional peptide modules. For example, optimal ligation of 4 and 11 occurred with the MPS peptide aldehyde 4 dissolved in dimethylformamide and the functional peptide 11 dissolved in sodium acetate buffer (0.2 M, pH 5.6). In all cases, the ligation was complete within 6 h as monitored by analytical HPLC (Fig. 2). In general, the product was readily soluble due to the hydrophilic cargo sequence and could be purified by either preparative HPLC or dialysis. This method was successfully used for synthesis of peptides 13, 14, 15, 16, and 17 (see Table 1). One exception was preparation of 18 by ligation of 9 and 11, which was performed in 6 M GnHCl (pH 5) solution. After removal of GnHCl by dialysis, the conjugate was sufficiently soluble in water to permit purification by RP-HPLC.
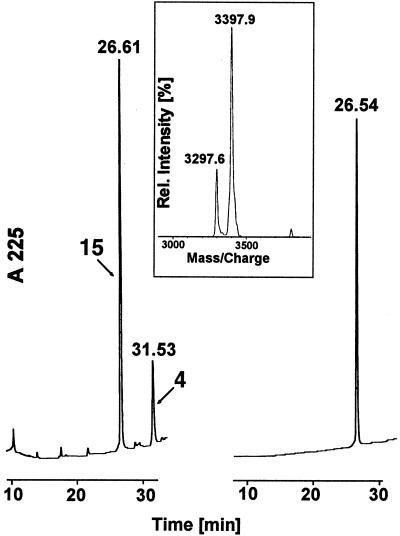
Figure 2.
Profile of the ligation reaction between peptides 4 and 11 to generate the functional cell-permeable peptide 15. Left panel shows RP-HPLC of the ligation reaction after 8 h. Right panel shows RP-HPLC of the purified product 15. The inset is MALDI-MS of the product showing a mass of 3397.9 (calc 3398.0).
Table 1.
Cell-permeable peptides used
| No. | Name | No. | Sequence | No. | Functional domain | Linkage |
|---|---|---|---|---|---|---|
| Cell-permeable peptides synthesized by modular method | ||||||
| 1 | ScN50A | 4 | AAVALLPAVLLALLA- | 1 | CYVQRKRQKLMP | A |
| 3 | NHCH2CHO | 0 | ||||
| 1 | ScN50B | 8 | AAVALLPAVLLALLAK(COCHO) | 1 | CYVQRKRQKLMP | B |
| 4 | -NH2 | 0 | ||||
| 1 | Scβ3-1A | 4 | AAVALLPAVLLALLA- | 1 | CYKEATSTFTNITY | A |
| 5 | NHCH2CHO | 1 | RGT | |||
| 1 | Scβ3-1B | 8 | AAVALLPAVLLALLAK(COCHO | 1 | CYKEATSTFTNITY | B |
| 6 | -NH2 | 1 | RGT | |||
| 1 | Scβ3- | 4 | AAVALLPAVLLALLA- | 1 | CYKEATPTFTNITY | A |
| 7 | 1P752A | NHCH2CHO | 2 | RGT | ||
| 1 | Scβ3-1 | 9 | VTVLALGALAGVGVGK(COCHO) | 1 | CYKEATSTFTNITY | B |
| 8 | -NH2 | 1 | RGT | |||
| Cell-permeable peptides synthesized by conventional method | ||||||
| 1 | SN50 | AAVALLPAVLLALLAPVQRKRQKLMP | ||||
| 9 | ||||||
| 2 | Sβ3-1 | VTVLALGALAGVGVGYKEATSTFTNITYRGT | ||||
| 0 | ||||||
Comparison of the Biological Activity of Peptides with Conventional Peptide Bond vs. Peptides with Thiazolidine Linkage.
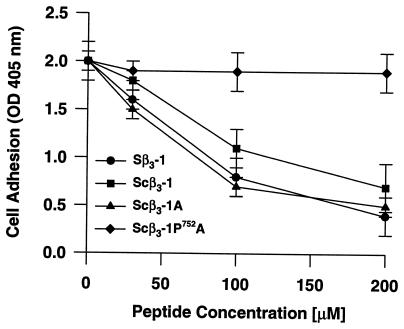
Integrin β3 cytoplasmic tail peptides. We have shown previously through structure-function analysis of the integrin β3 cytoplasmic tail that COOH-terminal residues 747–762 constitute a functionally important cell adhesion regulatory domain (CARD) and that a similar domain exists in integrin β1 (6). A cell-permeable peptide bearing the CARD sequence (Sβ3-1, 20), selectively inhibited adhesion of HEL cells to immobilized fibrinogen. For comparison, three thiazolidine-conjugated homologues of Sβ3-1, 20 (Scβ3-1A, 15; Scβ3-1B, 16; Scβ3-1, 18) were prepared and their functional activities were evaluated by cell adhesion assay. The extracellular concentration of each peptide required for 50% inhibition of adhesion (EC50) was determined by dose-response analysis. As shown in Fig. 3, the EC50 of the hybrid peptides, Scβ3-1A (15) and Scβ3-1 (18), was essentially equivalent to that of the conventionally prepared Sβ3-1 (20) peptide (≈60 μM). The inhibitory pattern of the hybrid Scβ3-1B (16) peptide was virtually identical to that of Scβ3-1A (15) (data not shown).
Figure 3.
Inhibition of HEL cell adhesion to immobilized fibrinogen by the cell-permeable peptides Sβ3-1, Scβ3-1, and Scβ3-1A is concentration dependent. The mutant Scβ3-1P752A did not inhibit HEL cell adhesion. Data are the mean ± SEM from three or more independent experiments, each performed in triplicate. The difference between the Scβ3-1 and Scβ3-1A peptides versus Scβ3-1P752A peptide at 200 μM was significant with p < 0.0001.
In the human, a loss-of-function point mutation S752 → P752 in the cytoplasmic segment of integrin β3 causes abnormal adhesive function of the platelet integrin αIIbβ3 (GPIIb–IIIa) and is responsible for the lifelong bleeding tendency of Glanzmann thrombasthenia patients (22). When this same point mutation was introduced into the CARD domain of the Sβ3-1 peptide, it lost inhibitory activity in the HEL cell adhesion assay (6). A homologue of the thiazolidine-ligated hybrid peptide Scβ3-1A (15) was prepared harboring the S752 → P752 mutation (Scβ3-1P752A, 17) and compared for functionality in the HEL cell adhesion assay. As shown in Fig. 3, this peptide had no significant inhibitory effect as predicted.
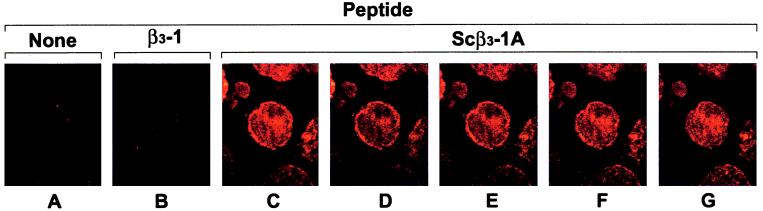
Import of the Sβ3-1 (20) peptide to the cytoplasm of HEL cells has previously been verified by immunofluorescent staining of peptide-treated cells with an antibody specific for the CARD region. Confocal microscopy of these samples demonstrated that the peptide was distributed throughout the cytoplasm and not simply membrane bound (6). Using the same technique, the thiazolidine-ligated Scβ3-1A (15) peptide was found to have a similar subcellular distribution (Fig. 4). In contrast, cells treated with the β3-1 (11) peptide, lacking a MPS, showed no staining. Together, these results document both the import competence and the functional activity of cell-permeable peptides synthesized by this new modular approach.
Figure 4.
Intracellular location of the cell-permeable Scβ3-1A peptide as demonstrated by confocal laser scanning microscopy. Intracellular peptide was detected as yellow fluorescence and analyzed by a four-step Z-position sectional scanning of the cell (1-μm sections). (A) Composite image of an untreated cell. (B) Composite image of a cell treated with β3-1 peptide (lacking a MPS). (C) Composite image of a cell treated with thiazolidine-ligated Scβ3-1A peptide. (D–G) Sequential Z-position sections of the Scβ3-1A peptide-treated cell showing distribution of the peptide throughout the cytoplasm and cell membrane.
Inhibition of transcription factor nuclear import.
One of the principle mechanisms of controlling gene expression in eukaryotic cells is the sequestration of transcription factors in the cytoplasmic compartment, away from their site of action in the nucleus. Signaling to the nucleus by these transcription factors in response to cell stimulation requires that they be shuttled from the cytoplasm to the nuclear compartment. The NLS motif present on transcription factors and other karyophilic proteins targets them to the nucleus by associating with a cytoplasmic NLS receptor complex (23). We have previously shown that a cell-permeable peptide (SN50, 19) bearing a NLS from the p50 subunit of transcription factor NF-κB was capable of blocking the nuclear import of NF-κB in endothelial and monocytic cell lines (5). Subsequently, we have demonstrated that this same peptide inhibited nuclear import of three different transcription factors (NF-κB, AP-1, and NFAT) in human Jurkat T cells (T.R.T., A.D.C., J. Donahue, Y.-Z. Lin, and J.H., unpublished results).
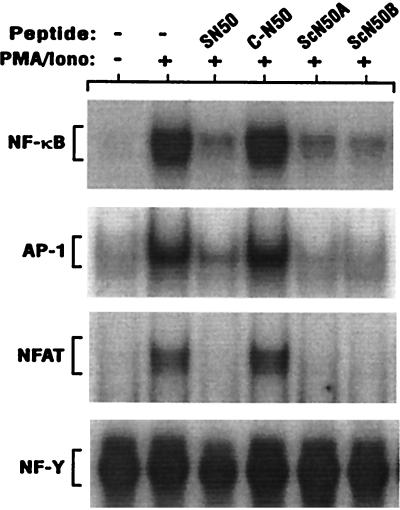
We have used the new modular synthesis approach described herein to prepare two SN50 homologues, ScN50A (13) and ScN50B (14), in which the MPS and functional NLS peptide modules are linked by two different thiazolidino linkages. These peptides were compared with SN50 (19) for their ability to inhibit the nuclear import of NF-κB, AP-1, and NFAT in Jurkat T cells stimulated with a combination of PMA and ionomycin. As shown in Fig. 5, the thiazolidine-ligated peptides, ScN50A (13) and ScN50B (14), blocked nuclear import of all three transcription factors as effectively as the previously described SN50 (19) peptide prepared by conventional peptide synthesis and having only peptide bonds in the backbone (5). The inhibitory effect of 13, 14, and 19 was limited to transcription factors rapidly translocated from cytoplasm to nucleus following cell stimulation as indicated by the lack of effect on NF-Y, a transcription factor constitutively present in the nucleus. This inhibitory effect requires that the functional peptide be cell permeable as shown by the lack of C-N50 (10) (the functional NLS without the MPS) effect on nuclear import. Thus, like SN50 (19), the thiazolidine-ligated ScN50 peptides (13, 14) are functionally active and may attenuate expression of genes regulated by these three transcription factors such as interleukin-2 (T.R.T., A.D.C., J. Donahue, Y.-Z. Lin, and J.H., unpublished results).
Figure 5.
Nuclear import inhibition by the cell-permeable p50 NLS peptides. Jurkat T cells were treated for 30 min with the peptides (75 μM) indicated above each lane (see Table 1). Cells were then stimulated with PMA (5 nM) and ionomycin (2 μM) for 60 min. Nuclear extracts were prepared and assayed for the presence of each transcription factor by electrophoretic mobility shift assay as described in Materials and Methods. Only the section of each autoradiogram that includes the specific transcription factor–32P-labeled DNA complex is shown as indicated for NF-κB, AP-1, NFAT and the constitutively nuclear transcription factor NF-Y.
DISCUSSION
Design Considerations.
To facilitate the rapid preparation of functional cell-permeable peptides, we have developed a site-specific modular synthesis approach that makes it possible to confer cell permeability on virtually any peptide containing an NH2-terminal cysteine. We established the following criteria for a method that would be generally useful for ligation of two peptide modules to make a hybrid peptide that is both cell permeable and bioactive: (i) The reaction should be site-specific to avoid possible interference with the biological activity of the functional peptide module. (ii) It should be performed in aqueous solutions using unprotected peptides. (iii) It should be synthetically straightforward and compatible with existing solid-phase peptide synthesis methods. Thiazolidine formation fulfills these requirements. It requires a 1,2-amino thiol which is derived from cysteine on one peptide and an aldehyde derived from mild periodate oxidation of serine or threonine on the other (7, 20). In our hands, there appears to be no discernible difference which moiety (the aldehyde or cysteine) is placed on which peptide module (hydrophobic or functional). In this study, the hydrophobic MPS was prepared with the aldehyde at the COOH terminus and the functional sequence with the cysteine at the NH2 terminus (Fig. 1). One advantage to this approach is the use of fully free peptides or proteins (synthetic or recombinant) which obviates any need for chemical modification that might affect oxidation- and acid-sensitive amino acids in the functional domain. In addition, the hydrophobic MPS can be pre-made in large quantities and stored as a shelf-ready reagent. In this study, we demonstrate the feasibility of a general method of synthesizing hybrid peptides made of two modules: a MPS module and a functional module bearing “cargo” derived from functional segments of intracellular proteins. Two distinct MPSs based on the core hydrophobic region of signal sequences from human integrin β3 and Kaposi’s fibroblast growth factor were modified and ligated to peptides representing two different functional domains: the CARD region of human integrin β3 consisting of residues 747–762 (6) and the NLS of NF-κB p50 (5).
This novel method of synthesizing cell-permeable bioactive peptides is highly efficient and convenient. A one-step ligation reaction employing pre-made, prepurified, and shelf-ready MPS allows rapid production of cell-permeable peptides carrying a wide variety of sequences derived from known intracellular proteins to conduct structure-function analysis. Thus, multiple analogs of short functional sequences could potentially be prepared and tested simultaneously for function without the need for separate synthesis of individual peptide analogs. Furthermore, shorter preparation time and high yields represent a definite advantage over conventional stepwise solid-phase synthesis of bimodular cell-permeable peptides.
The resulting hybrid peptides which contained a nonamide thiazolidino linkage instead of a peptide linkage between the MPS and functional modules entered cells efficiently and exerted an equally potent functional effect as homologous peptides prepared by conventional methods. These peptides inhibited the adhesion of HEL cells to fibrinogen and blocked nuclear import of three distinct transcription factors in Jurkat T cells.
Since efficient ligation methods of unprotected peptides through nonamide or amide linkage have recently been developed (24, 25), our results also point to the potential use of these facile ligation methods to prepare hybrid cellular import peptides. These results are significant in that the presence of a nonpeptide bond between two modules of a hybrid peptide does not hamper its cellular import (Figs. 3–5). It also indicates that the requirement for cellular import can be met by the hydrophobic threshold of the import sequence as illustrated by more effective import of Scβ3-1A peptide (Fig. 3). In this regard, our results are consistent with the recent report of using all d-amino acid in transport or antimicrobial peptide sequences as membrane-permeable sequences (26). The importance of hydrophobic threshold in the importing function is further illustrated in recent studies (27) which show that the MPS can be placed at either the carboxylic end of the cargo sequence with satisfactory results in intracellular transport of peptides.
Our new method of cell-permeable peptide synthesis will aid in the further elucidation of the mechanism of cellular import and in expanding the range of applications of cellular import of bioactive molecules. So far, available evidence points to the nonreceptor-mediated mechanism of cellular import based on MPS derived from the hydrophobic H region of the signal sequence (4). To study this mechanism further, chirally distinct analogs of MPS can be prepared apart from a module containing a functional cargo and antigenic epitope tag for tracking imported peptides in intracellular compartments. Moreover, the apparent versatility of the one-step ligation opens up the possibility of facile production of cell-permeable peptide libraries, mimetics, and other nonpeptide bioactive molecules.
Acknowledgments
We thank Traci Castleman for assistance in preparation of the manuscript. This work was supported by National Institutes of Health Grants HL45994 (to J.H.), HL30647 (to J.H.), CA36544 (to J.P.T.), and AI37965 (to J.P.T.).
ABBREVIATIONS
- MALDI-MS
matrix-assisted laser desorption ionization mass spectrometry
- MPS
membrane-permeable sequence
- HEL
human erythroleukemia
- RP
reverse phase
- CARD
cell adhesion regulatory domain
- PMA
phorbol 12-myristate 13-acetate
- AP-1
activator protein 1
- NFAT
nuclear factor of activated T cells
- NF-Y
nuclear factor-Y box binding
- NLS
nuclear localization signal
- GnHCl
guanidinum hydrochloride
References
- 1.Wang K, Feramisco J R, Ash J F. Methods Enzymol. 1982;85:514–562. doi: 10.1016/0076-6879(82)85050-7. [DOI] [PubMed] [Google Scholar]
- 2.Henkel T, Zabel U, Van Zee K, Muller J M, Fanning E, Baeuerle P A. Cell. 1992;68:1121–1133. doi: 10.1016/0092-8674(92)90083-o. [DOI] [PubMed] [Google Scholar]
- 3.Perlmutter R M, Alberola-Ila J. Curr Opin Immunol. 1996;8:285–290. doi: 10.1016/s0952-7915(96)80069-0. [DOI] [PubMed] [Google Scholar]
- 4.Hawiger J. Curr Opin Immunol. 1997;9:189–194. doi: 10.1016/s0952-7915(97)80134-3. [DOI] [PubMed] [Google Scholar]
- 5.Lin Y-Z, Yao S Y, Veach R A, Torgerson T R, Hawiger J. J Biol Chem. 1995;270:14255–14258. doi: 10.1074/jbc.270.24.14255. [DOI] [PubMed] [Google Scholar]
- 6.Liu X-Y, Timmons S, Lin Y-Z, Hawiger J. Proc Natl Acad Sci USA. 1996;93:11819–11824. doi: 10.1073/pnas.93.21.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang L, Tam J P. Anal Biochem. 1996;233:87–93. doi: 10.1006/abio.1996.0011. [DOI] [PubMed] [Google Scholar]
- 8.Liu C-F, Tam J P. J Am Chem Soc. 1994;116:4149–4153. [Google Scholar]
- 9.Merrifield R B. J Am Chem Soc. 1963;85:2149. [Google Scholar]
- 10.Tabilio A, Rosa J-P, Testa U, Kieffer N, Nurden A T, Del Canizo M C, Breton-Gorius J, Vainchenker W. EMBO J. 1984;3:453–459. doi: 10.1002/j.1460-2075.1984.tb01827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosa J-P, McEver R P. J Biol Chem. 1989;264:12596–12603. [PubMed] [Google Scholar]
- 12.Hawiger J, Timmons S. Methods Enzymol. 1992;215:228–243. doi: 10.1016/0076-6879(92)15067-m. [DOI] [PubMed] [Google Scholar]
- 13.O’Toole T E, Mandelman D, Forsyth J, Shattil S J, Plow E F, Ginsberg M H. Science. 1991;254:845–847. doi: 10.1126/science.1948065. [DOI] [PubMed] [Google Scholar]
- 14.Kajstura J, Reiss K. Folia Histochem Cytobiol. 1989;27:39–48. [PubMed] [Google Scholar]
- 15.Schreiber E, Mattias P, Muller M M, Schaffner W. Nucleic Acids Res. 1989;17:6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordle S R, Donald R, Read M A, Hawiger J. J Biol Chem. 1993;268:11803–11810. [PubMed] [Google Scholar]
- 17.Cook J S, Lucas J J, Sibley E, Bolanowski M A, Christy R J, Kelly T J, Lane M D. Proc Natl Acad Sci USA. 1988;85:2949–2953. doi: 10.1073/pnas.85.9.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jain J, Miiner Z, Rao A. J Immunol. 1993;151:837–848. [PubMed] [Google Scholar]
- 19.Boothby M, Liou H-C, Glimcher L H. J Immunol. 1989;142:1005–1014. [PubMed] [Google Scholar]
- 20.Rose U, Jones R M L, Sundaram G, Offord R E. In: Peptides 1988. Jung G, Bayer E, editors. New York: de Gruyter; 1989. pp. 274–276. [Google Scholar]
- 21.O’Shannessy D J, Wilchek M. Anal Biochem. 1990;191:1–8. doi: 10.1016/0003-2697(90)90377-l. [DOI] [PubMed] [Google Scholar]
- 22.Chen Y-P, Djaffar I, Pidard D, Steiner B, Cieutat A-M, Caen J P, Rosa J-P. Proc Natl Acad Sci USA. 1992;89:10169–10173. doi: 10.1073/pnas.89.21.10169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Görlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 24.Wallace C J. Curr Opin Biotechnol. 1995;6:403–410. doi: 10.1016/0958-1669(95)80069-7. [DOI] [PubMed] [Google Scholar]
- 25.Tam, J. P. & Spetzler, J. C. (1998) Curr. Protocols Immunol., in press.
- 26.Merrifield R B, Javvad P, Andreu D, Ubach J, Boman A, Boman H G. Proc Natl Acad Sci USA. 1995;92:3449–3453. doi: 10.1073/pnas.92.8.3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rojas M, Donahue J P, Tan Z, Lin Y Z. Nat Biotechnol. 1998;16:370–375. doi: 10.1038/nbt0498-370. [DOI] [PubMed] [Google Scholar]