Abstract
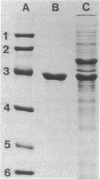
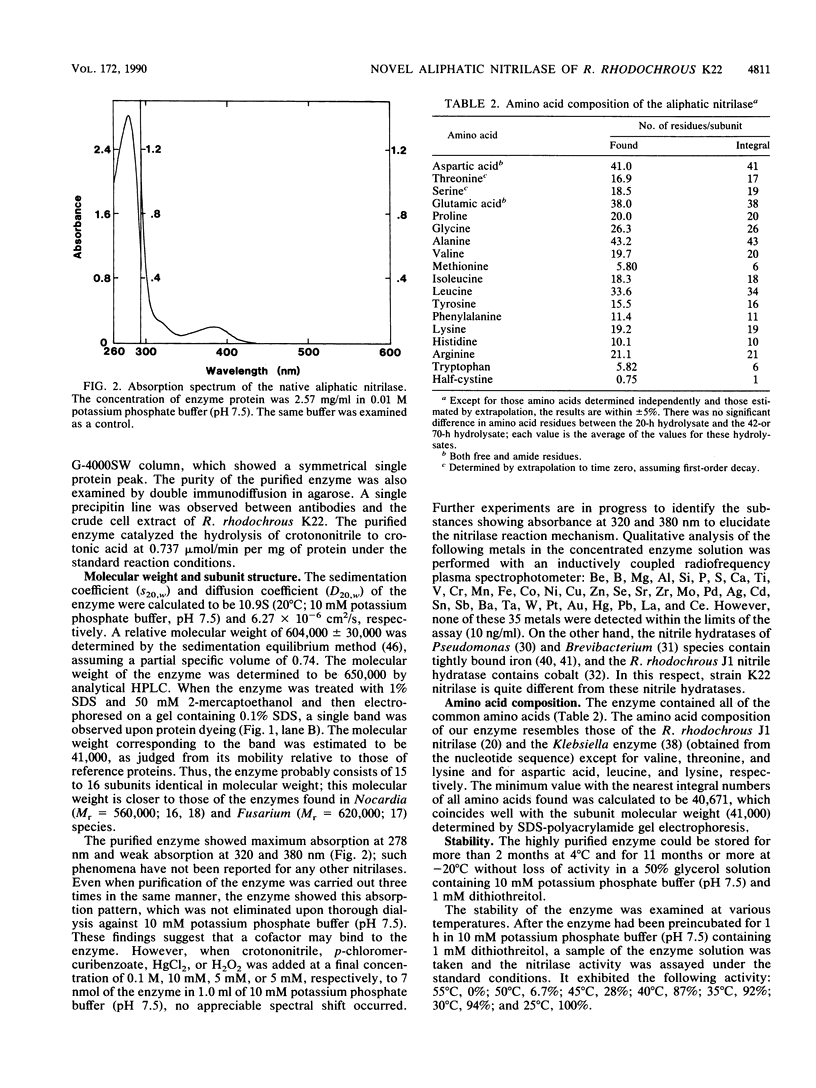
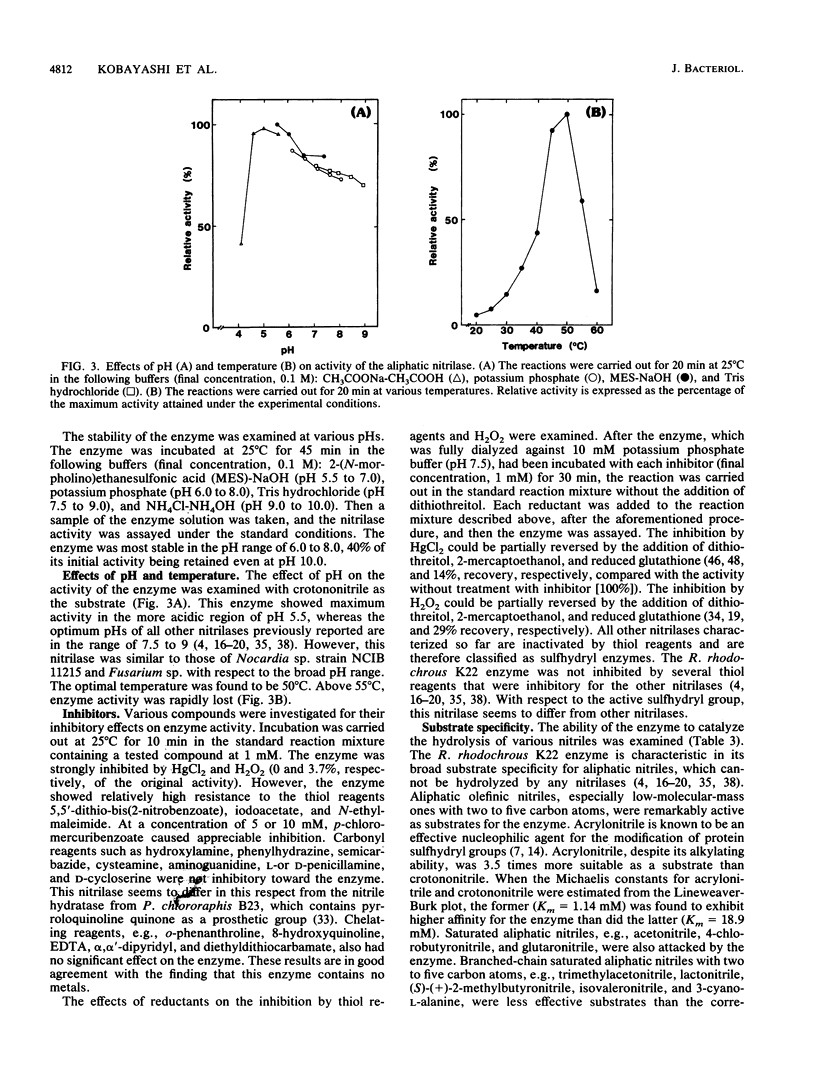
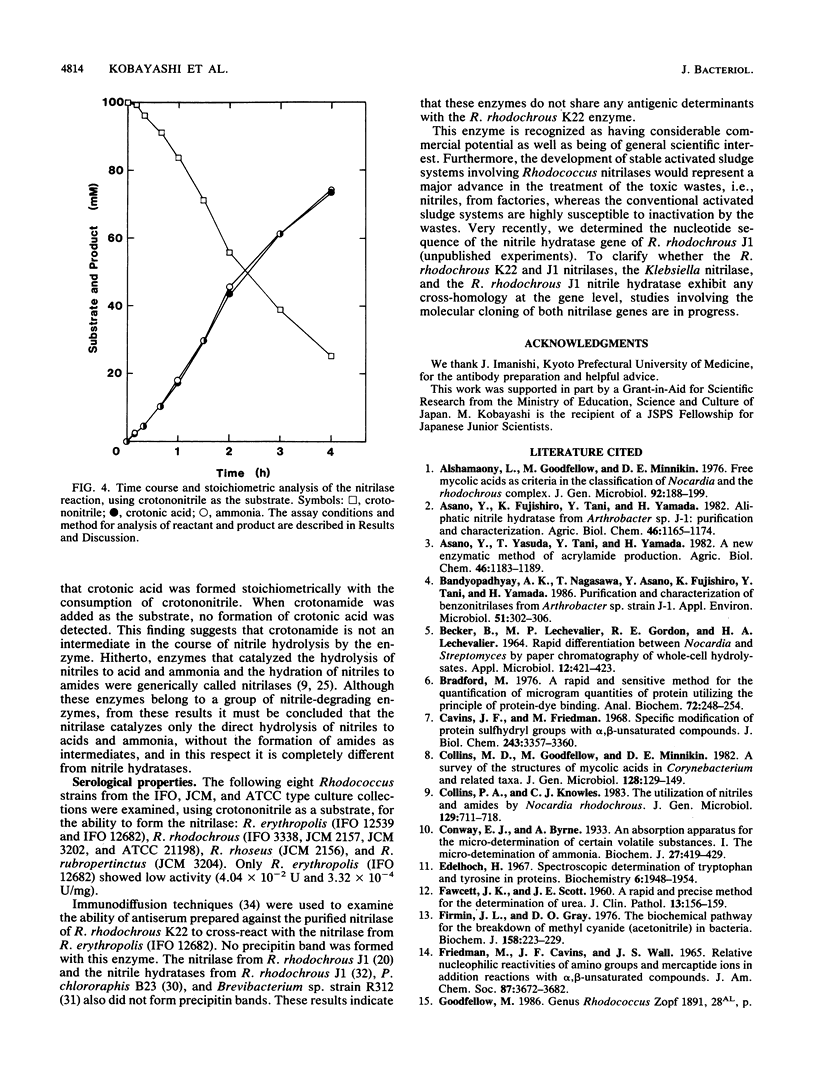
A novel nitrilase that preferentially catalyzes the hydrolysis of aliphatic nitriles to the corresponding carboxylic acids and ammonia was found in the cells of a facultative crotononitrile-utilizing actinomycete isolated from soil. The strain was taxonomically studied and identified as Rhodococcus rhodochrous. The nitrilase was purified, with 9.08% overall recovery, through five steps from a cell extract of the stain. After the last step, the purified enzyme appeared to be homogeneous, as judged by polyacrylamide gel electrophoresis, analytical centrifugation, and double immunodiffusion in agarose. The relative molecular weight values for the native enzyme, estimated from the ultracentrifugal equilibrium and by high-performance liquid chromatography, were approximately 604,000 +/- 30,000 and 650,000, respectively, and the enzyme consisted of 15 to 16 subunits identical in molecular weight (41,000). The enzyme acted on aliphatic olefinic nitriles such as crotononitrile and acrylonitrile as the most suitable substrates. The apparent Km values for crotononitrile and acrylonitrile were 18.9 and 1.14 mM, respectively. The nitrilase also catalyzed the direct hydrolysis of saturated aliphatic nitriles, such as valeronitrile, 4-chlorobutyronitrile, and glutaronitrile, to the corresponding acids without the formation of amide intermediates. Hence, the R. rhodochrous K22 nitrilase is a new type distinct from all other nitrilases that act on aromatic and related nitriles.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alashamaony L., Goodfellow M., Minnikin D. E. Free mycolic acids as criteria in the classification of Nocardia and the 'rhodochrous' complex. J Gen Microbiol. 1976 Jan;92(1):188–199. doi: 10.1099/00221287-92-1-188. [DOI] [PubMed] [Google Scholar]
- BECKER B., LECHEVALIER M. P., GORDON R. E., LECHEVALIER H. A. RAPID DIFFERENTIATION BETWEEN NOCARDIA AND STREPTOMYCES BY PAPER CHROMATOGRAPHY OF WHOLE-CELL HYDROLYSATES. Appl Microbiol. 1964 Sep;12:421–423. doi: 10.1128/am.12.5.421-423.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay A. K., Nagasawa T., Asano Y., Fujishiro K., Tani Y., Yamada H. Purification and Characterization of Benzonitrilases from Arthrobacter sp. Strain J-1. Appl Environ Microbiol. 1986 Feb;51(2):302–306. doi: 10.1128/aem.51.2.302-306.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Cavins J. F., Friedman M. Specific modification of protein sulfhydryl groups with alpha,beta-unsaturated compounds. J Biol Chem. 1968 Jun 25;243(12):3357–3360. [PubMed] [Google Scholar]
- Collins M. D., Goodfellow M., Minnikin D. E. A survey of the structures of mycolic acids in Corynebacterium and related taxa. J Gen Microbiol. 1982 Jan;128(1):129–149. doi: 10.1099/00221287-128-1-129. [DOI] [PubMed] [Google Scholar]
- Conway E. J., Byrne A. An absorption apparatus for the micro-determination of certain volatile substances: The micro-determination of ammonia. Biochem J. 1933;27(2):419–429. [PMC free article] [PubMed] [Google Scholar]
- Edelhoch H. Spectroscopic determination of tryptophan and tyrosine in proteins. Biochemistry. 1967 Jul;6(7):1948–1954. doi: 10.1021/bi00859a010. [DOI] [PubMed] [Google Scholar]
- FAWCETT J. K., SCOTT J. E. A rapid and precise method for the determination of urea. J Clin Pathol. 1960 Mar;13:156–159. doi: 10.1136/jcp.13.2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firmin J. L., Gray D. O. The biochemical pathway for the breakdown of methyl cyanide (acetonitrile) in bacteria. Biochem J. 1976 Aug 15;158(2):223–229. doi: 10.1042/bj1580223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HOOK R. H., ROBINSON W. G. RICININE NITRILASE. II. PURIFICATION AND PROPERTIES. J Biol Chem. 1964 Dec;239:4263–4267. [PubMed] [Google Scholar]
- Harper D. B. Characterization of a nitrilase from Nocardia sp. (Rhodochrous group) N.C.I.B. 11215, using p-hydroxybenzonitrile as sole carbon source. Int J Biochem. 1985;17(6):677–683. doi: 10.1016/0020-711x(85)90364-7. [DOI] [PubMed] [Google Scholar]
- Harper D. B. Fungal degradation of aromatic nitriles. Enzymology of C-N cleavage by Fusarium solani. Biochem J. 1977 Dec 1;167(3):685–692. doi: 10.1042/bj1670685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper D. B. Microbial metabolism of aromatic nitriles. Enzymology of C-N cleavage by Nocardia sp. (Rhodochrous group) N.C.I.B. 11216. Biochem J. 1977 Aug 1;165(2):309–319. doi: 10.1042/bj1650309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi M., Nagasawa T., Yamada H. Nitrilase of Rhodococcus rhodochrous J1. Purification and characterization. Eur J Biochem. 1989 Jun 15;182(2):349–356. doi: 10.1111/j.1432-1033.1989.tb14837.x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Mathew C. D., Nagasawa T., Kobayashi M., Yamada H. Nitrilase-Catalyzed Production of Nicotinic Acid from 3-Cyanopyridine in Rhodococcus rhodochrous J1. Appl Environ Microbiol. 1988 Apr;54(4):1030–1032. doi: 10.1128/aem.54.4.1030-1032.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milvy P., Wolff M. Mutagenic studies with acrylonitrile. Mutat Res. 1977 Jul;48(3-4):271–278. doi: 10.1016/0027-5107(77)90169-5. [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Nanba H., Ryuno K., Takeuchi K., Yamada H. Nitrile hydratase of Pseudomonas chlororaphis B23. Purification and characterization. Eur J Biochem. 1987 Feb 2;162(3):691–698. doi: 10.1111/j.1432-1033.1987.tb10692.x. [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Ryuno K., Yamada H. Nitrile hydratase of Brevibacterium R312--purification and characterization. Biochem Biophys Res Commun. 1986 Sep 30;139(3):1305–1312. doi: 10.1016/s0006-291x(86)80320-5. [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Takeuchi K., Yamada H. Occurrence of a cobalt-induced and cobalt-containing nitrile hydratase in Rhodococcus rhodochrous J1. Biochem Biophys Res Commun. 1988 Sep 15;155(2):1008–1016. doi: 10.1016/s0006-291x(88)80597-7. [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Yamada H. Nitrile hydratase is a quinoprotein. A possible new function of pyrroloquinoline quinone: activation of H2O in an enzymatic hydration reaction. Biochem Biophys Res Commun. 1987 Sep 15;147(2):701–709. doi: 10.1016/0006-291x(87)90987-9. [DOI] [PubMed] [Google Scholar]
- ROBINSON W. G., HOOK R. H. RICININE NITRILASE. I. REACTION PRODUCT AND SUBSTRATE SPECIFICITY. J Biol Chem. 1964 Dec;239:4257–4262. [PubMed] [Google Scholar]
- Stalker D. M., Malyj L. D., McBride K. E. Purification and properties of a nitrilase specific for the herbicide bromoxynil and corresponding nucleotide sequence analysis of the bxn gene. J Biol Chem. 1988 May 5;263(13):6310–6314. [PubMed] [Google Scholar]
- Sugiura Y., Kuwahara J., Nagasawa T., Yamada H. Significant interaction between low-spin iron(III) site and pyrroloquinoline quinone in active center of nitrile hydratase. Biochem Biophys Res Commun. 1988 Jul 29;154(2):522–528. doi: 10.1016/0006-291x(88)90171-4. [DOI] [PubMed] [Google Scholar]
- Venitt S., Bushell C. T., Osborne M. Mutagenicity of acrylonitrile (cyanoethylene) in Escherichia coli. Mutat Res. 1977 Nov;45(2):283–288. doi: 10.1016/0027-5107(77)90028-8. [DOI] [PubMed] [Google Scholar]