Abstract
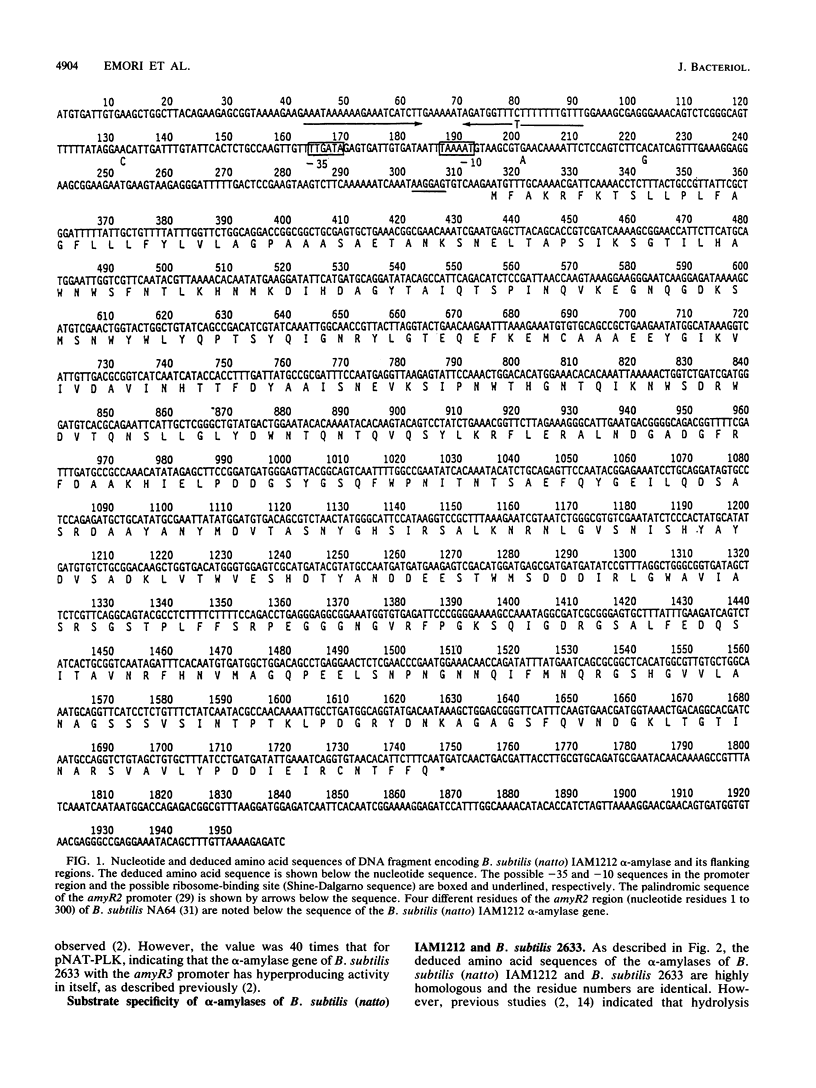
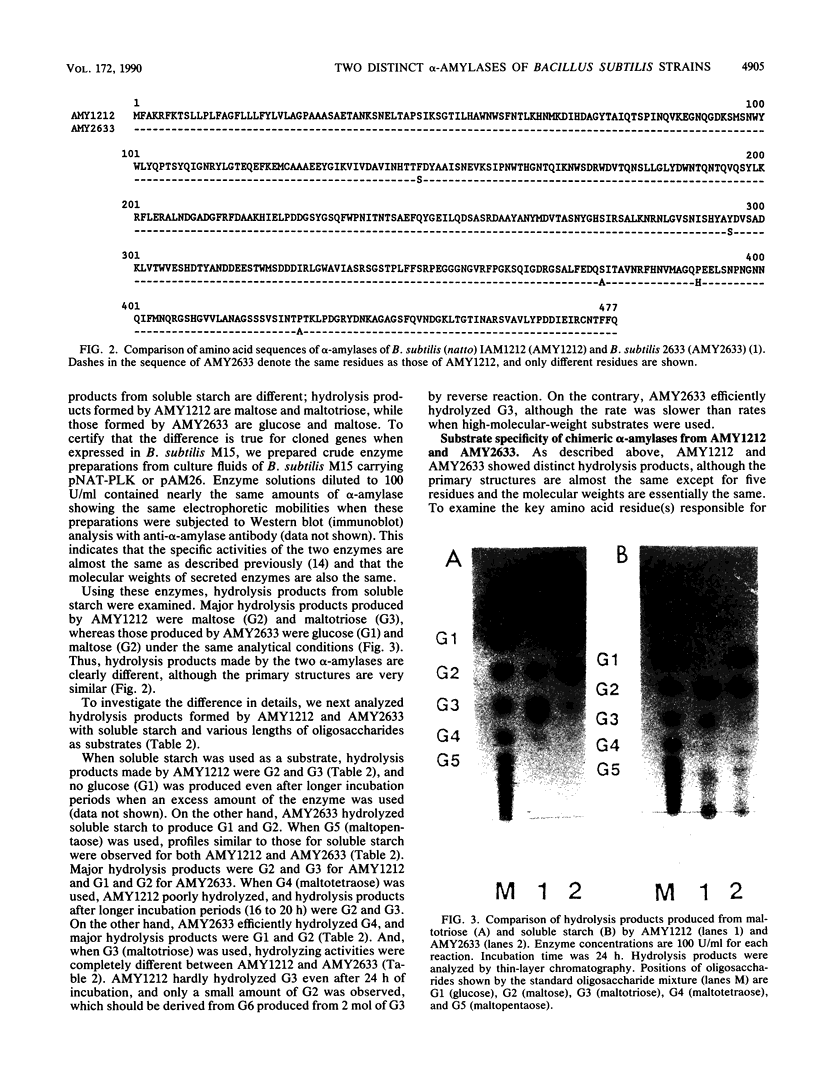
An alpha-amylase gene of Bacillus subtilis (natto) IAM1212 was cloned in a lambda EMBL3 bacteriophage vector, and the nucleotide sequence was determined. An open reading frame encoding the alpha-amylase (AMY1212) consists of 1,431 base pairs and contains 477 amino acid residues, which is the same in size as the alpha-amylase (AMY2633) of B. subtilis 2633, an alpha-amylase-hyperproducing strain, and smaller than that of B. subtilis 168, Marburg strain. The amino acid sequence of AMY1212 is different from that of AMY2633 at five residues. Enzymatic properties of these two alpha-amylases were examined by introducing the cloned genes into an alpha-amylase-deficient strain, B. subtilis M15. It was revealed that products of soluble starch hydrolyzed by AMY1212 are maltose and maltotriose, while those of AMY2633 are glucose and maltose. From the detailed analyses with oligosaccharides as substrates, it was concluded that the difference in hydrolysis products of the two similar alpha-amylases should be ascribed to the different activity hydrolyzing low-molecular-weight substrates, especially maltotriose; AMY1212 slowly hydrolyzes maltotetraose and cannot hydrolyze maltotriose, while AMY2633 efficiently hydrolyzes maltotetraose and maltotriose. Further analyses with chimeric alpha-amylase molecules constructed from the cloned genes revealed that only one amino acid substitution is responsible for the differences in hydrolysis products.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Emori M., Maruo B. Complete nucleotide sequence of an alpha-amylase gene from Bacillus subtilis 2633, an alpha-amylase extrahyperproducing strain. Nucleic Acids Res. 1988 Jul 25;16(14B):7178–7178. doi: 10.1093/nar/16.14.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischauf A. M., Lehrach H., Poustka A., Murray N. Lambda replacement vectors carrying polylinker sequences. J Mol Biol. 1983 Nov 15;170(4):827–842. doi: 10.1016/s0022-2836(83)80190-9. [DOI] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hattori M., Sakaki Y. Dideoxy sequencing method using denatured plasmid templates. Anal Biochem. 1986 Feb 1;152(2):232–238. doi: 10.1016/0003-2697(86)90403-3. [DOI] [PubMed] [Google Scholar]
- Hiromi K., Nitta Y., Numata C., Ono S. Subsite affinities of glucoamylase: examination of the validity of the subsite theory. Biochim Biophys Acta. 1973 Apr 12;302(2):362–375. doi: 10.1016/0005-2744(73)90164-2. [DOI] [PubMed] [Google Scholar]
- Kimura K., Kataoka S., Ishii Y., Takano T., Yamane K. Nucleotide sequence of the beta-cyclodextrin glucanotransferase gene of alkalophilic Bacillus sp. strain 1011 and similarity of its amino acid sequence to those of alpha-amylases. J Bacteriol. 1987 Sep;169(9):4399–4402. doi: 10.1128/jb.169.9.4399-4402.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H., Yamane K., Yamaguchi K., Nagata Y., Maruo B. Hybrid alpha-amylases produced by transformants of Bacillus subtilis. I. Purification and characterization of extracellular alpha-amylases produced by the parental strains and transformants. Biochim Biophys Acta. 1974 Sep 13;365(1):235–247. doi: 10.1016/0005-2795(74)90268-2. [DOI] [PubMed] [Google Scholar]
- Mielenz J. R. Bacillus stearothermophilus contains a plasmid-borne gene for alpha-amylase. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5975–5979. doi: 10.1073/pnas.80.19.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson W. L., Chambliss G. H. Molecular cloning of cis-acting regulatory alleles of the Bacillus subtilis amyR region by using gene conversion transformation. J Bacteriol. 1986 Mar;165(3):663–670. doi: 10.1128/jb.165.3.663-670.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poverenny A. M., Podgorodnichenko V. K., Bryksina L. E., Monastyrskaya G. S., Sverdlov E. D. Immunochemical approaches to DNA structure investigation--I. Immunochemical identification of the product of cytosine modification with bisulphite and O-methylhydroxylamine mixture. Mol Immunol. 1979 May;16(5):313–316. doi: 10.1016/0161-5890(79)90132-9. [DOI] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Takkinen K., Pettersson R. F., Kalkkinen N., Palva I., Söderlund H., Käriäinen L. Amino acid sequence of alpha-amylase from Bacillus amyloliquefaciens deduced from the nucleotide sequence of the cloned gene. J Biol Chem. 1983 Jan 25;258(2):1007–1013. [PubMed] [Google Scholar]
- Thoma J. A., Brothers C., Spradlin J. Subsite mapping of enzymes. Studies on Bacillus subtilis amylase. Biochemistry. 1970 Apr 14;9(8):1768–1775. doi: 10.1021/bi00810a016. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Weickert M. J., Chambliss G. H. Genetic analysis of the promoter region of the Bacillus subtilis alpha-amylase gene. J Bacteriol. 1989 Jul;171(7):3656–3666. doi: 10.1128/jb.171.7.3656-3666.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker N. E., Campbell L. L. Comparison of the alpha-amylase of Bacillus subtilis and Bacillus amyloliquefaciens. J Bacteriol. 1967 Oct;94(4):1131–1135. doi: 10.1128/jb.94.4.1131-1135.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K., Nagata Y., Maruo B. Isolation of mutants defective in alpha-amylase from Bacillus subtilis: genetic analyses. J Bacteriol. 1974 Aug;119(2):416–424. doi: 10.1128/jb.119.2.416-424.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K., Hirata Y., Furusato T., Yamazaki H., Nakayama A. Changes in the properties and molecular weights of Bacillus subtilis M-type and N-type alpha-amylases resulting from a spontaneous deletion. J Biochem. 1984 Dec;96(6):1849–1858. doi: 10.1093/oxfordjournals.jbchem.a135019. [DOI] [PubMed] [Google Scholar]
- Yamane K., Yamaguchi K., Maruo B. Purification and properties of a cross-reacting material related to -amylase and biochemical comparison with the parent -amylase. Biochim Biophys Acta. 1973 Jan 25;295(1):323–340. doi: 10.1016/0005-2795(73)90100-1. [DOI] [PubMed] [Google Scholar]
- Yang M., Galizzi A., Henner D. Nucleotide sequence of the amylase gene from Bacillus subtilis. Nucleic Acids Res. 1983 Jan 25;11(2):237–249. doi: 10.1093/nar/11.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuuki T., Nomura T., Tezuka H., Tsuboi A., Yamagata H., Tsukagoshi N., Udaka S. Complete nucleotide sequence of a gene coding for heat- and pH-stable alpha-amylase of Bacillus licheniformis: comparison of the amino acid sequences of three bacterial liquefying alpha-amylases deduced from the DNA sequences. J Biochem. 1985 Nov;98(5):1147–1156. doi: 10.1093/oxfordjournals.jbchem.a135381. [DOI] [PubMed] [Google Scholar]




