Abstract
The phospholipase D (PLD) superfamily includes enzymes of phospholipid metabolism, nucleases, as well as ORFs of unknown function in viruses and pathogenic bacteria. These enzymes are characterized by the invariant sequence motif, H(X)K(X)4D. The endonuclease member Nuc of the PLD family was over-expressed in bacteria and purified to homogeneity. Mutation of the conserved histidine to an asparagine in the endonuclease reduced the kcat for hydrolysis by a factor of 105, suggesting that the histidine residue plays a key role in catalysis. In addition to catalyzing hydrolysis, a number of phosphohydrolases will catalyze a phosphate (oxygen)-water exchange reaction. We have taken advantage of this observation and demonstrate that a 32P-labeled protein could be trapped when the enzyme was incubated with 32P-labeled inorganic phosphate. The phosphoenzyme intermediate was stable in 1 M NaOH and labile in 1 M HCl and 1 M hydroxylamine, suggesting that the enzyme forms a phosphohistidine intermediate. The pH-stability profile of the phosphoenzyme intermediate was consistent with phosphohistidine and the only radioactive amino acid found after alkaline hydrolysis was phosphohistidine. These results suggest that the enzymes in the PLD superfamily use the conserved histidine for nucleophilic attack on the substrate phosphorus atom and most likely proceed via a common two-step catalytic mechanism.
Phospholipase D (PLD)1 appears to play a pivotal role in a number of signal transduction pathways. PLD is one of a group of phospholipid-modifying enzymes that are activated by receptors that regulate cellular growth (1, 2). PLD hydrolyzes phosphatidylcholine to produce phosphatidic acid and choline. Phosphatidic acid can be further hydrolyzed to lysophosphatidic acid by phospholipase A2 or to diacylglycerol by phosphatidic acid phosphohydrolase. Lysophosphatidic acid is a known mitogen that interacts with G protein-coupled receptors in many cell types (3), and diacylglycerol activates protein kinase C (4). In addition, the stimulation of PLD by small G proteins such as ARF and Rho suggests that some forms of PLD may be involved in intermembrane protein trafficking and cytoskeletal rearrangement (1, 2).
Recently, PLD has been identified as a member of a large superfamily of proteins that includes eukaryotic and prokaryotic enzymes of phospholipid metabolism, such as cardiolipin synthases, phosphatidylserine synthases (PSS), pox viral envelope proteins, and a small family of nucleases found in bacteria (5, 6). Both the PLD and the nucleases recognize phosphodiester substrates and catalyze their cleavage by water, whereas cardiolipin synthases and PSS substitute a specific alcohol for water in the hydrolysis generating a new, substituted phosphodiester. The hallmark of the PLD superfamily is the consensus motif, H(X)K(X)4D (Fig. 1), embedded within a more loosely conserved region that is usually present in two separate copies in PLD, PSS, and cardiolipin synthases but found as a single copy in the nucleases (5, 6). When two copies of the motif are present, a Ser or Thr residue is often conserved eight amino acids C-terminal to the Asp. The nucleases also contain a Ser at this position, but PSS lacks the Ser/Thr. Frohman and coworkers (7) mutagenized a number of residues in human PLD1 and measured PLD activity after transfection of these mutants into COS cells. Mutagenesis of the conserved His in either motif, or the Lys in the second motif, resulted in complete loss of PLD activity. Mutation of Ser-911 to Thr resulted in a 60% loss of activity, whereas mutation of Ser-911 to Ala abolished enzyme activity. The small amounts of enzyme available in the cell lysates generated in this study made it difficult to carry out a more detailed kinetic analysis of these mutants.
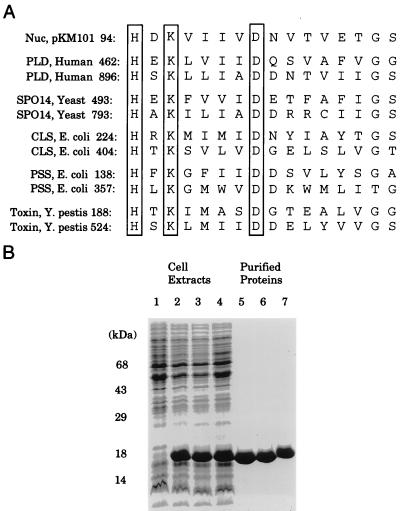
Figure 1.
Consensus motif for the PLD superfamily. (A) The conserved H, K, and D residues are boxed. (B) SDS gel electrophoresis of proteins present in the bacterial extracts (lane 1), proteins present after induction of wild-type Nuc, the K96S and H94N mutants of Nuc (lanes 2–4, respectively), and the corresponding purified proteins (lanes 5–7).
To understand the catalytic mechanism used by the PLD superfamily, we have over-expressed, purified, and crystallized the recombinant endonuclease Nuc (8). In contrast to human and plant PLDs that contain two H(X)K(X)4D domains and are >100 kDa, Nuc contains a single active site domain and is only 17 kDa (155 aa). Here, we describe a detailed kinetic analysis of Nuc and demonstrate that the catalytic mechanism of this enzyme proceeds via a phosphohistidine intermediate. Our results suggest that this mechanistic strategy will likely be used by the entire PLD superfamily.
EXPERIMENTAL PROCEDURES
Materials.
Bis(p-nitrophenyl) phosphate, p-nitrophenyl phenylphosphonate (pNP-PPh), and diethylpyrocarbonate (DEPC), were purchased from Sigma. 32P-labeled inorganic phosphate (9,000 Ci/mmol; 150 mCi/ml) (1 Ci = 37 GBq) was purchased from New England Nuclear.
Site-Directed Mutagenesis, Protein Purification, and Analysis.
H94N, K96S, D101N, D101E, S109A, and H140N mutations were introduced into the Nuc sequence that had been cloned in the plasmid pT7–7 as described (8) by using the Clontech Transformer Site-Directed Mutagenesis Kit. Mutations were confirmed by DNA sequencing. Wild-type and mutant proteins were purified as described (8).
Enzyme Assays and Data Analysis.
The hydrolysis of artificial substrates was carried out in assay buffer (100 mM Tris/100 mM Bis⋅Tris/200 mM acetic acid). This buffer maintains a constant ionic strength between the pH ranges of ≈3.6–9.0 (9). All reactions were carried out at 30°C in a temperature-controlled Perkin–Elmer Lambda 6 spectrophotometer. The appearance of p-nitrophenolate ion was measured continuously at 405 nm. The nuclease assay was performed as described (8). Calf thymus DNA (7.5 μg) was incubated with 0–3 μg of enzyme for 20 min at 37°C in assay buffer with 100 μM EDTA. Samples were resolved on an agarose gel and visualized using a UV light source.
Reaction of Nuc with DEPC.
Reactions were carried out essentially as described by Miles (10). Nuc (initially in an enzyme storage buffer composed of 15 mM Tris⋅Cl, pH 7.5/75 mM NaCl/50% glycerol) was dialyzed 15,000-fold into 50 mM sodium acetate (pH 4.5) containing 5% glycerol to exchange the buffer and avoid DEPC reaction with Tris⋅Cl. Concentrated (25-fold) DEPC stock solutions were prepared in ethanol. The endonuclease (12 μM) was mixed with DEPC on ice in the presence or absence of 1 mM sodium tungstate and then transferred to a 30°C water bath at various times. The endonuclease also was inactivated with DEPC until 50% of the original activity remained and hydroxylamine was added at a final concentration of 0.5 M and reacted for an additional 35 min. The reactions were quenched, and the enzyme activity was assayed by a 10-fold dilution into assay buffer containing 100 mM pNP-PPh at pH 7.0, as described above.
32P Labeling, Phosphoamino Acid Stability, and Phosphoamino Acid Analysis.
Nuc (50 μM) was mixed with 1 mM potassium [32P]phosphate (ranging in specific activity from 1–10 Ci/mmol), in 50 mM sodium acetate (pH 4.5) and 5% glycerol, at room temperature. Samples were removed at intervals, and reactions were quenched by the addition of an equal volume of 1% SDS and 1 M sodium carbonate. Sample buffer was added (12 mM Tris⋅Cl, pH 6.8/5% glycerol/0.4% SDS/2.9 mM β-mercaptoethanol/0.02% Bromphenol Blue), and samples were loaded, unheated, in the wells of 12% SDS-polyacrylamide gels and subjected to electrophoresis. Gels were dried and subjected to autoradiography. For assessment of phosphoamino acid stability, Nuc was labeled with 32P-inorganic phosphate at room temperature for 1 min as described; sample buffer was added, and each sample was split into four gel lanes and subjected to electrophoresis as described above. The gel was soaked in buffer containing 12.5 mM Tris base, 96 mM glycine, and 20% methanol, and the protein was transferred to an Immobilon P (Millipore) membrane by electrophoresis at 100 V for 1 hr in the same buffer. The four lanes containing equal amounts of radioactivity were cut into strips and counted in a Beckman scintillation counter. The strips were wet subsequently with methanol, rinsed with water, and placed into sealed polypropylene tubes containing either 1 M HCl, 1 M NaOH, 1 M hydroxylamine in 200 mM Tris⋅Cl (pH 7.4), or 1 M Tris⋅HCl (pH 7.4) at 37°C for 45 min. The filters were removed, dipped in 1 M Tris (pH 7.0), dried, counted, and subjected to autoradiography. The data are expressed as a percentage of starting counts on the filter. The filter strips were subsequently stained with Coomassie Brilliant Blue (Sigma) and destained in 50% methanol/10% acetic acid to insure that approximately equal amounts of protein remained. For quantitation of the pH stability of Nuc, labeled enzyme prepared as described above was transferred to an Immobilon P (Millipore, Bedford, MA) membrane and counted. The strips were then incubated at 80°C for 30 min in 10 ml of 50 mM sodium citrate, pH 2.5, or in solutions of 25 mM Tris, 25 mM Bis⋅Tris, and 50 mM acetic acid at pH values ranging from 3.5 to 8.8. The filter strips were rinsed in water and dried, and the radioactivity remaining was quantitated. Acrylamide slices containing radiolabeled endonuclease were incubated for 5 hr with 3 M potassium hydroxide. The alkaline hydrolysate was diluted 400-fold with water containing the phosphoamino acids noted in Fig. 5. The sample was subjected to ion-exchange chromatography as described by Wei and Matthews (11). The eluate was incubated with o-pthalaldehyde and the resulting fluorescence was detected on-line (11). Radioactivity was quantitated by liquid scintillation counting. Phospholysine and phosphohistidine standards were synthesized as described (11). All other standards were purchased from Sigma.
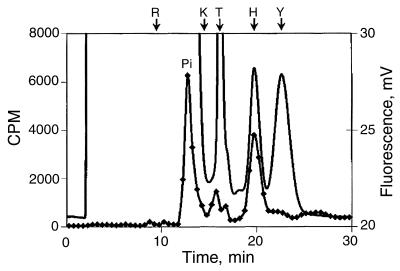
Figure 5.
Phosphoamino acid analysis. Chromatography of 32P-labeled amino acids after alkaline hydrolysis of 32P-labeled endonuclease. The position of inorganic phosphate (Pi), phosphoarginine, phospholysine, phosphothreonine, phosphohistidine, and phosphotyrosine are noted on the chromatogram. Phosphoserine elutes 30 sec after phosphothreonine (11). Fluorescence is plotted on the right axis and is represented by a continuous line. Radioactivity is plotted on the left axis and is represented by the data points.
RESULTS AND DISCUSSION
Although DNA is rapidly hydrolyzed by the recombinant endonuclease (8), it cleaves DNA at multiple sites making quantitation and kinetic analysis difficult. For this reason, we sought a substrate that would cleave a single site and a substrate whose hydrolysis yielded a quantifiable product. Because the phosphodiester linkage is the feature common to all known substrates hydrolyzed by the PLD superfamily of enzymes, we tested two compounds (12), bis(p-nitrophenyl) phosphate and pNP-PPh as possible substrates. Both substrates yield p-nitrophenolate ion when hydrolyzed by the purified endonuclease (8). A Km of 10 mM and kcat of 0.1 sec−1 was observed for the endonuclease and bis(p-nitrophenyl) phosphate, whereas a Km of 30 mM and a kcat of 3.5 were noted for pNP-PPh. Because of the more favorable kcat value, the phosphonate substrate was used for kinetic analysis. To address the function of the conserved amino acids within the H(X)K(X)4D motif, we produced several site-directed mutants and determined their kinetic constants with pNP-PPh. Site-directed mutagenesis was used to change His-94 to Asn and Lys-96 to Ser. The two mutant proteins, H94N and K96S, were purified to homogeneity as shown in Fig. 1. (The Asp at position 101 was also altered, but the D101N and D101E proteins could not be purified because Escherichia coli cells producing these proteins underwent lysis.) Kinetic analysis by using the artificial substrate pNP-PPh revealed that both mutants were markedly impaired in activity, with kcat values that were reduced by a factor of 105 for H94N and 104 for K96S when compared with the recombinant endonuclease. In contrast, the Km values that were difficult to measure accurately because of the extremely low enzyme activity did not appear to be markedly altered. The functional impact of these mutations also was evaluated by using the native substrate, DNA. Wild-type Nuc effectively hydrolyzes nucleic acids (8); however, substitution of the conserved histidine or lysine reduces the turnover rate to an undetectable level (data not shown). These results underscore the importance of both the His and Lys residue in the H(X)K(X)4D motif and suggest that they play key roles in catalysis; however, the results obtained with the site-directed mutants do not allow us to assign specific functions to these two residues.
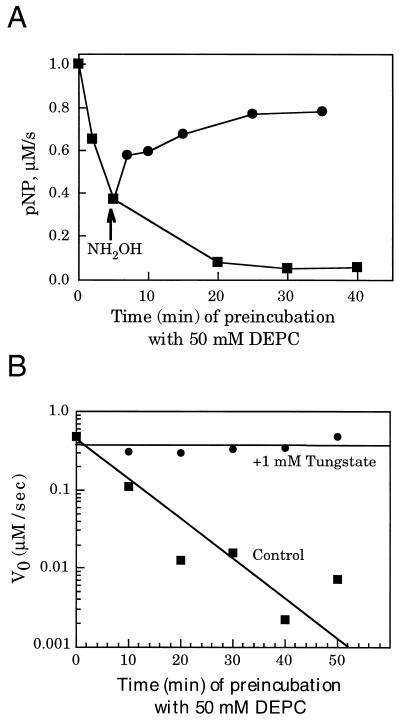
We also attempted to selectively modify the His residue in the H(X)K(X)4D sequence by using DEPC. DEPC has been used as a selective reagent for both histidine and lysine residues in proteins; however, if the reaction is carried out at an acid pH, it shows a preference for inactivation of hisidine (10). When recombinant Nuc was treated with DEPC at pH 4.5, a loss in endonuclease activity was observed with increasing DEPC concentrations (Fig. 2A). The inactivation was time dependent and could be prevented by the addition of 1 mM tungstate, a competitive inhibitor of the endonuclease (Fig. 2B). The KI for tungstate is ≈1 mM (data not shown). When DEPC modifies histidine, this reaction can be reversed with hydroxylamine (10). This result is in contrast to the lysine modification, which is irreversible (10). When the endonuclease was inactivated 50% with DEPC and then treated with hydroxylamine, we could demonstrate that the endonuclease regained 80% of its activity, suggesting that the majority of the DEPC modification was occurring at histidine (Fig. 2A).
Figure 2.
DEPC inactivation of Nuc. (A) Inactivation of Nuc by increasing concentrations of DEPC. Nuc (12 μM) was incubated for 5 min at pH 4.5 with 50 mM DEPC. Hydroxylamine was added to one-half of the reaction at a final concentration of 0.5 M. At the indicated times, the reaction mixtures were diluted 10-fold into assay buffer containing 100 mM pNP-PPh at pH 7.0. Activity was measured for 1 min as described in Experimental Procedures. (B) Time course of inactivation, in presence and absence of tungstate. Nuc (12 μM) was mixed with 50 mM DEPC in the presence or absence (“Control”) of 1 mM tungstate at pH 4.5. Aliquots were removed at the indicated times and diluted 10-fold into assay buffer containing 100 mM pNP-PPh, at pH 7.0, and activity was measured as described.
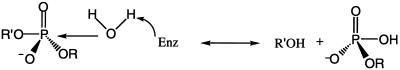
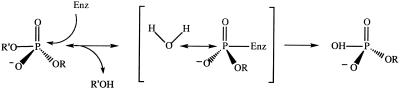
To assign specific functions to the amino acids in the H(X)K(X)4D motif, we took advantage of the fact that hydrolysis of a phosphodiester substrate can proceed by two possible mechanisms: a displacement of the leaving (-OR′) group which does not involve a phospho-enzyme intermediate (Scheme S1),
Scheme 1.
or by a two step mechanism which proceeds via the formation of a covalent enzyme intermediate (Scheme S2).
Scheme 2.
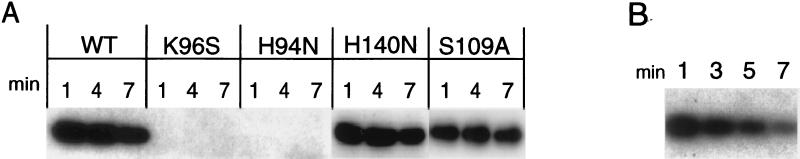
A number of phosphohydrolases also have been shown to catalyze phosphate (oxygen)-water exchange (13). We asked whether the endonuclease would catalyze phosphate (oxygen)-water exchange by using 32P-inorganic phosphate. If Scheme S1 is used by the endonuclease, 32P-labeled phosphate would not form a covalent intermediate with the enzyme. In contrast, if the endonuclease catalyzes a phosphate (oxygen)-water exchange via Scheme S2, one might expect to trap 32P-labeled phosphate covalently bound to the enzyme. When the recombinant endonuclease was mixed with 32P-labeled phosphate at pH 4.5 and the reaction quickly adjusted to pH 11, a 32P-labeled protein was visible after SDS gel electrophoresis (Fig. 3). The reaction of Nuc with 32P-labeled phosphate proceeded rapidly (i.e., within 1 min), and the label could be chased out of the enzyme with unlabeled inorganic phosphate (Fig. 3B), suggesting that the phospho-enzyme intermediate is on the reaction pathway. Neither the H94N nor the K96S mutant was capable of forming the intermediate (Fig. 3A), confirming their importance in the catalytic mechanism. In contrast, substitution of the only remaining histidine H140N or the conserved serine S109A had limited impact on intermediate formation. In three separate experiments, the amount of 32P-label incorporated in the endonuclease was 1.8, 9.9, and 10.8%. Because the phosphate (oxygen)-water exchange is a reversible reaction, the stoichiometry of incorporated 32P-label would not be expected to be 1. The variability noted in the 32P covalently bound to the endonuclease in the three trapping experiments most likely reflects differences in our ability to rapidly quench the reaction in the three experiments.
Figure 3.
Formation and analysis of a covalent phosphoenzyme intermediate. (A) Wild-type, K96S, H94N, H140N, or S109A Nuc (50 μM) was mixed with 1 mM [32P]phosphate at pH 4.5 and incubated for the indicated times at room temperature, where upon the reactions were quenched and subjected to SDS/PAGE and autoradiography. (B) Nuc was phosphorylated for 1 min with [32P]phosphate, followed by the addition of 100 mM unlabeled potassium phosphate. The reaction was quenched at the indicated time points, and samples were subjected to SDS/PAGE and autoradiography.
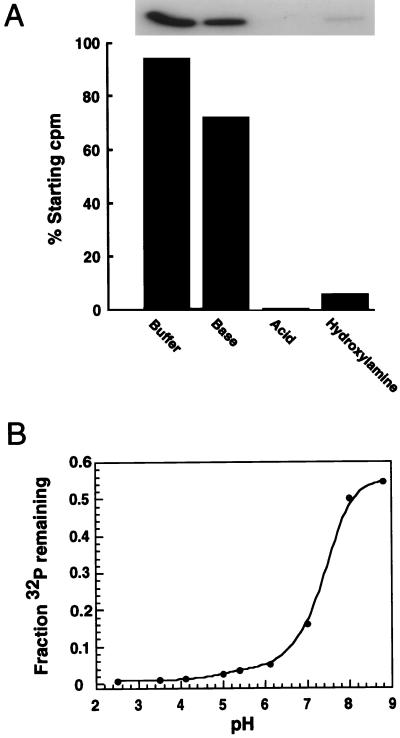
To form the covalent phosphoenzyme intermediate, a nucleophilic amino acid on the enzyme must attack the phosphorus atom on the substrate. The same nucleophile would be expected to catalyze the phosphate (oxygen)-water exchange reaction carried out with 32P-labeled phosphate. Possible candidate nucleophiles in Nuc are His-94, Lys-96, Asp-101, and Ser-109. To determine the nature of the covalently modified 32P-labeled endonuclease, the labeled enzyme was transferred to an Immobilon filter that was treated with 1 M HCl, 1 M NaOH, 1 M hydroxylamine, or 1 M Tris⋅Cl, pH 7.0. The 32P-labeled enzyme was quite stable in base and at neutrality but labile under acidic conditions and in the presence of hydroxylamine (Fig. 4A). In addition, the stability of the phosphoryl group increased progressively under more basic conditions. The pH-stability profile of the 32P-labeled endonuclease is similar to the pH-stability of phosphohistidine (14) (Fig. 4B). This pH-lability profile is not characteristic of phosphoproteins labeled on Ser, Thr, Tyr, Cys, Asp, or Lys (15). Collectively, the physical characteristics of the 32P-labeled phosphoprotein are consistent with the properties displayed by phosphohistidine (14, 15). To identify the labeled amino acid directly, we used HPLC to analyze the products after alkaline hydrolysis of 32P-labeled endonuclease (Fig. 5). The 32P-labeled product from the endonuclease comigrates with an authentic sample of phosphohistidine. This analysis unambiguously identified the labeled amino acid in Nuc as histidine with the other radiolabel being inorganic phosphate. The small amounts of radiolabeled material eluting between 15 and 18 min are likely caused by unhydrolyzed radiolabeled peptides. The elution times of these phosphopeptides do not correspond to the authentic samples of phosphoserine, phosphothreonine, or phospholysine. It is noteworthy that phospholysine was not present in the amino acid analysis of the 32P-labeled endonuclease (Fig. 3A). These observations would suggest that K96 does not participate as the nucleophile in catalysis. More likely K96 plays a key role in binding and orientation of the substrate and/or other key residues within the active site.
Figure 4.
(A) Chemical properties of the phosphoenzyme intermediate. 32P-labeled, phosphorylated Nuc was transferred to Immobilon P. Filter strips were counted, then incubated in: (i) 1 M HCl; (ii) 1 M NaOH; (iii) 1 M hydroxylamine in 200 mM Tris⋅Cl, pH 7.4; and (iv)1 M Tris⋅Cl, pH 7.4, for 45 min at 37°C. Radioactivity remaining on each filter strip was visualized by autoradiography and quantitated by counting. (B) Filter strips were incubated in 50 mM sodium citrate, pH 2.5, or in solutions of 25 mM Tris, 25 mM Bis·Tris, and 50 mM acetic acid at pH values ranging from 3.5 to 8.8, for 30 min at 80°C. The amount of radioactivity on each filter strip before and after incubation was quantitated by scintillation spectrometry.
Members of the PLD superfamily are likely to use a common catalytic mechanism to hydrolyze a phosphodiester substrate in light of the fact that the superfamily contains the conserved motif H(X)K(X)4D. Two of the enzymes in the superfamily, PLD from cabbage (16) and PSS from E. coli (17), have been shown to cleave their phospholipid substrates with retention of configuration at phosphorus, suggesting a phosphatidyl enzyme intermediate is formed in both cases. This result is consistent with our finding that the endonuclease also forms a phosphoenzyme intermediate and further substantiates the idea that these enzymes use a common mechanism.
Sung et al. (7) have suggested that human PLD also uses a phosphoenzyme intermediate. They have proposed that a conserved Ser near the active site may be responsible for formation of a phosphoenzyme intermediate. These conclusions were based on mutagenesis studies of residues within the active site, and their results did not use trapping a phosphoenzyme intermediate. Mutagenesis of this serine to threonine reduced PLD activity by 60%, and substitution to alanine resulted in a mutant with 1% activity compared with wild-type PLD (7). The conserved Ser in human PLD corresponds to S109 in the endonuclease. Substitution of this residue to alanine only modestly reduced the activity of Nuc. For example, the rate of pNP-PPh hydrolysis is 30% that of wild-type Nuc (data not shown), and this mutant labeled almost as well as the wild-type enzyme on incubation with labeled phosphate (Fig. 3A). The fact that turnover rates are modestly altered and the S109A mutant still effectively formed the phosphoenzyme intermediate suggests that the conserved serine does not function as the nucleophile in the catalytic mechanism. In addition, the properties of the intermediate are inconsistent with those of a phosphoserine. Our results show that the intermediate can be trapped only when His-94 is present in the enzyme. When His-94 is mutated, no 32P-intermediate is observed, whereas mutation of the only other histidine in the protein (H140N) had no effect on intermediate formation. In addition, phosphoamino acid analysis suggests that the majority of the radiolabel is present on histidine and not on serine (Fig. 5). These data strongly suggests that the catalytic histidine embedded in the H(X)K(X)4D active-site motif is playing a role as an essential nucleophile in the catalytic mechanism.
The role of the conserved residues as defined by biochemical analysis is corroborated by structural studies of Nuc (J. A. Stuckey and J.E.D., unpublished data). The conserved aspartate Asp-101, for example, was shown to be critical for the structural integrity of Nuc. The backbone atoms of several residues in a central loop of the structure form hydrogen bonds with Asp-101. This loop is composed of residues that form one of the four structural motifs that were defined for the superfamily by Ponting and Kerr (6). These motifs form the core of the Nuc structure, which is likely to be preserved among the superfamily enzymes; therefore, it is not surprising that mutations in Asp-101 render the enzyme insoluble. A structure of Nuc complexed with tungstate at the active site also has been determined. This structure shows that Lys-96 forms hydrogen bonds with the nucleophilic histidine and to oxygen atoms of tungstate. The contributions of this residue to the hydrogen bond network of the Nuc-catalytic pocket and the role of Lys-96 in substrate binding are critical for Nuc function. It is probable that the structural and functional significance of these highly conserved residues, as demonstrated for Nuc, is a unifying characteristic of all superfamily catalysts.
In summary, our results suggest that the endonuclease forms a phosphoenzyme intermediate. The intermediate is formed via the histidine residue in the conserved H(X)K(X)4D motif found in all members of the PLD family. Our results suggest that it is highly likely that this mechanism will be used by the entire family of PLDs. It is particularly striking that members of this superfamily include enzymes involved in lipid synthesis and lipid breakdown, nucleases, and proteins with no known functions. It is also noteworthy to mention that homology between these diverse proteins is limited to such a small motif and similar motifs may be overlooked by commonly used sequence analysis techniques.
Although the substrates for this diverse family of enzymes vary widely, the common theme uniting this group of proteins is their catalytic mechanism. As more and more deduced protein sequences appear from genomic sequencing efforts, it is likely that common catalytic strategies will be used by increasingly large superfamilies of proteins and these catalytic mechanisms may provide new insights into biological functions.
Acknowledgments
We thank Jeanne Stuckey for providing information about the crystal structure of the endonuclease and Jin Zhou for technical assistance. We also thank Karina Chan for assistance with the phosphoamino acid analysis. This work was supported by National Institutes of Health Grant National Institute of Diabetes and Digestive and Kidney Diseases 18849 to J.E.D and by the Walther Cancer Institute, Indianapolis, IN. E.B.G. was supported by a Walther Cancer Institute postdoctoral fellowship. Y.Z. was supported by a National Institutes of Health (National Institute of Diabetes and Digestive and Kidney Diseases) National Research Service Award postdoctoral fellowship.
ABBREVIATIONS
- PLD
phospholipase D
- PSS
phosphatidylserine synthase
- pNP-PPh
p-nitrophenyl phenylphosphonate
- DEPC
diethylpyrocarbonate
References
- 1.Exton J H. Physiol Rev. 1997;77:303–320. doi: 10.1152/physrev.1997.77.2.303. [DOI] [PubMed] [Google Scholar]
- 2.Singer W D, Brown H A, Sternweis P C. Annu Rev Biochem. 1997;66:475–509. doi: 10.1146/annurev.biochem.66.1.475. [DOI] [PubMed] [Google Scholar]
- 3.Moolenaar W H. J Biol Chem. 1995;270:12949–12952. doi: 10.1074/jbc.270.22.12949. [DOI] [PubMed] [Google Scholar]
- 4.Berridge M J. Nature (London) 1993;361:315–326. doi: 10.1038/361315a0. [DOI] [PubMed] [Google Scholar]
- 5.Koonin E V. Trends Biochem Sci. 1996;21:242–243. [PubMed] [Google Scholar]
- 6.Ponting C P, Kerr I D. Protein Sci. 1996;5:914–922. doi: 10.1002/pro.5560050513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sung T-C, Roper R L, Zhang Y, Rudge S A, Temel R, Hammond S M, Morris A J, Moss B, Engebrecht J, Frohman M A. EMBO J. 1997;16:4519–4530. doi: 10.1093/emboj/16.15.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao Y, Stuckey J A, Lohse D L, Dixon J E. Protein Sci. 1997;6:1–4. doi: 10.1002/pro.5560061221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis K J, Morrison J F. Methods Enzymol. 1982;87:405–427. doi: 10.1016/s0076-6879(82)87025-0. [DOI] [PubMed] [Google Scholar]
- 10.Miles E W. Methods Enzymol. 1977;47:431–432. doi: 10.1016/0076-6879(77)47043-5. [DOI] [PubMed] [Google Scholar]
- 11.Wei Y, Matthews H. Methods Enzymol. 1991;200:388–414. doi: 10.1016/0076-6879(91)00156-q. [DOI] [PubMed] [Google Scholar]
- 12.Landt M, Everard R A, Butler L G. Biochemistry. 1980;19:138–143. doi: 10.1021/bi00542a021. [DOI] [PubMed] [Google Scholar]
- 13.Van Etten R L, Risley J M. Proc Natl Acad Sci USA. 1978;75:4784–4787. doi: 10.1073/pnas.75.10.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hultquist D E, Moyer R W, Boyer P D. Biochemistry. 1966;5:322–331. doi: 10.1021/bi00865a041. [DOI] [PubMed] [Google Scholar]
- 15.Duclos B, Marcandier S, Cozzone A J. Methods Enzymol. 1991;201:10–28. doi: 10.1016/0076-6879(91)01004-l. [DOI] [PubMed] [Google Scholar]
- 16.Bruzik K, Tsai M-D. Biochemistry. 1984;23:1656–1661. doi: 10.1021/bi00303a012. [DOI] [PubMed] [Google Scholar]
- 17.Raetz C R H, Carman G M, Dowhan W, Jiang R-T, Waszkuc W, Loffredo W, Tsai M-D. Biochemistry. 1987;26:4022–4027. doi: 10.1021/bi00387a042. [DOI] [PubMed] [Google Scholar]