Abstract
Glucose-6-phosphatase catalyzes the terminal step in the gluconeogenic and glycogenolytic pathways. Transcription of the gene encoding the glucose-6-phosphatase catalytic subunit (G6Pase) is stimulated by cAMP and glucocorticoids whereas insulin strongly inhibits both this induction and basal G6Pase gene transcription. Previously, we have demonstrated that the maximum repression of basal G6Pase gene transcription by insulin requires two distinct promoter regions, designated A (from −271 to −199) and B (from −198 to −159). Region B contains an insulin response sequence because it can confer an inhibitory effect of insulin on the expression of a heterologous fusion gene. By contrast, region A fails to mediate an insulin response in a heterologous context, and the mutation of region B within an otherwise intact promoter almost completely abolishes the effect of insulin on basal G6Pase gene transcription. Therefore, region A is acting as an accessory element to enhance the effect of insulin, mediated through region B, on G6Pase gene transcription. Such an arrangement is a common feature of cAMP and glucocorticoid-regulated genes but has not been previously described for insulin. A combination of fusion gene and protein-binding analyses revealed that the accessory factor binding region A is hepatocyte nuclear factor-1. Thus, despite the usually antagonistic effects of cAMP/glucocorticoids and insulin, all three agents are able to use the same factor to enhance their action on gene transcription. The potential role of G6Pase overexpression in the pathophysiology of MODY3 and 5, rare forms of diabetes caused by hepatocyte nuclear factor-1 mutations, is discussed.
Type II, non-insulin-dependent diabetes mellitus (NIDDM) is characterized by defects in insulin secretion, insulin-dependent peripheral glucose utilization, and hepatic glucose production (HGP) (1). The ability of insulin to stimulate peripheral glucose utilization and repress HGP in patients with NIDDM is reduced, a phenomenon known as insulin resistance (1). The elevated HGP is a consequence of an increased rate of gluconeogenesis, rather than glycogenolysis (2, 3). Various investigators have speculated that an alteration in the insulin-regulated expression of the genes encoding key gluconeogenic/glycolytic enzymes, as a consequence of insulin resistance, may contribute to the increased HGP (1, 4, 5).
The gene encoding the glucose-6-phosphatase catalytic subunit (G6Pase) (see refs. 6 and 7 for review) is one such candidate because it catalyses the terminal step in the gluconeogenic pathway, the hydrolysis of glucose-6-phosphate, allowing for the release of glucose into the bloodstream. In animal models of diabetes, G6Pase activity (8–10) and mRNA levels (9–12) are increased, and this increase contributes to the elevated HGP (13). Multiple hormones and signaling molecules regulate G6Pase gene expression. Thus, glucocorticoids, cAMP, glucose, and fatty acids all increase G6Pase mRNA levels (10, 12, 14–16) whereas insulin, tumor necrosis factor-α, and interleukin-6 all repress basal G6Pase gene expression (10, 14, 17, 18). Insulin treatment also overrides the stimulatory effects of cAMP and glucocorticoids on G6Pase gene transcription (19, 20).
We have recently identified two regions of the mouse G6Pase promoter designated A (from −271 to −199) and B (from −198 to −159), which together are required for the maximal repression of basal G6Pase gene transcription by insulin (20). Region B contains an insulin response sequence (IRS) because it can confer an inhibitory effect of insulin on the expression of a heterologous fusion gene (20). Within region B, there are three copies of the consensus inhibitory insulin response sequence, T(G/A)TTT(T/G)(G/T), found in several insulin regulated genes (4, 20). Thus, the IRSs identified in the phosphoenolpyruvate carboxykinase and tyrosine aminotransferase promoters both contain this same motif whereas the IRS in the insulin-like growth factor-binding protein-1 (IGFBP-1) promoter has two copies of this motif arranged as an inverted palindrome (21–25). Whether region A in the G6Pase promoter contains an independent IRS, or acts as an accessory element that enhances the action of the IRS located in region B, was not determined (20). The experiments described in this paper were designed to address that question and to identify the accessory factor or insulin-responsive transcription factor binding region A.
MATERIALS AND METHODS
Plasmid Construction.
The full length mouse G6Pase promoter, spanning nucleotides −751 to +66 relative to the transcription start site (20), was ligated into the pCAT(An) expression vector, a gift from Howard Towle (26). A series of truncated G6Pase-chloramphenicol acetyltransferase (CAT) fusion genes was constructed by using either PCR or restriction enzyme digestion (27). All of the promoter fragments generated by PCR were completely sequenced, using the United States Biochemical Sequenase kit, to verify the absence of polymerase errors. Plasmid constructs were purified by centrifugation twice through cesium chloride gradients (27).
The hepatocyte nuclear factor-1 (HNF-1)-binding site in the G6Pase promoter was mutated by site-directed mutagenesis within the context of the −231 to +66-promoter fragment by using PCR and the following oligonucleotide as the 5′ primer: 5′-TTCCTCGAG(-231)GTGTGCCCAAGTTAATACGCGGTTCTGCCAATGGCGATC-3′. An XhoI site used for cloning purposes and the mutated HNF-1-binding site (compare wild-type sequence shown in Fig. 2A) are underlined. The 3′ PCR primer (5′-CCGCTCGAGATCCAGATCCTC-3′), with XhoI-cloning site underlined, was designed to conserve the junction between the G6Pase promoter and CAT reporter gene to be the same as that in all other G6Pase-CAT fusion gene constructs.
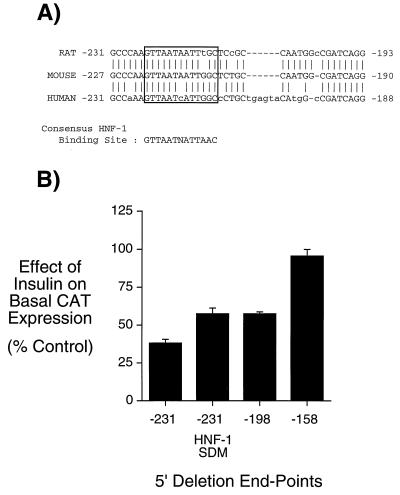
Figure 2.
Mutation of an HNF-1-binding site in region A of the G6Pase promoter reduces the inhibitory effect of insulin on basal G6Pase-CAT fusion gene expression. (A) Comparison of the mouse G6Pase-promoter sequence between −227 to −190 with the equivalent sequence in the rat and human G6Pase genes. The potential HNF-1-binding site is boxed. (B) HepG2 cells were transiently cotransfected, as described (20, 30), with various G6Pase-CAT fusion genes (15 μg) and an expression vector (5 μg) encoding the insulin receptor. The G6Pase-CAT fusion genes contained either distinct lengths of wild-type-promoter sequence, as indicated by the 5′ deletion endpoint or a site-directed mutation of the putative region A HNF-1-binding site, designated −231 HNF-1 SDM. After transfection, cells were incubated for 18–20 hr in serum-free medium in the presence or absence of 100 nM insulin. The cells were then harvested, and CAT activity and protein concentration was assayed as described in Materials and Methods. Results are presented as the ratio of CAT activities in insulin-treated vs. control cells (expressed as percent control) and represent the mean ± SEM of three experiments in which each construct was assayed in duplicate.
A previously described three-step PCR strategy (28) was used to create a site-directed mutant (SDM) of the three region B IRS motifs. The resulting construct, designated −751 RB SDM (Fig. 5), was generated within the context of the −751 to +66 G6Pase-promoter fragment. Briefly, two complementary PCR primers were designed to mutate single nucleotides within each of the three region B IRS motifs (20). The sequence of the sense strand oligonucleotide was as follows (mutated nucleotides are underlined): 5′-GGCGATCAGGCTCTTTTTGTGTGCCTCTTTTGCTCTTTTACGTAAATCAC-3′. This sense strand oligonucleotide was used in conjunction with the same 3′ PCR primer described above to generate the 3′ half of the G6Pase promoter whereas the complementary antisense strand oligonucleotide was used in conjunction with a 5′ PCR primer to generate the 5′ half of the G6Pase promoter. This 5′ primer was designed to maintain the 5′ junction of the G6Pase-promoter fragment to be the same as that in the wild-type −751 to +66 G6Pase-CAT fusion gene construct. The PCR products from these two reactions were then combined and used themselves as both primer and template in a second PCR reaction to generate a small amount of the full-length, mutated G6Pase-promoter fragment. Finally, the 5′ and 3′ PCR primers were then used to amplify this fragment.
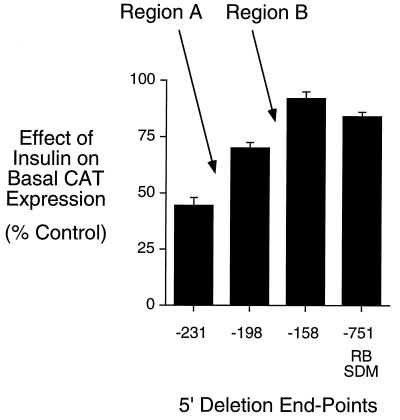
Figure 5.
Mutation of the region B IRS in the context of the full length G6Pase promoter substantially reduces the inhibitory effect of insulin on basal G6Pase-CAT fusion gene expression. HepG2 cells were transiently cotransfected, as described (20, 30), with various G6Pase-CAT fusion genes (15 μg) and an expression vector (5 μg) encoding the insulin receptor. The G6Pase-CAT fusion genes contained either distinct lengths of wild-type-promoter sequence, as indicated by the 5′ deletion endpoint or a site-directed mutation of the region B IRS, designated −751 RB SDM. After transfection, cells were incubated for 18–20 hr in serum-free medium in the presence or absence of 10 nM insulin. The cells were then harvested and CAT activity and protein concentration assayed as described in Materials and Methods. Results are presented as the ratio of CAT activities in insulin-treated vs. control cells (expressed as percent control) and represent the mean ± SEM of four to five experiments in which each construct was assayed in duplicate.
The plasmid TKC-VI contains the herpes simplex virus thymidine kinase (TK) promoter ligated to the CAT reporter gene (21, 29). The TK-promoter sequence extends from −480 to +51 and contains a BamHI linker between positions −40 and −35 (21, 29). Complementary oligonucleotides representing the G6Pase-promoter sequence between −227 and −190 (Fig. 2A) were synthesized with BamHI compatible ends and were cloned in either orientation into BamHI-cleaved TKC-VI by standard techniques (27).
Cell Culture and Transient Transfection.
(i) Human HepG2 hepatoma cells were grown in DMEM containing 2.5% (vol/vol) newborn calf serum, 2.5% (vol/vol) fetal calf serum, and 5% (vol/vol) Nu serum IV (Collaborative Research) and transiently transfected in suspension by using the calcium phosphate-DNA coprecipitation method as described (30). An expression vector encoding the insulin receptor (5 μg), courtesy of Jonathan Whittaker (Hagedorn Research Institute, Gentofte, Denmark) was cotransfected with the reporter gene construct (15 μg). (ii) Rat H4IIE hepatoma cells were grown in DMEM containing 2.5% (vol/vol) fetal calf serum and 2.5% (vol/vol) newborn calf serum and transiently transfected in suspension by using the calcium phosphate-DNA coprecipitation method as described (22).
CAT Assay.
CAT assays were performed exactly as described (30). Because β-galactosidase is very poorly expressed in H4IIE cells (22, 30) and because insulin stimulates Rous sarcoma virus-β galactosidase expression ≈2-fold (data not shown) in HepG2 cells, CAT activity was corrected for the protein concentration in the cell lysate, as measured by the Pierce BCA assay. Each construct was analyzed in duplicate in multiple transfections, as specified in the figure legends, using several independent plasmid preparations.
Gel Retardation Assay.
HepG2 or H4IIE nuclear extracts were prepared as described (22, 31) except that the nuclear pellet was extracted with 20 mM Hepes (pH 7.8), 0.75 mM spermidine, 0.15 mM spermine, 0.2 mM EDTA, 2 mM EGTA, 2 mM DTT, and 25% glycerol containing 200 mM NaCl, instead of 0.4 M ammonium sulfate, and the supernatant was used directly in gel retardation assays. The protein concentration of the nuclear extract was determined by the Bio-Rad assay and was typically ≈1 μg μl−1.
Complementary oligonucleotides representing the G6Pase-promoter sequence between −231 and −199 (Fig. 3A) were synthesized with HindIII compatible ends, gel purified, annealed, and then labeled with [α-32P]dATP by using the Klenow fragment of Escherichia coli DNA polymerase I to a specific activity of ≈2.5 μCi/pmol (1 Ci = 37 GBq). Labeled oligonucleotide (7.5 fmol, ≈30,000 cpm) was incubated with HepG2 or H4IIE nuclear extract (5 μg) in a final reaction volume of 20 μl containing 20 mM Hepes (pH 7.8), 100 mM NaCl, 50 mM MgCl2, 0.38 mM spermidine, 0.08 mM spermine, 0.1 mM EDTA, 1 mM EGTA, 2 mM DTT, 12.5% glycerol (vol/vol), and 1 μg of poly(dI-dC)⋅poly(dI-dC). For competition experiments, a 25-fold molar excess of various unlabeled double-stranded oligonucleotides, as shown in Fig. 3A, were incubated with the labeled oligomer before the addition of nuclear extract. After incubation for 10 min at room temperature, the reactants were loaded onto a 6% polyacrylamide gel and electrophoresed at room temperature for 120 min at 150 V in 0.5× TBE (1× TBE = 90 mM Tris base/9 mM boric acid/2 mM EDTA) (27). After electrophoresis, the gels were dried, exposed to Kodak XAR5 film, and binding was analyzed by autoradiography.
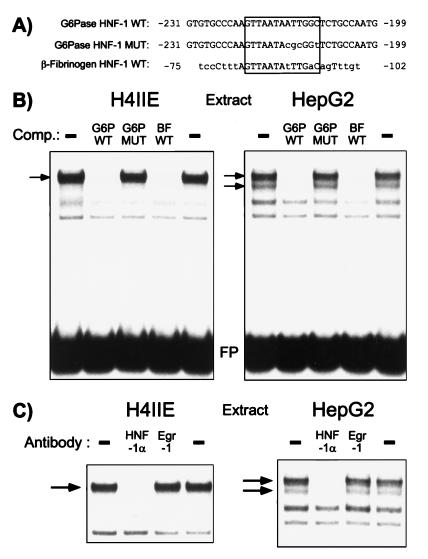
Figure 3.
HNF-1 binds to region A in the G6Pase promoter. (A) Oligonucleotides used in the protein-DNA binding studies. The lower case letters indicate differences between the G6Pase wild-type (WT) and the G6Pase mutant (MUT) and β-fibrinogen wild-type (WT) sequences. The HNF-1-binding sites in the G6Pase and β-fibrinogen promoters are boxed. (B) The labeled wild-type G6Pase region A HNF-1-binding site, designated G6P WT, was incubated in the absence (−) or presence of a 25-fold molar excess of the unlabeled competitor DNAs shown, representing the wild-type (G6P WT) or mutated (G6P MUT) G6Pase region A HNF-1-binding site and the wild-type β-fibrinogen HNF-1-binding site (BF WT). Nuclear extract from either H4IIE or HepG2 cells was then added and protein binding analyzed by using the gel retardation assay as described in Materials and Methods. The specific protein-DNA complexes are indicated by the arrows. The autoradiograph shown is representative of three experiments. FP, free probe; Comp., competitor. (C) Nuclear extract from H4IIE or HepG2 cells was incubated with the indicated antisera for 10 min on ice, before the addition of the labeled G6P WT oligonucleotide probe and binding buffer and an incubation for an additional 10 min at room temperature. Protein binding was then analyzed by using the gel retardation assay as described in Materials and Methods. In the representative autoradiograph shown only the retarded complexes are visible and not the free probe, which was present in excess. The specific protein–DNA interactions are indicated by the arrows.
Gel supershift assays were carried out by incubating HepG2 or H4IIE nuclear extract with antisera (1 μl) for 10 min on ice, before the addition of the labeled oligonucleotide probe and binding buffer and an incubation for an additional 10 min at room temperature. Then binding was analyzed by acrylamide gel electrophoresis as described above. The HNF-1α-specific antiserum, designated TC284 (32, 33), was a generous gift from Tanguy Chouard and Moshe Yaniv (Institute Pasteur, France), whereas the Egr-1-specific antiserum was purchased from Santa Cruz Biotechnology.
RESULTS
Deletion of the G6Pase-Promoter Sequence Between −231 and −159 Abolishes the Inhibitory Effect of Insulin on Basal G6Pase-CAT Fusion Gene Expression.
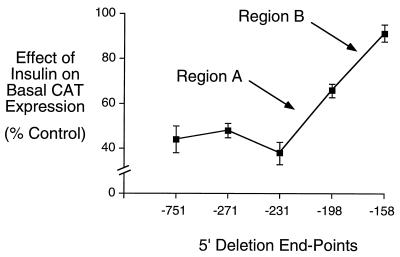
It has previously been demonstrated that the maximum repression of basal mouse G6Pase gene transcription by insulin requires two distinct promoter regions, designated A (from −271 to −199) and B (from −198 to −159) (20). Region B contains an IRS because it can confer an inhibitory effect of insulin on the expression of a heterologous fusion gene. Whether region A also contains an independent IRS, that directly mediates an effect of insulin, or only acts as an accessory element that enhances the action of the IRS located in region B, was not determined (20). To begin to address this question, we initially sought to more precisely localize the cis-acting element within region A that mediates, directly or indirectly, the action of insulin. A 5′-truncated G6Pase-CAT fusion gene was therefore generated with an end-point of −231 that bisects region A. The ability of insulin to repress the basal expression of this and other 5′-truncated G6Pase-CAT fusion genes was analyzed by transient transfection of HepG2 cells. Fig. 1 shows that deletion of the G6Pase-promoter sequence between −271 and −232 had no effect on the ability of insulin to repress basal G6Pase-CAT gene expression, whereas deletion of the G6Pase-promoter sequence between −231 and −199 partially reduced this response. As previously reported, deletion of additional sequence between −198 and −159 (region B) completely abolishes the remaining effect of insulin on basal G6Pase-CAT fusion gene expression (Fig. 1; ref. 20). These results redefine the location of region A as the sequence between −231 and −199 in the G6Pase promoter. It is important to note that deletion of regions A and B has no effect on basal G6Pase-CAT expression in the absence of insulin (20).
Figure 1.
Deletion of the G6Pase-promoter sequence between −231 and −159 abolishes the inhibitory effect of insulin on basal G6Pase-CAT fusion gene expression. HepG2 cells were transiently cotransfected, as described (20, 30), with various G6Pase-CAT fusion genes (15 μg) containing distinct lengths of promoter sequence, as indicated by the 5′ deletion endpoint, and an expression vector (5 μg) encoding the insulin receptor. After transfection, cells were incubated for 18–20 hr in serum-free medium in the presence or absence of 100 nM insulin. The cells were then harvested and CAT activity and protein concentration assayed as described in Materials and Methods. Results are presented as the ratio of CAT activities in insulin-treated vs. control cells (expressed as percent control) and represent the mean ± SEM of 9–12 experiments in which each construct was assayed in duplicate. Similar basal G6Pase-CAT expression is obtained with the constructs with 5′ endpoints of −271, −198 and −158 (20) and −231 (data not shown) in the absence of insulin.
Mutation of an HNF-1-Binding Site in Region A of the G6Pase Promoter Reduces the Inhibitory Effect of Insulin on Basal G6Pase-CAT Fusion Gene Expression.
Examination of the residues between −231 and −199 for known transcription factor-binding sites revealed a strong hepatocyte nuclear factor-1 (HNF-1) consensus-binding sequence (34, 35) located between −221 and −209. This binding site is conserved in the equivalent location within both the human and rat G6Pase promoters (Fig. 2A; refs. 10, 19) and Chou and coworkers (36) have recently shown that HNF-1 binds this sequence in the human G6Pase promoter. To assess the role of this HNF-1-binding site in mediating the effect of insulin through region A, this element was mutated by site-directed mutagenesis within the context of the −231 to +66 G6Pase-promoter fragment; the shortest sequence that confers a maximal repression of basal G6Pase-CAT fusion gene transcription by insulin (Fig. 1). The construct generated, designated −231 HNF-1 SDM, was analyzed by transient transfection in HepG2 cells (Fig. 2B). Mutation of the HNF-1-binding site in region A reduced the insulin-mediated repression of basal G6Pase-CAT fusion gene expression compared with the wild-type −231 G6Pase-CAT construct (Fig. 2B). Importantly, this inhibitory effect of insulin on basal CAT expression directed by the −231 HNF-1 SDM construct was equivalent to that seen when region A was completely deleted, as is the case with the −198 5′ deletion end-point construct (Fig. 2B). This result suggests that HNF-1 could be the factor that is mediating the action of insulin through region A.
HNF-1 Binds to Region A in the G6Pase Promoter.
To provide evidence that HNF-1 is indeed the factor that is mediating the action of insulin through region A, protein binding to this site was analyzed by using the gel retardation assay. When a labeled double-stranded oligonucleotide, designated G6P WT, representing the wild-type G6Pase region A promoter sequence from −231 to −199 (Fig. 3A) was incubated with nuclear extract prepared from H4IIE cells several protein-DNA complexes were detected (Fig. 3B Left). Competition experiments, in which a 25-fold molar excess of unlabeled DNA was included with the labeled probe, were used to correlate protein binding with the insulin response. The G6P WT oligonucleotide, representing the wild-type G6Pase region A HNF-1-binding site, competed effectively for the binding of a single protein-DNA complex (Fig. 3B, see arrow) indicating that the other two bands detected in the assay represent nonspecific binding. By contrast, an oligonucleotide, designated G6P MUT, that contains a mutation identical to that described in the −231 HNF-1 SDM construct (Figs. 2B and 3A) failed to compete with the labeled probe for protein binding (Fig. 3B). Thus, the binding of the specific protein-DNA complex correlates with the insulin response.
The HNF-1-binding site in the mouse β-fibrinogen gene (37) has a similar core sequence to that of the G6Pase HNF-1-binding site but a distinct flanking sequence (Fig. 3A). Nevertheless, an unlabeled oligonucleotide representing this HNF-1-binding site in the mouse β-fibrinogen gene, designated BF WT, competed strongly for the specific protein-DNA complex detected by using H4IIE cell nuclear extract and the G6P WT oligonucleotide as the labeled probe (Fig. 3B Left). To confirm that the specific protein–DNA interaction detected represents HNF-1, a gel retardation assay was performed in which H4IIE cell nuclear extract was preincubated with either an antiserum specific for the α isoform of HNF-1 or, as a control, an antiserum specific for Egr-1 (Fig. 3C Left). The HNF-1α antiserum abolished binding of the specific protein-DNA complex whereas the Egr-1 antiserum had no effect (Fig. 3C Left). Thus, the combination of these protein-binding experiments and the functional data shown in Fig. 2B support the hypothesis that HNF-1 mediates the action of insulin through region A.
Similar results were obtained by using HepG2 nuclear extract in the gel retardation assay except that two specific protein–DNA interactions were detected (Fig. 3B Right, see arrows). Repeating the same competition experiment as described above, but now using HepG2 cell nuclear extract, revealed that the binding of both complexes correlates with the insulin response (Fig. 3B Right). Both these protein–DNA interactions represent HNF-1α because preincubation with the HNF-1α antiserum, but not the control Egr-1 antiserum, abolished protein binding (Fig. 3C Right). The fact that the pattern of HNF-1α binding in the gel retardation assay differs when isolated from different sources has been previously reported (38) and may be explained by differential splicing of the gene encoding HNF-1α (39) or post-translational modification of the HNF-1α protein (33). In liver, the level of HNF-1β (vHNF-1) mRNA expression is ≈20-fold less than HNF-1α (40) and HNF-1β protein is barely detectable on Western blotting (41). Thus, it appears most likely that HNF-1α mediates the effect of insulin through region A although a role for HNF-1β cannot be precluded.
To address the question as to whether insulin might directly inhibit basal G6Pase gene transcription by regulating the binding of HNF-1 to region A, nuclear extracts were prepared from control HepG2 cells and cells treated with insulin for 1 or 4 hr. An identical protein-binding pattern was obtained with these extracts in a gel retardation assay when the G6P WT oligonucleotide was used as the labeled probe (data not shown). This result is consistent with those of Wong and coworkers (42) who showed that although the abundance of HNF-1 protein, though not mRNA, is decreased in streptozotocin-treated diabetic rats and this represents an effect of hyperglycemia rather than hypoinsulinemia because it is reversible upon phlorizin treatment (43). Although these results suggest that insulin treatment does not modify HNF-1 expression or DNA-binding activity, they do not preclude an action of insulin on the transactivation potential of HNF-1.
The HNF-1-Binding Site in Region A Fails to Confer an Inhibitory Effect of Insulin on the Expression of a Heterologous Fusion Gene.
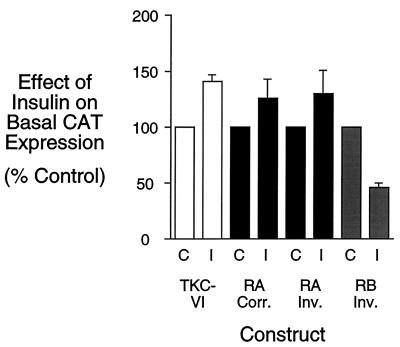
To determine whether the HNF-1-binding site in region A is an IRS that directly mediates an effect of insulin on basal G6Pase gene transcription, this element was analyzed in the context of the TKC-VI vector. This vector contains the heterologous TK promoter ligated to the CAT reporter gene (20, 29). Insulin treatment of H4IIE hepatoma cells transiently transfected with the basic TKC-VI vector alone resulted in a small increase in CAT expression (Fig. 4). When a double-stranded oligonucleotide representing the G6Pase region A HNF-1-binding site was ligated, in either orientation, into the TKC-VI vector it was unable to confer an insulin-dependent inhibition of CAT expression (Fig. 4). In contrast, as a positive control, when a double-stranded oligonucleotide representing the wild-type region B sequence was ligated, in an inverted orientation, into the TKC-VI vector, it conferred an insulin-dependent inhibition of basal CAT expression after transient transfection into H4IIE cells (Fig. 4). These results suggest that region A, unlike region B, is not an independent IRS and that insulin treatment neither modifies the transactivation potential (Fig. 4) nor DNA-binding activity (see above) of HNF-1. Instead, we now investigated the alternative hypothesis that HNF-1 is acting as an accessory factor to enhance the effect of insulin mediated through the region B IRS.
Figure 4.
The HNF-1-binding site in region A fails to confer an inhibitory effect of insulin on the expression of a heterologous fusion gene. H4IIE cells were transiently transfected, as described (20, 22) with either the basic TKC-VI vector or constructs in which double-stranded oligonucleotides representing either the region A (RA) HNF-1-binding site (G6Pase-promoter sequence from −227 to −190; see Fig. 2A) or the region B (RB) IRS (G6Pase-promoter sequence from −197 to −159; see ref. 20) had been ligated into the BamHI site of the TK promoter in either the correct (Corr.) or inverted (Inv.) orientation, relative to that in the G6Pase promoter. After transfection, cells were incubated for 18–20 hr in serum-free medium in the presence (I) or absence (C) of 10 nM insulin. The cells were then harvested and CAT activity was assayed as described in Materials and Methods. Results are presented as the ratio of CAT activities in insulin-treated vs. control cells (expressed as percent control) and represent the mean of ± SEM of four experiments in which each construct was assayed in duplicate.
Mutation of the Region B IRS in the Context of the Full Length G6Pase Promoter Substantially Reduces the Inhibitory Effect of Insulin on Basal G6Pase-CAT Fusion Gene Expression.
To directly determine whether HNF-1 is acting as an accessory factor, we mutated the region B IRS, in the context of the −751 to +66 G6Pase-promoter fragment, by introducing single point mutations in all three copies of the T(G/A)TTT(T/G)(G/T) motif at a site known to abolish the effect of insulin through this sequence when analyzed in a heterologous context (20, 22). This construct, designated −751 RB SDM, in which the sequence of the HNF-1-binding site is unaltered, was analyzed by transient transfection of HepG2 cells (Fig. 5). The ability of insulin to suppress basal G6Pase-CAT expression directed by the −751 RB SDM construct was substantially reduced, beyond the level that would be predicted if region A contained an independent IRS. Rather, this result, consistent with the heterologous fusion gene data shown in Fig. 4, again suggests that HNF-1 bound to region A acts as an accessory factor to enhance the action of insulin mediated through the three IRS motifs in region B.
DISCUSSION
Two segments of the mouse G6Pase promoter, regions A and B, are required for the full inhibitory effect of insulin on basal G6Pase gene transcription (20). These sequences were originally termed a multicomponent insulin response sequence to reflect the fact that, although region B was shown to contain an IRS, the role of region A was undetermined (20). The data presented in this paper suggest that region A is acting as an accessory element to enhance the effect of insulin, mediated through the IRS in region B, on G6Pase gene transcription. The accessory factor binding region A is HNF-1 whereas the factor(s) mediating the effect of insulin through the region B IRS are unknown (4, 20).
Accessory elements are a common feature in the regulation of gene transcription by cAMP and glucocorticoids where their cognate-binding factors often enhance the action of CREB bound to a CRE and the glucocorticoid receptor bound to a GRE, respectively (see ref. 44 for review). A specific example of an accessory factor enhancing the effect of insulin on gene transcription had not been previously delineated; however, the available data suggests that accessory factors may be important in the regulation of several other genes by insulin. For example, deletion of the acetyl CoA carboxylase gene-promoter sequence between −340 and −182 abolishes the stimulatory action of insulin, but this sequence fails to mediate an insulin response in a heterologous context (45). By contrast, a larger construct containing the promoter region located between −340 and −34 does mediate an insulin response in a heterologous context, suggesting that additional cis-acting elements located between −182 and −34 in the acetyl CoA carboxylase promoter are required to manifest the action of insulin (45). Whether this latter segment of the promoter binds an insulin-responsive transcription factor or an accessory factor remains to be determined (45).
HNF-1 also acts as an accessory factor to enhance the stimulatory effects of both glucocorticoids and cAMP on the transcription of specific genes. Rechler and coworkers (46, 47) demonstrated that an HNF-1-binding site within the rat IGFBP-1 promoter is required for the full effect of dexamethasone on IGFBP-1 gene transcription. Mutation of this HNF-1-binding site decreased dexamethasone-stimulated IGFBP-1 gene transcription by >80% (46, 47). Similarly, Weiss and coworkers (48) have shown that an HNF-1-binding site in the mouse phenylalanine hydroxylase promoter is critical for obtaining the maximal induction of gene transcription by both dexamethasone and cAMP. Thus, despite the usually antagonistic effects of cAMP/glucocorticoids and insulin, all three agents are able to use the same factor to enhance their action on gene transcription. Moreover, it appears that HNF-1 can enhance both positive and negative effects of hormones on the expression of various genes. Thus, although HNF-1 is necessary for the full induction of both IGFBP-1 and phenylalanine hydroxylase gene transcription by dexamethasone, it also is required for the maximal repression of G6Pase gene transcription by insulin.
MODY3 and 5 are both rare forms of diabetes, related to non-insulin-dependent diabetes mellitus, that are characterized by an early age of onset (<25 years) and autosomal dominant inheritance with a high penetrance (49–52). MODY3 and 5 result from mutations within the HNF-1α and β genes, respectively (52, 53). Although the primary cause of the hyperglycemia found in MODY3 patients is believed to be because of insulin secretory defects (49, 50), other factors, such as overexpression of the gene encoding G6Pase, could contribute to the associated pathophysiology. Thus, based on the data presented here, low plasma insulin concentrations coupled with mutant hepatic HNF-1 gene expression will likely result in the elevated expression of G6Pase in MODY3 and 5 patients, which may lead to increased HGP (13, 54), contributing to the hyperglycemia. Interestingly, HNF-1α homozygous null mutant mice have a constitutively active gluconeogenic pathway (32). Whether this constitutively active gluconeogenesis parallels increased G6Pase expression is unknown and awaits a more detailed analysis of the metabolic perturbations found in these mice.
In summary, this paper describes the identification of HNF-1 as an accessory factor required for the full effect of insulin on mouse G6Pase gene transcription. Future studies will examine whether HNF-1 manifests this action through an interaction with the factor(s) binding the region B IRS or with the basal transcriptional machinery.
Acknowledgments
We thank Rebecca Taub for providing the mouse G6Pase promoter and for useful discussions, Tanguy Chouard and Moshe Yaniv for the generous gift of the HNF-1α antiserum, and Howard Towle and Jonathan Whittaker for the pCAT(An) and insulin receptor expression vectors, respectively. We also thank Cathy Caldwell for providing the H4IIE cell line and Daryl Granner, Rob Hall, Don Scott, and Norman C. W. Wong for useful suggestions regarding the manuscript. Data analysis was performed in part through the use of the Vanderbilt University Medical Center Cell Imaging Resource (CA68485 and DK20593), and the research was supported by grants from the Vanderbilt University Research Council, the Mark Collie Foundation, the Juvenile Diabetes Foundation, and a Career Development Award from the American Diabetes Association (to R.M.O.).
ABBREVIATIONS
- G6Pase
glucose-6-phosphatase catalytic subunit
- HNF-1
hepatocyte nuclear factor-1
- IRS
insulin response sequence
- IGFBP-1
insulin-like growth factor binding protein-1
- HGP
hepatic glucose production
- CAT
chloramphenicol acetyltransferase
- TK
thymidine kinase
- RB
region B
- SDM
site-directed mutant
References
- 1.DeFronzo R A, Bonadonna R C, Ferrannini E. Diabetes Care. 1992;15:318–368. doi: 10.2337/diacare.15.3.318. [DOI] [PubMed] [Google Scholar]
- 2.Consoli A. Diabetes Care. 1992;15:430–441. doi: 10.2337/diacare.15.3.430. [DOI] [PubMed] [Google Scholar]
- 3.Cline G W, Rothman D L, Magnusson I, Katz L D, Shulman G I. J Clin Invest. 1994;94:2369–2376. doi: 10.1172/JCI117602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Brien R M, Granner D K. Physiol Rev. 1996;76:1109–1161. doi: 10.1152/physrev.1996.76.4.1109. [DOI] [PubMed] [Google Scholar]
- 5.Granner D K, O’Brien R M. Diabetes Care. 1992;15:369–395. doi: 10.2337/diacare.15.3.369. [DOI] [PubMed] [Google Scholar]
- 6.Mithieux G. Eur J Endocrinol. 1997;136:137–145. doi: 10.1530/eje.0.1360137. [DOI] [PubMed] [Google Scholar]
- 7.Foster J D, Pederson B A, Nordlie R C. Proc Soc Exp Biol Med. 1997;215:314–332. doi: 10.3181/00379727-215-44142. [DOI] [PubMed] [Google Scholar]
- 8.Burchell A, Cain D I. Diabetologia. 1985;28:852–856. doi: 10.1007/BF00291077. [DOI] [PubMed] [Google Scholar]
- 9.Liu Z, Barrett E J, Dalkin A C, Zwart A D, Chou J Y. Biochem Biophys Res Commun. 1994;205:680–686. doi: 10.1006/bbrc.1994.2719. [DOI] [PubMed] [Google Scholar]
- 10.Argaud D, Zhang Q, Pan W, Maitra S, Pilkis S J, Lange A J. Diabetes. 1996;45:1563–1571. doi: 10.2337/diab.45.11.1563. [DOI] [PubMed] [Google Scholar]
- 11.Haber B A, Chin S, Chuang E, Buikhuisen W, Naji A, Taub R. J Clin Invest. 1995;95:832–841. doi: 10.1172/JCI117733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Massillon D, Barzilai N, Chen W, Hu M, Rossetti L. J Biol Chem. 1996;271:9871–9874. doi: 10.1074/jbc.271.17.9871. [DOI] [PubMed] [Google Scholar]
- 13.Barzilai N, Rossetti L. J Biol Chem. 1993;268:25019–25025. [PubMed] [Google Scholar]
- 14.Lange A J, Argaud D, El-Maghrabi M R, Pan W, Maitra S R, Pilkis S J. Biochem Biophys Res Commun. 1994;201:302–309. doi: 10.1006/bbrc.1994.1702. [DOI] [PubMed] [Google Scholar]
- 15.Argaud D, Kirby T L, Newgard C B, Lange A J. J Biol Chem. 1997;272:12854–12861. doi: 10.1074/jbc.272.19.12854. [DOI] [PubMed] [Google Scholar]
- 16.Massillon D, Barzilai N, Hawkins M, Prus-Wertheimer D, Rossetti L. Diabetes. 1997;46:153–157. doi: 10.2337/diab.46.1.153. [DOI] [PubMed] [Google Scholar]
- 17.Metzger S, Goldschmidt N, Barash V, Peretz T, Drize O, Shilyansky J, Shiloni E, Chajek-Shaul T. Am J Physiol. 1997;36:E262–E267. doi: 10.1152/ajpendo.1997.273.2.E262. [DOI] [PubMed] [Google Scholar]
- 18.Metzger S, Begleibter N, Barash V, Drize O, Peretz T, Shiloni E, Chajek-Shaul T. Metabolism. 1997;46:579–583. doi: 10.1016/s0026-0495(97)90197-9. [DOI] [PubMed] [Google Scholar]
- 19.Schmoll D, Allan B B, Burchell A. FEBS Lett. 1996;383:63–66. doi: 10.1016/0014-5793(96)00224-4. [DOI] [PubMed] [Google Scholar]
- 20.Streeper R S, Svitek C A, Chapman S, Greenbaum L E, Taub R, O’Brien R M. J Biol Chem. 1997;272:11698–11701. doi: 10.1074/jbc.272.18.11698. [DOI] [PubMed] [Google Scholar]
- 21.O’Brien R M, Lucas P C, Forest C D, Magnuson M A, Granner D K. Science. 1990;249:533–537. doi: 10.1126/science.2166335. [DOI] [PubMed] [Google Scholar]
- 22.O’Brien R M, Noisin E L, Suwanichkul A, Yamasaki T, Lucas P C, Wang J-C, Powell D R, Granner D K. Mol Cell Biol. 1995;15:1747–1758. doi: 10.1128/mcb.15.3.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganss R, Weih F, Schütz G. Mol Endocrinol. 1994;8:895–903. doi: 10.1210/mend.8.7.7984151. [DOI] [PubMed] [Google Scholar]
- 24.Suwanickul A, Morris S L, Powell D R. J Biol Chem. 1993;268:17063–17068. [PubMed] [Google Scholar]
- 25.Goswami R, Lacson R, Yang E, Sam R, Unterman T. Endocrinology. 1994;134:736–743. doi: 10.1210/endo.134.2.7507835. [DOI] [PubMed] [Google Scholar]
- 26.Jacoby D B, Zilz N D, Towle H C. J Biol Chem. 1989;264:17623–17626. [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Higuchi R, Krummel B, Saiki R. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sudhof T C, Russell D W, Brown M S, Goldstein J L. Cell. 1987;48:1061–1069. doi: 10.1016/0092-8674(87)90713-6. [DOI] [PubMed] [Google Scholar]
- 30.O’Brien R M, Lucas P C, Yamasaki T, Noisin E L, Granner D K. J Biol Chem. 1994;269:30419–30428. [PubMed] [Google Scholar]
- 31.Shapiro D J, Sharp P A, Wahli W W, Keller M J. DNA. 1988;7:47–55. doi: 10.1089/dna.1988.7.47. [DOI] [PubMed] [Google Scholar]
- 32.Pontoglio M, Barra J, Hadchouel M, Doyen A, Kress C, Bach J P, Babinet C, Yaniv M. Cell. 1996;84:575–585. doi: 10.1016/s0092-8674(00)81033-8. [DOI] [PubMed] [Google Scholar]
- 33.Chouard T, Jeannequin O, Rey-Campos J, Yaniv M, Traincard F. Biochimie. 1997;79:707–715. doi: 10.1016/s0300-9084(97)86928-3. [DOI] [PubMed] [Google Scholar]
- 34.Mendel D B, Crabtree G R. J Biol Chem. 1991;266:677–680. [PubMed] [Google Scholar]
- 35.Tronche F, Yaniv M. BioEssays. 1992;14:579–587. doi: 10.1002/bies.950140902. [DOI] [PubMed] [Google Scholar]
- 36.Lin B, Morris D W, Chou J Y. Biochemistry. 1997;36:14096–14106. doi: 10.1021/bi9703249. [DOI] [PubMed] [Google Scholar]
- 37.Courtois G, Morgan J G, Campbell L A, Fourel G, Crabtree G R. Science. 1987;238:688–692. doi: 10.1126/science.3499668. [DOI] [PubMed] [Google Scholar]
- 38.Mendel D B, Hansen L P, Graves M K, Conley P B, Crabtree G R. Genes Dev. 1991;5:1042–1056. doi: 10.1101/gad.5.6.1042. [DOI] [PubMed] [Google Scholar]
- 39.Bach I, Yaniv M. EMBO J. 1993;12:4229–4242. doi: 10.1002/j.1460-2075.1993.tb06107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bach I, Mattei M-G, Cereghini S, Yaniv M. Nucleic Acids Res. 1991;19:3553–3559. doi: 10.1093/nar/19.13.3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Simone V, De Magistris L, Lazzaro D, Gerstner J, Monaci P, Nicosia A, Cortese R. EMBO J. 1991;10:1435–1443. doi: 10.1002/j.1460-2075.1991.tb07664.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barrera-Hernandez G, Wanke I E, Wong N C W. J Biol Chem. 1996;271:9969–9975. doi: 10.1074/jbc.271.17.9969. [DOI] [PubMed] [Google Scholar]
- 43.Barrera-Hernandez G, Wanke I E, Wong N C W. Diabetes. 1996;45:1217–1222. doi: 10.2337/diab.45.9.1217. [DOI] [PubMed] [Google Scholar]
- 44.Lucas P C, Granner D K. Annu Rev Biochem. 1992;61:1131–1173. doi: 10.1146/annurev.bi.61.070192.005411. [DOI] [PubMed] [Google Scholar]
- 45.Park K, Kim K-H. J Biol Chem. 1991;266:12249–12256. [PubMed] [Google Scholar]
- 46.Suh D-S, Zhou Y, Ooi G T, Rechler M M. Mol Endocrinol. 1996;10:1227–1237. doi: 10.1210/mend.10.10.9121490. [DOI] [PubMed] [Google Scholar]
- 47.Suh D-S, Rechler M M. Mol Endocrinol. 1997;11:1822–1831. doi: 10.1210/mend.11.12.0021. [DOI] [PubMed] [Google Scholar]
- 48.Faust D M, Catherin A-M, Barbaux S, Belkadi L, Imaizumi-Scherrer T, Weiss M C. Mol Cell Biol. 1996;270:3125–3137. doi: 10.1128/mcb.16.6.3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lehto M, Tuomi T, Mahtani M M, Widen E, Forsblom C, Sarelin L, Gullstrom M, Isomaa B, Lehtovirta M, Hyrkko A, et al. J Clin Invest. 1997;99:582–591. doi: 10.1172/JCI119199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Byrne M M, Sturis J, Menzel S, Yamagata K, Fajans S S, Dronsfield M J, Bain S C, Hattersley A T, Vehlo G, Froguel P, et al. Diabetes. 1996;45:1503–1510. doi: 10.2337/diab.45.11.1503. [DOI] [PubMed] [Google Scholar]
- 51.Velho, G. & Froguel, P. (1997) Diabetes Metab.23, Suppl. 2, 34–37. [PubMed]
- 52.Horikawa Y, Iwasaki N, Hara M, Furuta H, Hinokio Y, Cockburn B N, Lindner T, Yamagata K, Ogata M, Tomonaga O, et al. Nat Genet. 1997;17:384–385. doi: 10.1038/ng1297-384. [DOI] [PubMed] [Google Scholar]
- 53.Yamagata K, Oda N, Kaisaki P J, Menzel S, Furuta H, Vaxillaire M, Southam L, Cox R D, Lathrop G M, Boriraj V V, et al. Nature (London) 1996;384:455–458. doi: 10.1038/384455a0. [DOI] [PubMed] [Google Scholar]
- 54.Efendic S, Karlander S, Vranic M. J Clin Invest. 1988;81:1953–1961. doi: 10.1172/JCI113543. [DOI] [PMC free article] [PubMed] [Google Scholar]