Abstract
Despite extensive studies have shown that increased endothelial nitric oxide synthase (NOS3) expression in the uterine artery endothelial cells (UAEC) plays a key role in uterine vasodilatation, the molecular mechanism controlling NOS3 expression in UAEC is unknown. According to the sheep NOS3 promoter sequence isolated in our laboratory, we hypothesize that the activator protein-1 (AP-1) site in the proximal sheep NOS3 promoter (TGAGTCA, -682 to -676) is important for NOS3 expression. We developed a c-Jun adenoviral expression system to overexpress c-Jun protein into UAEC to investigate the effects of c-Jun/AP-1 on NOS3 expression. Basal levels of c-Jun protein and mRNA were detected in UAEC. C-Jun protein was overexpressed in a concentration and time-dependent fashion in UAEC infected with sense c-Jun (S-c-Jun), but not sham and antisense c-Jun (A-c-Jun) adenoviruses. Infection with S-c-Jun adenovirus (25 MOI, multiplicity of infection) resulted in efficient c-Jun protein overexpression in UAEC up to 3 days. In S-c-Jun, but not sham and A-c-Jun adenovirus infected UAEC, NOS3 mRNA and protein levels were increased (P<0.05) compared to noninfected controls. Increased NOS3 expression was associated with increased total NOS activity. Transient transfections showed that c-Jun overexpression augmented the transactivation of the sheep NOS3 promoter-driven luciferase/reporter constructs with the AP-1 site but not of deletion constructs without the AP-1 site. When the AP-1 site was mutated, c-Jun failed to trans-activate the sheep NOS3 promoter. AP-1 DNA binding activity also increased in c-Jun overexpressed UAEC. Lastly, the pharmacological AP-1 activator phorbol myristate acetate increased AP-1 binding, trans-activated the wild-type but not the AP-1 mutant NOS3 promoter and dose-dependently stimulated UAEC NOS3 and c-Jun protein expression. Hence, our data show that c-Jun/AP-1 regulates NOS3 transcription involving the proximal AP-1 site in the 5′-regulatory region of the sheep NOS3 gene.
Keywords: c-Jun/AP-1, endothelial nitric oxide synthase expression, uterine artery endothelial cells
INTRODUCTION
The endothelial isoform of nitric oxide synthase (NOS3, also termed as eNOS) is one of three NOS isoenzymes (NOS1/nNOS, NOS2/iNOS, and NOS3/eNOS) that catalyze the conversion of L-arginine to L-citrulline for the biosynthesis of the potent vasodilator nitric oxide (NO) (Nathan and Xie, 1994; Palmer et al., 1988). In the uterine circulation, numerous studies have shown that NO produced locally by uterine artery endothelial cells (UAEC) is the most important, though not the only, factor to regulate uterine vasodilatation during normal pregnancy and the follicular phase of the estrous cycle as well as estrogen replacement therapy (Chen et al., 2006; Magness et al., 1997; Vagnoni et al., 1998; Weiner et al., 1994). Uterine vasodilatation as shown by dramatic rises in uterine blood flow during pregnancy is required for delivering nutrients and oxygen to the developing fetus and thus is detrimental to fetal development (Lang et al., 2003). The primary NOS responsible for NO/endothelium-dependent mechanism for uterine vasodilatation is endothelial NOS3 (Byers et al., 2005; Rupnow et al., 2001), although iNOS and nNOS may play a minor role, but none of them are expressed in the uterine artery endothelium in vivo (Mershon et al., 2002; Rosenfeld et al., 2003). The importance of the NOS3-NO system has been recently further emphasized to be critical for pregnancy outcomes in the NOS3-deficient mice (van der Heijden et al., 2005).
The NOS3 protein was initially identified as a constitutively expressed enzyme in endothelial cells (Fleming and Busse, 2003; Forstermann et al., 1998). However, many studies have also demonstrated that the expression level of NOS3 gene can be up-regulated by many physiological and pathophysiological factors, including estrogen (Chen et al., 2006; Vagnoni and Magness, 1998; Weiner et al., 1994; Zheng et al., 2005), growth factors (Inoue et al., 1995; Zheng et al., 1999), shear stress (Li et al., 2003), and protein kinase C activators (Navarro-Antolin et al., 2000), whereas the inflammatory cytokines tumor necrosis factor-α (Yoshizumi et al., 1993) and bacterial toxin lipopolysaccharide (Schwartz et al., 1997) down-regulates the expression of this vasodilatory enzyme. The regulation of NOS3 expression in endothelial cells can take place at the levels of transcription (i.e., NOS3 promoter activity) and post-transcription (i.e., mRNA stability) (Fleming and Busse, 2003). Despite a large body of evidence accumulated showing that increased uterine artery endothelial NOS3 protein coincides with the pregnancy-associated and estrogen-induced uterine vasodilatation (Chen et al., 2006; Magness et al., 1997; Vagnoni et al., 1998; Weiner et al., 1994), the mechanisms by which NOS3 expression is controlled in the UAEC are generally unknown. Uterine artery endothelial NOS3 mRNA is increased in ovariectomized sheep receiving estrogen replacement therapy (Rosenfeld et al., 2003), suggesting that uterine artery endothelial NOS3 expression might be regulated at the level of gene transcription.
The human NOS3 promoter contains consensus sequences for the binding of various transcription factors, including activator protein (AP)-1, AP-2, nuclear factors (NF-1, IL6, NF-κB), Ets protein polyomavirus enhancer activator 3, and CACCC-, CCAAT-, heavy metal-, acute-phase response-, shear stress-, cAMP-response-, interferon-γ response and sterol-regulatory cis elements, as well as several half-sites of the estrogen-responsive element (ERE) but no bona fide EREs, etc (Zhang et al., 1995). Similarly, the bovine and canine NOS3 promoters also have an AP-1 binding site (Schwemmer and Bassenge, 1999; Venema et al., 1994). We have recently isolated a 1305-bp sheep NOS3 promoter sequence. Sequence analysis revealed that sheep NOS3 promoter also possesses a consensus AP-1 binding site (5′-TGAGTCA-3′, −682 to −676) and many other cis-regulatory sequences similar to these of the human and bovine NOS3 promoters (Venema et al., 1994; Wariishi et al., 1995; Zhang et al., 1995).
The AP-1 transcription factors consist of Jun (c-Jun, Jun-B, Jun-D, and v-Jun) homodimers or heterodimers of Jun with Fos (c-Fos, Fos-B, Fra-1, and Fra-2), which is known to mediate the induction and/or inhibition of numerous genes by the tumor promoter 12-O-tetradecanoylphorbol-13-acetate (TPA) and hence the name TPA response element (TRE) for their recognition sites [-TGA(C/G)TCA-] (Karin et al., 1997). Of specific interest to endothelial cells the role of AP-1 in NOS3 expression can be stimulatory or inhibitory according to the stimuli surveyed. For example, in cyclosporine A-treated human umbilical vein endothelial cells (Navarro-Antolin et al., 2000) and shear stress-treated pulmonary artery endothelial cells (Wedgwood et al., 2003), increased c-Jun/AP-1 expression and AP-1 DNA binding activity on the NOS3 promoter AP-1 site are associated with the upregulation of NOS3 mRNA and protein. By contrast, in high glucose-treated human aortic endothelial cells c-Jun/AP-1 binding to NOS3 promoter is accompanied by decreased NOS3 mRNA (Srinivasan et al., 2004). In the present study, we developed an adenoviral gene delivery system to overexpress c-Jun in UAEC in vitro to determine if the AP-1 transcription factors play a critical role in the transcriptional control of NOS3 expression in UAEC. Our data consistently demonstrate that c-jun/AP-1 is involved in the transcriptional regulation of NOS3 mRNA and protein expression in UAEC.
MATERIALS & METHODS
Materials
Rabbit anti-c-Jun polyclonal antibody (pAb) which was raised against a peptide corresponding to a highly conserved DNA binding domain of c-Jun p39 of mouse origin was purchased Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Mouse anti-NOS3 monoclonal antibody (mAb, N20030) was from BD Transduction Labs (CA). Anti-β-actin mAb was from Ambion, Inc. (Austin, TX). Fetal calf serum (FCS), medium-199 (M-199), and DMEM were from Life Technologies Inc. (Grand Island, NY). Tissue culture plasticware was from Corning (Corning, NY). Immobilon-p [polyvinyl difluoride (PVDF)] membrane was from Millipore (Bedford, MA). All other reagents were from Sigma (St. Louis, MO) unless otherwise specified.
Generation of recombinant adenoviral c-Jun vectors
The E1 region-deleted recombinant adenoviral vectors carrying either sense (S-c-jun) or antisense (A-c-jun) c-jun cDNA were constructed as described previously (Yu et al., 2001). Briefly, a 2.6-kb-pair fragment of full-length c-jun cDNA was subcloned into the pACCMVpLpA shuttle vector in sense or antisense orientation to yield the sense or antisense constructs, e.g., pSR–sense-c-jun and pSR–antisense-c-jun. Both pSR-sense-c-jun and pSR-antisense-c-jun were then independently co-transfected with pJM17 into HEK-293 cells by calcium phosphate/DNA coprecipitation. For viral plaque assays, the co-transfected HEK-293 cells were overlaid with 0.65% agarose (prepared with 1x DMEM) every 3 to 4 days. The growth of these E1-deleted adenoviruses was limited to the HEK-293 cells. The polymerase chain reaction (PCR) assay was used to identify the recombinant adenoviral vectors carrying c-Jun. The adenoviral vectors expressing sense and antisense c-jun were used to infect UAEC. Prior to infection, the concentration of the viral particles of each viral stock were determined and tittered according to plaque forming units per cell, also termed as multiplicity of infection (MOI).
Cell culture, adenoviral infection, preparation of total cell extracts and total RNA
UAEC were isolated from pregnant (day 120–130) ewes under nonsurvival surgery by collagenase digestion, cultured in growth medium (DMEM with 20% FCS, 100 U/ml penicillin, and 100 μg/ml streptomycin), and propagated as previously described (Bird et al., 2000; Chen et al., 2004). The animal use protocol was approved by the University of California San Diego Animal Subjects Committee, and we followed the National Research Council’s Guide for the Care and Use of Laboratory Animals throughout the study. Frozen UAEC aliquots (passage 3) were thawed and plated in 100-mm dishes to grow to about 80% confluence in growth medium, and subcultured in DMEM containing 20% FBS, 0.1% BSA, 25 mM 4-(2-hydroxyethyl)-piperazine-1-ethanesulfonic acid (HEPES), 1% antibiotics for experimental use at passages 4–6. For viral infection, subconfluent (approximately 70–80%) cells were incubated with the appropriate amounts of viral particles in M-199 containing 0.5% FBS for 18 hrs. The medium was replaced with fresh DMEM-0.5% FBS and the cells were lysed at different time points post-infection for preparing either total protein extracts or total RNA samples as below. For protein extraction, after rinsing twice with ice-cold PBS, the cells were lysed with 100 μl of a nondenaturing lysis buffer (Chen et al., 2004) per well of a 6-well plate on ice with continuous shaking for 15 min. The total cell extracts were collected using a disposable cell scraper, vortexed, and clarified by centrifugation (13,000 × g, 15 min). The protein content of the samples was measured by a Bio-Rad procedure using BSA as the standard. Aliquots of the extracts were stored at −20ºC until Western blot analysis could be performed. Total RNA samples were extracted from cells in 60-mm dishes by using the guanidium acid-isothiocyanate-phenol-chloroform method using TriZOL reagent (Invitrogen, San Diego, CA) following the manufacturer’s protocol. The total RNA samples were dissolved in diethyl pyrocarbonate (DEPC)-treated water, quantified by measuring absorbance at 260 and 280 nm, and stored at −80 °C until RT-PCR assay was performed.
Quantitative reverse transcription-polymerase chain reaction (RT-PCR) analysis of NOS3, c-Jun, and the ribosomal protein L-19 mRNAs
The steady state levels of “target” mRNAs encoding NOS3, c-Jun and L19 were analyzed by competitive/quantitative RT-PCR developed according to a previously described method (Liao et al., 2005). Oligonucleotides used for RT-PCR were custom-ordered from Gene Link, Inc. (Hawthorne, NY). PCR primer pairs were designed from different exons of NOS3, c-Jun, or L-19 to discriminate PCR products that might arise from possible chromosomal DNA contaminants. Specifically, they were derived from the cDNA clones at the following nucleotides positioned at: 2029–2049 and 2359–2379 for rat c-jun (Sakai et al., 1989), 409–430 and 685–704 for sheep NOS3 (Cale et al., 2005) and 38–56 and 370–388 for sheep L19 (GenBank A# AY158223). The internal controls for NOS3 and L-19 were 171–190 and 561–580 of the pGL3-basic vector from Promega (Madison, WI).
Prior to PCR reaction, each internal control DNA (~250 or 500 base pairs) possessing target-specific primer pairs were generated by PCR using the pGL3-basic vector, purified by QIAquick Gel Extraction Kit (Qiagen, TX) and quantified by OD260/280. The extracted RNA samples (2 μg) were subjected to a RT reaction (20 μl) using 15 units of Cloned AMV Reverse Transcriptase (Invitrogen) with random hexamer (10 ng/μl), and deoxynucleotide triphosphates (dNTPs; 1 mM) at 45 °C for 60 min, and terminated at 70 °C for 10 min. The resultant single strand cDNA was brought to 100 μl of volume with DEPC-water for PCR. The linear portion of PCR amplification between each target cDNA and its corresponding internal control DNA was determined separately for all targets. Briefly, a fixed amount (20 ng) of cDNA derived from UAEC was mixed with a series of dilutions of the internal control DNA. The target and the internal control DNAs were co-amplified by PCR using a specific primer set for each individual target. PCR was performed in a reaction mixture (25 μl) containing MgCl2 (1.5 mM), dNTP (0.2 mM), and 2.5 units of Platinum Taq DNA polymerase (Invitrogen) under the following conditions: 28 cycles for L19, 32 cycles for NOS3 and c-Jun, denaturation at 94°C for 30s, annealing at 60°C (for L19 and NOS3), 62°C (for c-Jun), and extension at 72°C for 30s. Aliquots of PCR products were electrophoresed on 2% agarose gel, stained with GelStar nucleic acid gel stain (BioWittaker Molecular Applications, Rockland, ME), and visualized under a UV light. Digital images were captured and analyzed by using the FluorChem™ Imaging Systems (Alpha Innotech Crop., San Leandro, CA). The PCR products were sequenced and analyzed using the DS-Gene 1.5 software (Accelrys, San Diego, CA). The relative integrated density of each band was multiplied by the absorbance of the surface area. Finally, the logarithm (log) transformation of the ratios of the densitometric readings of the amplified target cDNA and internal control DNA were plotted on the ordinate against the log concentrations of internal control DNA on the abscissa. The concentrations of NOS3 and L19 target mRNAs in the UAEC were determined when the log transformation of a ratio between target and internal control signals equals to 0.
Western blot analysis
Proteins were mixed with SDS sample buffer and boiled for 10 minutes, separated on SDS-PAGE, and transferred to nitrocellulose membranes by using a Fisher semi-dry blotter. After nonspecific binding were blocked in a buffer (0.1% Tween-20 in PBS, TBST) containing 5% nonfat dry milk powder and 1%BSA, the membranes were incubated with the rabbit anti–c-Jun pAb (1:500) or mouse anti-NOS3 (1:750) or anti-β-actin (1:20,000) mAb. The membranes were then washed and incubated with anti-mouse or anti-rabbit horseradish peroxidase–conjugated IgG for 1 hr at room temperature. Bound antibodies were visualized by using the Chemi-Glow™ Chemiluminescent substrate (Alpha Innotech Corp, CA). Digital images were captured by using the Alpha Innotech ChemiImager Imaging System with a high-resolution CCD camera and analyzed with the ChemiImager™ 4400 software.
Total NOS activity assay
Total cell extracts in UAEC were prepared in the non-denaturing buffer and NOS activity was measured by the ability of converting [3H]-L-arginine to [3H]citrulline. Briefly, equal amounts of total cell extracts (>200ug/sample) were mixed with 50 μl of assay cocktail containing 1 mM nicotinamide adenine dinucleotide phosphate reduced, 3 μM tetrahydrobiopterin, 100 nM calmodulin, 2.5 mM CaCl2, 10 μM L-arginine, and 0.2 μCi [3H]-L-arginine (40–70 Ci/mmol, Amersham Biosciences), the reaction mixture was incubated at 37ºC for 30 min (mixed every other 5 min). The reaction was stopped by the addition of 1 ml of cold stop buffer (20 mM Hepes, pH 5.5, 2 mM of each EDTA and EGTA), chilled on ice for 5 min, and then centrifuged (12,000 × g, 3 min). The supernatant was applied on 1-ml columns of Dowex AG 50WX-8 (Na+ form; preequilibrated in stop buffer). [3H]-citrulline formed was eluted with 1 ml water and quantified by liquid scintillation spectrometry. Reactions using buffer without cellular proteins were run in parallel and used to determine assay background. Total NOS activity was expressed as radioactivity (DPM) of each group after subtraction of the background.
Nuclear extract preparation and electrophoretic mobility shift assay (EMSA)
EMSA was performed to determine if increased NOS3 mRNA/protein expression in c-Jun overexpressed UAEC is associated with increased AP-1 DNA binding activity in the sheep NOS3 promoter. EMSA was carried out by using a LightShift Chemiluminescent EMSA kit from Pierce according to the manufacturer’s protocol. Briefly, nuclear extracts were prepared from adenovirus infected UAEC or control cells as follows: cells were washed in cold PBS, collected with a cell scraper into 1 ml PBS. The cells were pelleted by low speed centrifugation (1000 rpm, 5 min) and then lysed for 15 min under constant shaking with 100 μl of 10 mM HEPES, 0.1 mM EDTA, 0.1 mM EGTA, 10 mM KCl, 1mM DTT and 1% NP-40 containing protease inhibitors. The DNA/protein pellet was obtained by centrifugation for 5 min at 13,000 rpm at 4°C. The nuclear pellet was resuspended in 15 μl of 20 mM HEPES, 1 mM EDTA, 0,1 mM EGTA, 420 mM NaCl, 1 mM DTT and protease inhibitors and vortexed vigorously for 15 min at 4°C to release nuclear proteins. After centrifugation (13,000 rpm, 5min) at 4°C, the supernatants (nuclear extracts) were transferred to a tube containing 22.5 μl of Buffer D (20 mM HEPES, 1 mM EDTA, 0,1 mM EGTA, 100 mM KCl, 1% NP-40, 20 % glycerol, 1 mM DTT and protease inhibitors). Protein content was measured using Bradford reagent (Pierce). Nuclear extracts were kept at −70°C and used within 1 month. The sense and antisense sheep NOS3 promoter AP-1 oliogonucleotide probe (5′-CCCCAACTTGAGTCACAGGGG-3′, at −690 to −670, AP-1 is underlined) were biotinylated using the Biotin 3′-end DNA labeling kit from Pierce according to the manufacturer’s protocol. Biotinylation efficiency was calculated using dot blotting. The probe with more than 70% labeling efficiency was used for the EMSA. Both sense and antisense probes were labeled and then annealed in 1 mM Tris-HCl (pH 7.4), 0.5 mM EDTA and 100 mM NaCl at 65°C for 5 min and then cooled at RT before use. The biotinylated double strand AP-1 oligonucleotide was incubated with nuclear extracts (5 μg/reaction) in a binding buffer (10 mM Tris, 50 mM KCl, 1 mM DTT, 1 μg Poly-dIdC, 2.5% Glycerol, 5 mM MgCl2, 2 μg BSA, 0.1 mM EDTA) for 20 min at room temperature. Binding reactions were also performed with unlabeled NOS3 wild-type and mutant (5′-CCCCAACTTagaTCtCAGG GG-3′, mutations are indicated in small case) AP-1 oligonucleotides to show AP-1 DNA binding specificity. Addition of anti-c-Jun antibody (1 μg) to the binding reactions of EMSA was also performed to determine the involvement of c-Jun protein in the AP-1 complex formed.
Cloning of sheep NOS3 promoter and reconstruction of NOS3 promoter luciferase reporter constructs
The 5′-flanking regulatory region of sheep NOS3 gene was isolated by PCR amplification. Briefly, a primer set of 5′-GGCAGACCCCACCTTCTTGG-3′ and 5′-CCACAC TCTTCAAGTTGCCCAT-3′, designed based on the 5′-flanking regulatory region of bovine NOS3 gene at −1283 and + 22 (Genebank A# L27056) (Venema et al., 1994), was used for NOS3 promoter cloning. PCR was carried out using the Expand High Fidelity PCR System (Roche, Indianapolis, IN) in a reaction mixture (100 μl) of 0.25μg ovine genomic DNA polymerase under extracted from UAEC, 1.5mM MgCl2, 0.2mM dNTPs and 4.5units Taq DNA the conditions of 94°C for 2min, 30 cycles of denaturation (94°C, 15sec), annealing (60°C, 30sec), and extension (72°C, 2min). The expected 1305 base pair (bp) sheep NOS3 promoter PCR product was gel purified and quantified by OD260/280. The purified PCR product was subcloned into the pCR2.1-TOPO cloning vector using the Invitrogen TOPO TA Cloning Kit. Three clones of NOS3 promoter pCR2.1-TOPO vectors were then selected for sequencing by using the core service at University of California San Diego Cancer Center. Sequence comparisons and analyses were performed using the DS-Gene 1.5 software (Accelrys, San Diego, CA).
For the construction of NOS3 promoter luciferase reporter constructs, the NOS3 promoter in pCR2.1-TOPO vector was released by double digestion with KpnI and XhoI, then subcloned into the same sites of the promoter-less luciferase reporter vector pGL3-Basic (Promega, WI), then designated the plasmid as -1305pNOS3/pGL3-Luc. Deletions of the promoter from the 5′ at positions of −779, −658, and −50 to +22 were PCR amplified by using the wild-type 1305pNOS3/pGL3-Luc vector as the template. The PCR products of promoter deletions were confirmed by sequencing and then subcloned into the pGL3-Basic vector. The reconstructed plasmids were designed as -779pNOS3/pGL3-Luc (+AP-1), -658pNOS3/pGL3-Luc (-AP-1), and -50pNOS3/pGL3-Luc, respectively.
Site-directed mutagenesis of AP-1 site in sheep NOS3 promoter
To obtain a construct of the sheep NOS3 promoter with mutations in the proximal AP-1 cis-regulatory element (-TGAGTCA-, −685 to −679), site-directed mutagenesis was carried out by PCR using the 1305pNOS3/pGL3 plasmid as a template as described previously (Liao et al., 2003). Briefly, a primer set derived from the sequence of pGL3-Basic vector and the primer AP-1 sense (5′-CAACTAGATCTACAGGGGGTGGCG-3′; mutations positioned at −685, and −682 to −680 are underlined, these mutations created a BglII site) were used to generate the downstream PCR fragment. Primers AP-1 antisense (5′-CCCTGTAGATCTAGTTGGGGGAC ACAAAAGG-3′, complementary of AP-1 sense) and a primer from pGL3-Basic were used to generate the upstream PCR fragment. After appropriate restriction enzyme digestions, the PCR fragments were ligated into the KpnI/HindIII sites of the pGL3-Basic vector. This AP-1 mutant construct was named as 1305pNOS3AP-1M/pGL3-Luc and confirmed by sequencing.
Luciferase assay
UAEC were grown to 70-80% confluency in DMEM + 20% FBS + antibiotics in 24 well plates. Transfection of NOS3 promoter-firefly luciferase constructs was carried out using Targefect F-2 reagent (Targeting Systems, San Diego, CA) according to the manufacturer’s protocol. Briefly, liposomes were assembled using 1.9 μg of NOS3-firefly luciferase pGL3 vectors, 0.1 μg of TK-renilla luciferase vector (internal control) and 6 μl of Targefect F-2 reagent per ml of high glucose (4.5 g/liter) DMEM. The liposome/DNA complex was mix well by gently flicking the tubes 12 times and incubated at 37°C for 25 min. After being washed with high glucose DMEM, the cells were transfected with 0.15 ml medium containing the liposome/DNA complex. The transfection was carried out at 37°C for 4 hr and then the cells were cultured in complete UAEC culture medium to recover for 20–24 hr. Confluent and transfected UAEC were then infected with 25 MOI adenoviruses of sham, A-c-Jun or S-c-Jun in M199 containing 25mM HEPES, 0.1% BSA and 1% FBS. After 24 hr incubation at 37°C, luciferase activities of both firefly and renilla reporters were measured by using the Dual Luciferase Assay Kit (Promega, Madison, WI) in a same reaction tube and recorded by using the 20/20n Luminometer (Turner BioSystems, CA). The NOS3 promoter luciferase activity was calculated as firefly/renilla luciferase activities. Each treatment was tested in quadruplicate and each set of treatments was tested in three independent experiments.
Statistics
Data are presented as means ± SEM and analyzed by one-way ANOVA (SigmaStat, Jandel Scientific, San Rafael, CA). When an F-test was significant (P<0.05), treatment responses were compared with their corresponding controls by Student-Newman-Keuls multiple comparisons. For some experiments, student t-test was used to compare treatment responses to untreated controls.
RESULTS
Cloning and analysis of sheep NOS3 promoter sequence
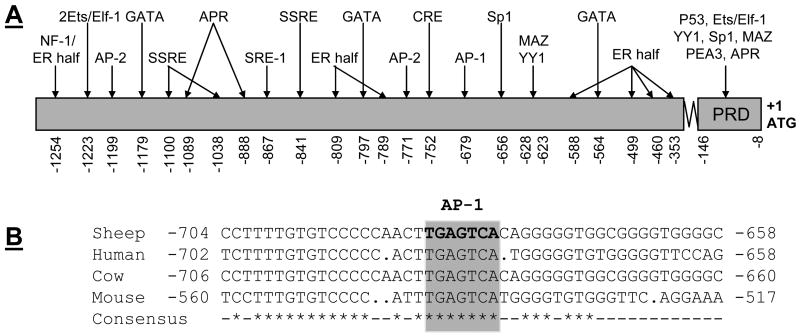
We isolated a distinct 1305-bp DNA fragment of sheep NOS3 5′-flanking regulatory region and deposited the sequence into Genbank with an accession # AY684193. Sequence comparison indicated that the homology of sheep NOS3 promoter to that of bovine (Venema et al., 1994), human (Marsden et al., 1993), and mouse (Gnanapandithen et al., 1996) is 99.0%, 67.0%, and 56.1%, respectively. The putative regulatory sequences of the sheep NOS3 promoter (see Supplemental data) are diagramed in Fig. 1A. The sheep NOS3 promoter did not contain a TATA box, a characteristic of typical constitutively expressed gene. A number of putative regulatory elements were identified, including a consensus sequence (TGAGTCA) for activator protein (AP)-1 binding, 2 AP-2 binding sites (CCCCCACCC), a cAMP response element (TGCGTCA), 7 estrogen half-palindromic motifs (TGACC or GGTCA), 3 GAGA motifs (GATA or TATC) and 12 CACCC boxes (CACCC or GGGTG). In addition, a potential site for the binding of the NF-1 transcription factor (TGGNNNNNNNCCA), a sterol regulatory element (SRE-1, CACCCCAC), 3 acute-phase response (APR) elements (CTGGGA or TCCCAG), 3 shear stress-responsive element (SSRE, GGTCTC), 26 Ets (GGAA/T or A/TTCC, not included) were also identified. Five Sp1 binding sites (GGGCGG or CCGCCC) were identified, 4 of which were present in the so-called “positive regulatory domain” (Karantzoulis-Fegaras et al., 1999; Wariishi et al., 1995; Zhang et al., 1995), which also contained a p53 half site (GGGCTAGTTC), a Ets/Elf site, 1 myc-associated zine-finger protein (MAZ, CCCTCCC), 1 Ying Yang (CCATT, YY1), one PAX (GTTCC), and a Ets family transcription factor PEA3 (CCCTTCCT) (Fleming and Busse, 2003; Karantzoulis-Fegaras et al., 1999). Of note, the AP-1 site was conserved in the NOS3 promoter of all species surveyed (Fig. 1B).
Fig. 1. Diagram of the putative -acting elements within the sheep endothelial nitric oxide (NOS3) promoter.
A: A 1283-bp 5′flanking region of NOS3 gene plus the beginning of the coding region (22-bp) was isolated and sequenced, then deposited to GenBank with an accession number AY684193. The ATG initiation codon is numbered as +1. Potential cis-regulatory elements are indicated. Abbreviations for cis-elements are as follows: AP-1, activator protein 1; SRE-1, sterol regulatory element-1; AP-2, activator protein 2; SSRE, shear stress response element; NF-1, nuclear factor-1; and CRE, cAMP response element; ER half: estrogen half-palindromic motif; SRE: sterol regulatory element; APR: acute-phase response; MAZ: myc-associated zine-finger protein; YY: Ying-yang; PAX: paired box containing genes; and PEA3: Ets family transcription factor. B: Alignment of the sheep, human, bovine, and mouse NOS3 promoter regions surrounding the AP-1 binding site. The AP-1 consensus element is boxed and shade indicated. The ATG initiation codon is numbered as +1.
Overexpression of c-Jun in UAEC
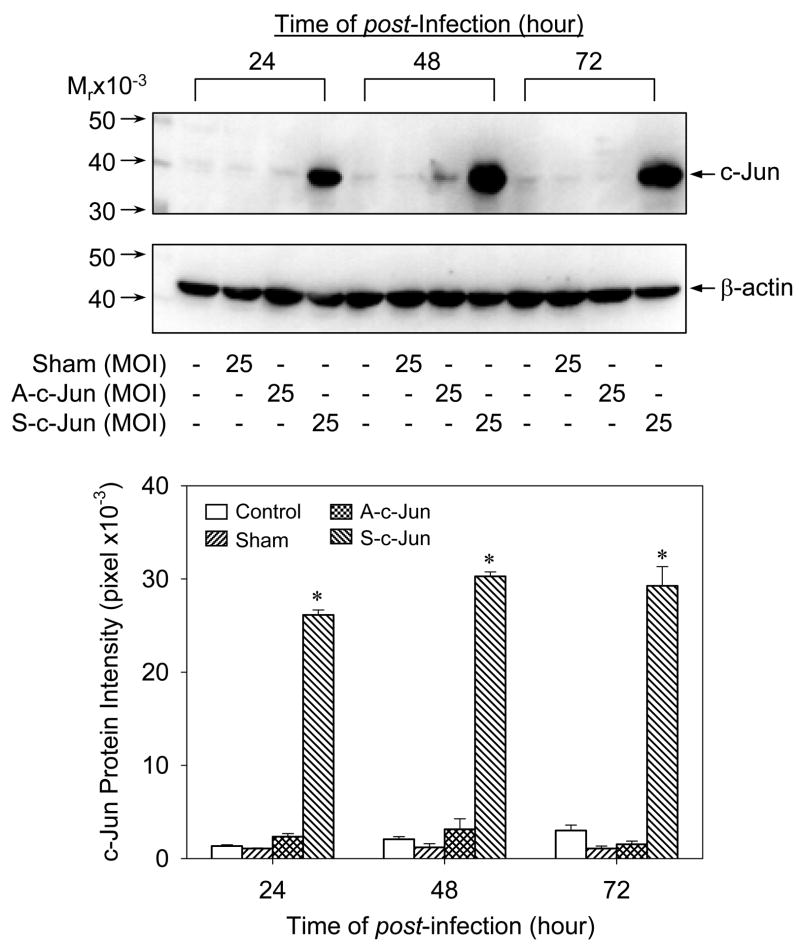
To directly assess the potential requirement for AP-1 in NOS3 expression in UAEC, we developed a c-Jun overexpression system using adenovirus carrying full-length c-Jun cDNA. UAEC expressed basal c-Jun protein under the experimental conditions. In subconfluent UAEC infected with 25 MOI of sham (without c-Jun cDNA), A-c-Jun and S-c-Jun adenoviruses, significant increase (7–10 fold) in c-Jun protein expression was detected in S-c-Jun, but not sham and A-c-Jun infected nor non-infected controls at 24, 48, and 72 hr post-infection. In addition, c-Jun protein levels did not change significantly in the 3-day time course studies of the S-c-Jun adenovirus infection experiments (Fig. 2). When cells were infected with increasing concentrations of S-c-Jun adenoviruses, a concentration-dependent increase in c-Jun protein expression were achieved at 48 hr post-infection. In cells infected with sham and A-c-Jun adenoviruses or in uninfected controls, c-Jun protein levels were unchanged. At concentrations of 25–75 MOI, S-c-Jun adenoviruses stimulated 7–10 fold increases in c-Jun protein expression (Fig. 3). As an internal control, we measured β-actin protein levels, which were not altered in any of the cells infected with any adenoviruses (lower panels in Fig. 2–3). According to these data, it was concluded that the adenoviruses at a concentration of 25 MOI is sufficient for overexpression of c-Jun in UAEC, which was chosen for the following experiments.
Fig. 2. Time-dependent adenoviral delivery of c-Jun into UAEC.
Subconfluent UAEC were infected with 25 plaque forming units/cell (multiplicity of infection, MOI) of sham, A-c-Jun and S-c-Jun adenoviral particles for 4hr in M-199 containing 0.5%FCS, 0.1%BSA, 25 mM HEPES. The medium was replaced with DMEM with 0.5%FCS, 0.1%BSA, 25 mM HEPES and the cells were cultured for up to 3 days. Cellular proteins (40ug/sample) were extracted for size fractionation on 10% SDS-PAGE and the proteins were transferred to PVDF membrane. c-Jun protein was determined by Western blotting by using a rabbit anti-c-Jun polyclonal antibody. *P<0.05 vs. noninfected control.
Fig. 3. Dose dependency of adenoviral delivery of c-Jun into UAEC.
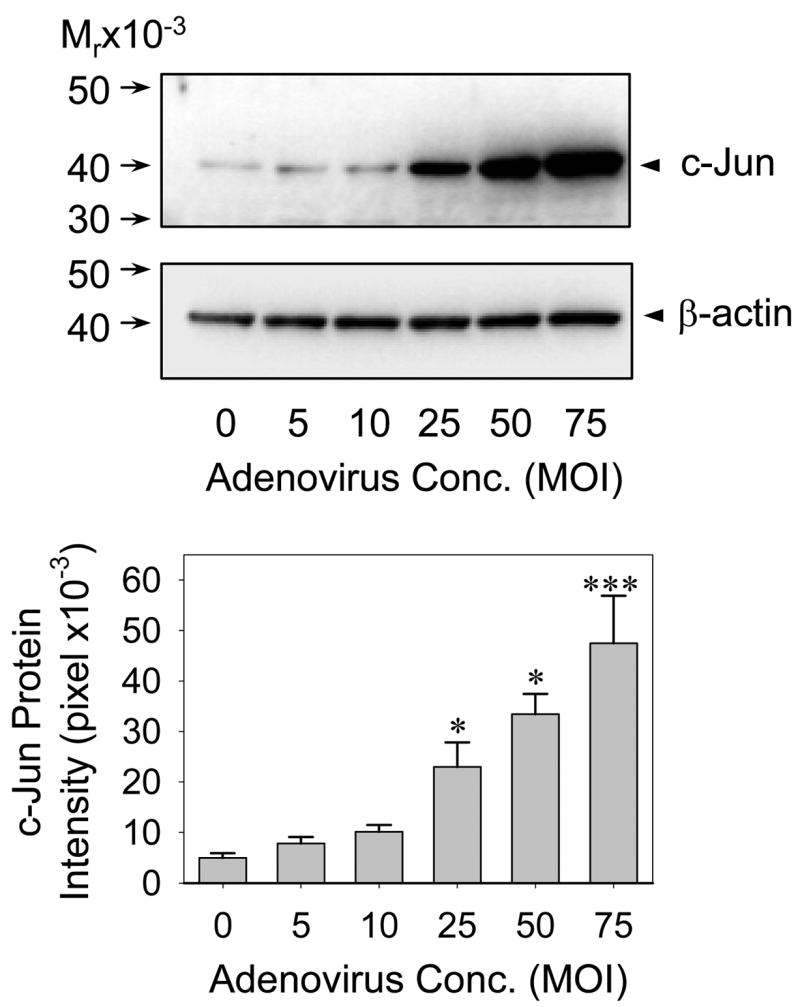
Subconfluent UAEC were infected with increasing concentrations of S-c-Jun adenoviral particles for 4hr in M-199 containing 0.5%FCS, 0.1% BSA, 25 mM HEPES. The medium was replaced with DMEM containing 0.5%FCS, 0.1%BSA, 25 mM HEPES and the cells were cultured for 24hr. Cellular proteins were extracted and c-Jun protein levels were measured as described in Fig. 2. *P<0.05; ***P<0.001 vs. noninfected control.
We also measured c-Jun mRNA levels by RT-PCR in control, sham, A-c-Jun and S-c-Jun infected UAEC to further confirm this adenoviral c-Jun overexpression system. As shown in Fig. 4, basal c-Jun mRNA levels were detected in non-infected control and sham adenoviruses infected cells. As expected, S-c-Jun adenoviruses induced a dramatic increase in c-Jun mRNA in UAEC. In addition, infection with A-c-Jun adenovirus also resulted in increased c-Jun mRNA to levels similar to that of S-c-Jun adenovirus infected cells. In this experiment, we were able to detect both sense and antisense c-Jun mRNA derived from adenovirus infections by RT-PCR using the same primer set. This is because after the sense and antisense mRNAs were reverse transcribed, they were able to be amplified by PCR using the same primer set. This was shown by the equal signal intensities of c-Jun mRNAs in both sense and antisense c-Jun adenovirus infected cells. Thus, the antisense c-Jun mRNA was apparently induced by the infection with the A-c-Jun adenoviruses in UAEC. However, the antisense c-Jun mRNA did not encode the protein compared to Fig. 2.
Fig. 4. Reverse transcription-polymerase chain reaction (RT-PCR) detection of c-Jun mRNA in UAEC infected with or without adenoviruses.
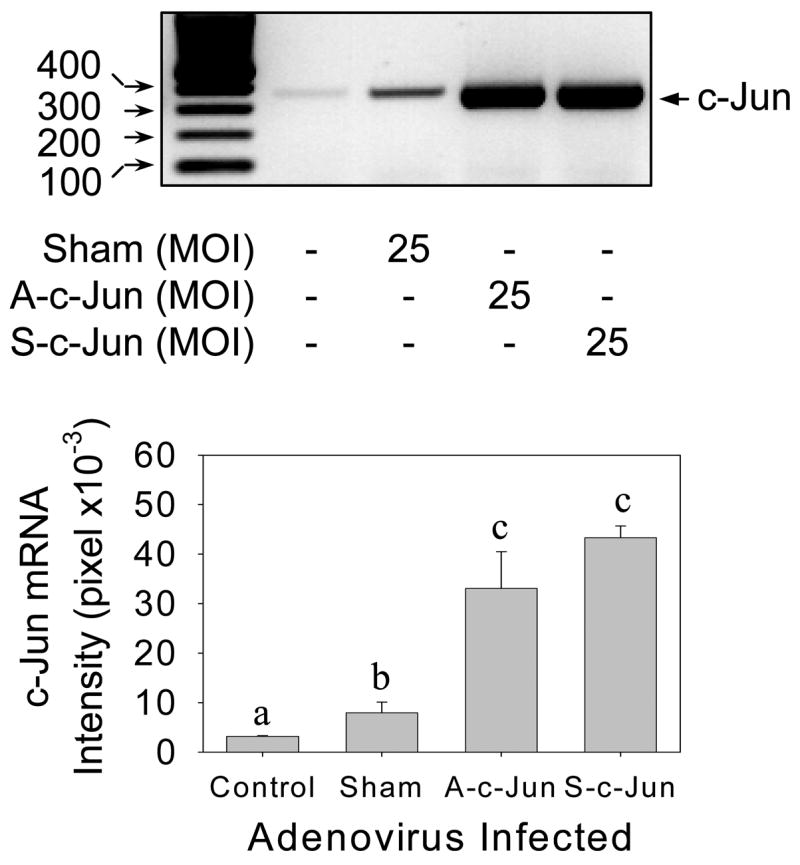
Subconfluent UAEC were infected with or without sham, A-c-Jun, or S-c-Jun adenoviruses (25 MOI) for 4hr. After 24hr incubation, total RNA samples were extracted and c-Jun mRNA levels were measured by RT-PCR. The ribosomal protein L-19 mRNA levels were also measured by using a quantitative RT-PCR assay ascribed in Fig. 5 for monitoring sample loading in the RT-PCR reaction. Letters of different superscripts differ significantly (P<0.05).
Effects of c-Jun overexpression on UAEC NOS3 mRNA expression
To investigate the effects of c-Jun overexpression on NOS3 mRNA expression, a competitive RT-PCR assay was developed by the addition of an exogenous competitor for the assay as described previously (Liao et al., 2005). In these experiments, L-19 mRNA was measured similarly as a sample loading control. The competitive RT-PCR assays for NOS3 and L-19 mRNAs are illustrated in their respective standard curves in Fig. 5A. The correlation coefficients (r2) for both NOS3 and L-19 assays were >0.98 (P < 0.001), indicating that PCR amplification was linear. According to a negative relationship between the competitor (Internal control, IC) and the target cDNA concentrations in the PCR reaction, we first determined the optimal amount of competitor added to achieve a Log [ratio, target/IC] = 0. Then a fixed amount of competitor, i.e., NOS3 = 0.582pM and L19 = 10fM, was added into its respective PCR reaction for measuring NOS3 or L19 mRNA levels in all the experimental samples. Based on the standard curves, NOS3 mRNA levels in UAEC were >1000 fold less than that of L-19. By using this assay system, NOS3 mRNA was found to be constitutively expressed in non-infected control, sham, A-c-Jun and S-c-Jun infected UAEC. However, significant increased NOS3 but not L-19 mRNA levels were detected in S-c-Jun compared to non-infected control, sham and A-c-Jun adenoviruses infected UAEC (Fig. 5B).
Fig. 5. Competitive reverse transcription-polymerase chain reaction (RT-PCR) analysis of NOS3 and the ribosomal protein L19 mRNAs mRNAs in UAEC infected with or without adenoviruses.
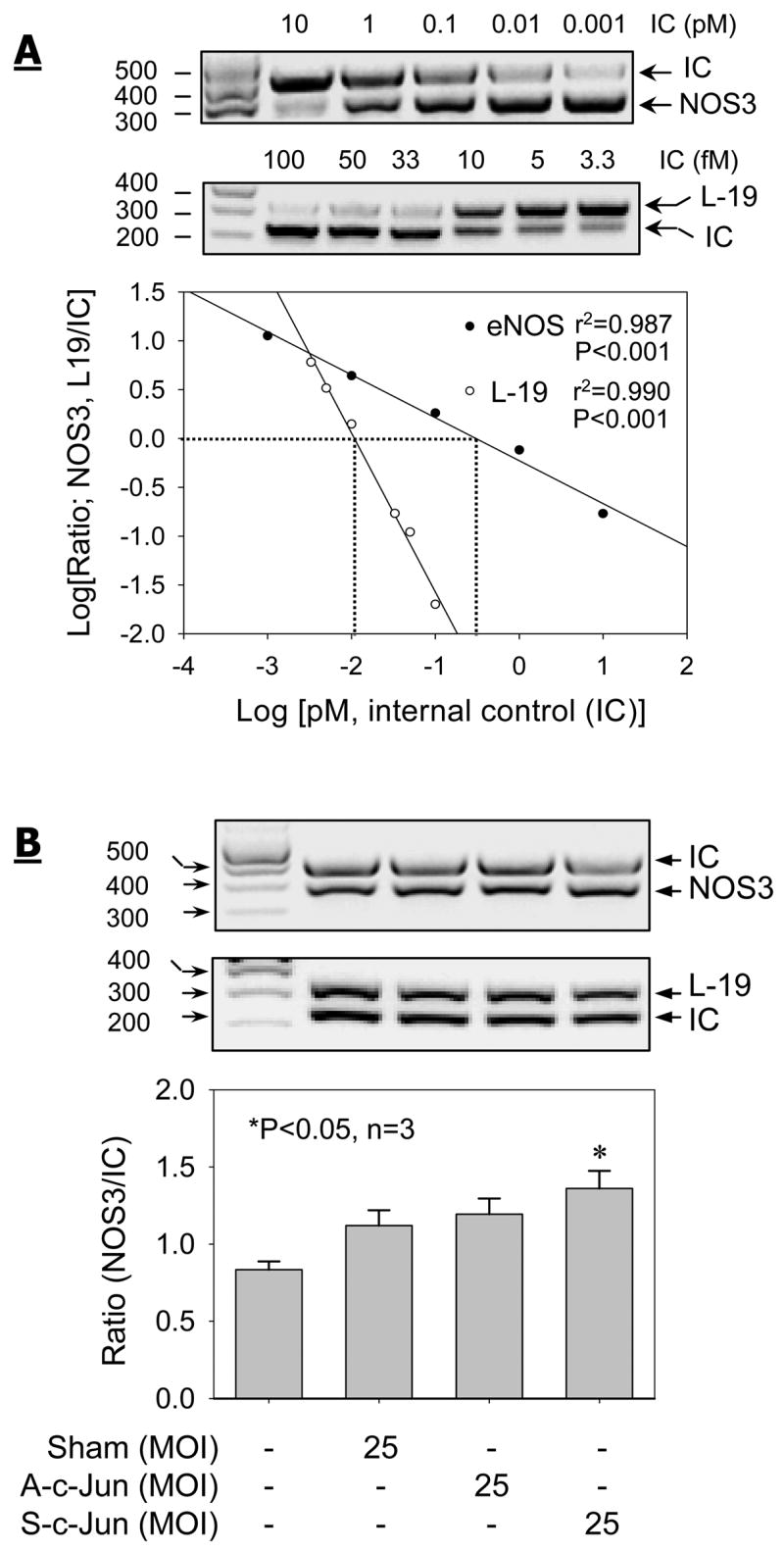
Subconfluent UAEC were infected with or without sham, A-c-Jun, or S-c-Jun adenoviruses (25 MOI) for 4hr. After 24hr incubation, total RNA samples were extracted and NOS3 and the ribosomal protein L-19 mRNA levels were measured by competitive RT-PCR. (A) Increasing concentrations of respective internal control (IC) were added to compete with the target (NOS3 or L-19) molecules the PCR reactions. The signal intensities of NOS3 or L-19 PCR product and its corresponding IC were both Log transformed for the generation of standard curves. The optimal concentrations of NOS3 and L-19 ICs were determined at which the Log-Log transformation of the ratio in PCR product signal intensity versus each target and its corresponding internal control equals to 0. (B) The optimal concentration of the IC for NOS3 and L-19 determined in (A) was added to respective RT-PCR assays for determine the relative changes in NOS3 and L-19 mRNA levels in UAEC infected with or without sham, A-c-Jun, or S-c-Jun adenoviruses. Results represent data (mean ± SEM) using different cell preparations derived from three pregnant ewes. *P<0.05 vs. noninfected control.
Effects of c-Jun overexpression on UAEC NOS3 protein expression
To this end, subconfluent UAEC were infected with or without 25 MOI of sham, A-c-Jun or S-c-Jun adenoviruses for 48 hr, cellular proteins were extracted for examining NOS3 protein expression by Western analysis with anti-NOS3 or β-actin antibodies. As shown in Fig. 6, the ~135-kDa NOS3 protein was detected in all samples, indicating the constitutive expression of NOS3 protein in endothelial cells. In comparison to non-infected control, sham and A-c-Jun infected cells, NOS3 protein levels were significantly increased (~100%) in S-c-Jun infected UAEC. Infection with all the adenoviruses used did not result in a change in β-actin protein levels in UAEC.
Fig. 6. NOS3 protein expression in UAEC infected with or without adenoviruses.
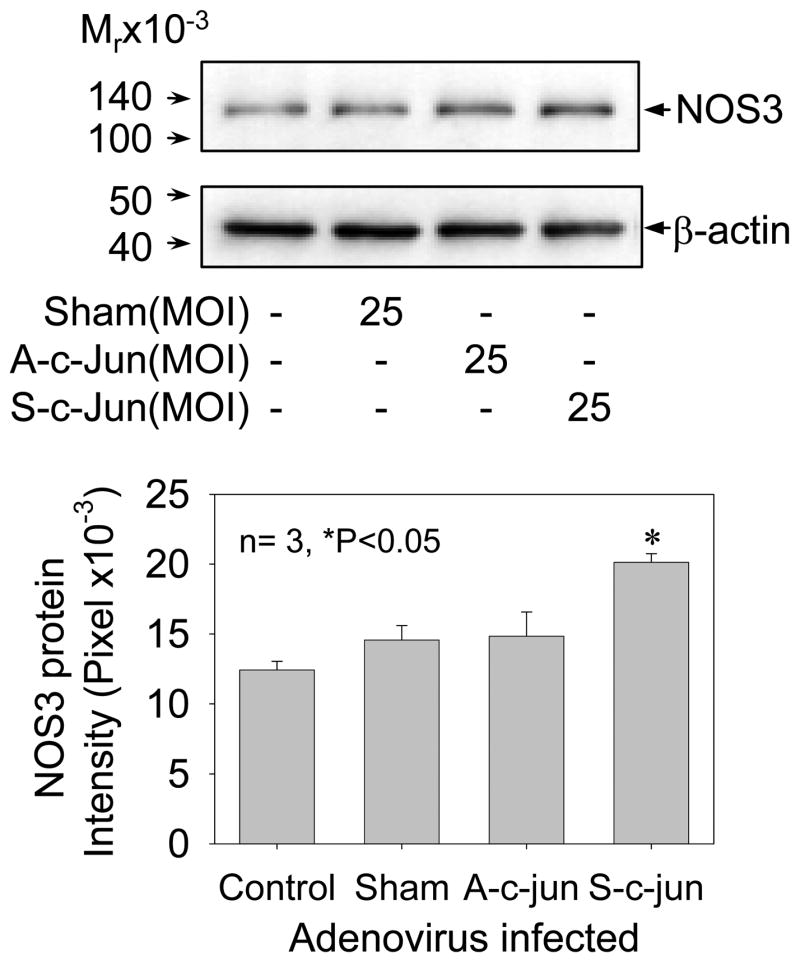
Subconfluent UAEC were infected with or without sham, A-c-Jun, or S-c-Jun adenoviruses (25 MOI) for 4hr. After 24hr incubation, total protein samples were extracted and NOS3 and β-actin protein levels were measured by Western blot analysis with their corresponding specific antibody. Results represent data (mean ± SEM) using different cell preparations derived from three pregnant ewes. *P<0.05 vs. controls.
Effects of c-Jun overexpression on total NOS activity
We determined if increased NOS3 mRNA and protein expression in UAEC overexpressing with c-Jun is associated with an increase in NOS activity. Total NOS activity was measured by using the [3H]-L-citrulline conversion to [3H]-L-citrulline assay. Compared to non-infected cells, total NOS activity in cells infected with 25 MOI of S-c-Jun, but not with sham or A-c-Jun adenoviruses, was significantly increased (Fig. 7).
Fig. 7. Total NOS activity in UAEC infected with or without adenoviruses.
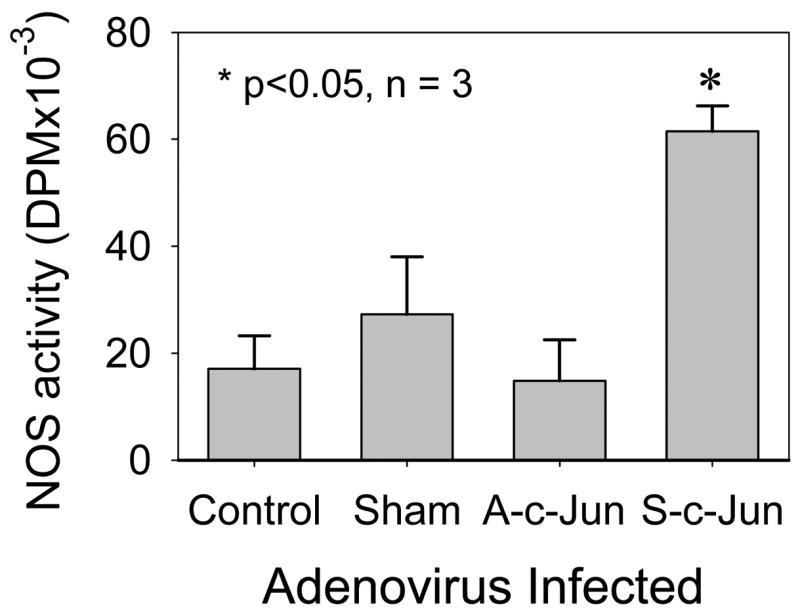
Subconfluent UAEC were infected with or without sham, A-c-Jun, or S-c-Jun adenoviruses (25 MOI) for 4hr. After 24hr incubation, total cell extracts were prepared. Total NOS activity were measured by the ability of total extracts to convert [3H]-L-arginine to [3H]-citrulline as described in Materials and Methods. Reactions using buffer without cellular proteins were run in parallel and used as assay background. Total NOS activity was expressed as [3H]-citrulline formed (DPM) of each group after subtraction of the background. Results represent data (mean ± SEM) run in duplicate using different cell preparations derived from three pregnant ewes. *P<0.05 vs. controls.
Effects of c-Jun overexpression on NOS3 promoter AP-1 binding activity
Since c-Jun overexpression results in an increase in NOS3 mRNA and protein expression in UAEC as shown above, and there is a perfect AP-1 site in sheep NOS3 promoter, we then assessed the binding activity of c-Jun/AP-1 to the AP-1 site in the sheep NOS3 promoter in sham, A-c-Jun, S-c-Jun and non-infected UAEC by EMSA. The EMSA assay was conducted with nuclear extracts prepared from UAEC transfected with or without sham, A-c-Jun, S-c-Jun adenoviruses by using biotinylated double-stranded sheep NOS3 promoter oligonucleotide containing the AP-1 site as the probe. A retarded predominant band was detected in the nuclear extracts of UAEC infected with or without sham, A-c-Jun, S-c-Jun adenoviruses and also cells treated with phorbol myristate acetate (PMA, 100nM for 1hr). This band was blocked by the addition of 500x molar access of the same unlabeled oligonucleotide probe, but not by its AP-1 mutated counterpart, suggesting the specificity and formation of an AP-1 DNA binding complex. In S-c-Jun, but not sham and A-c-Jun adenovirus infected cells, the intensity of this band was dramatically increased compared to non-infected controls. In addition, when a specific anti-c-Jun antibody was added into the EMSA reaction using nuclear extracts of both A-c-Jun and S-c-Jun infected UAEC, the AP-1 binding complex was abolished, suggesting the involvement of c-Jun protein in the formation of the AP-1 complex. Moreover, in the well-known AP-1 activator PMA (100nM for 1hr) treated UAEC (positive control), the AP-1 binding activity was dramatically increased in comparison to the non-infected cells (Fig. 8).
Fig. 8. AP-1 DNA binding complex in UAEC treated with PMA or infected with or without adenoviruses.
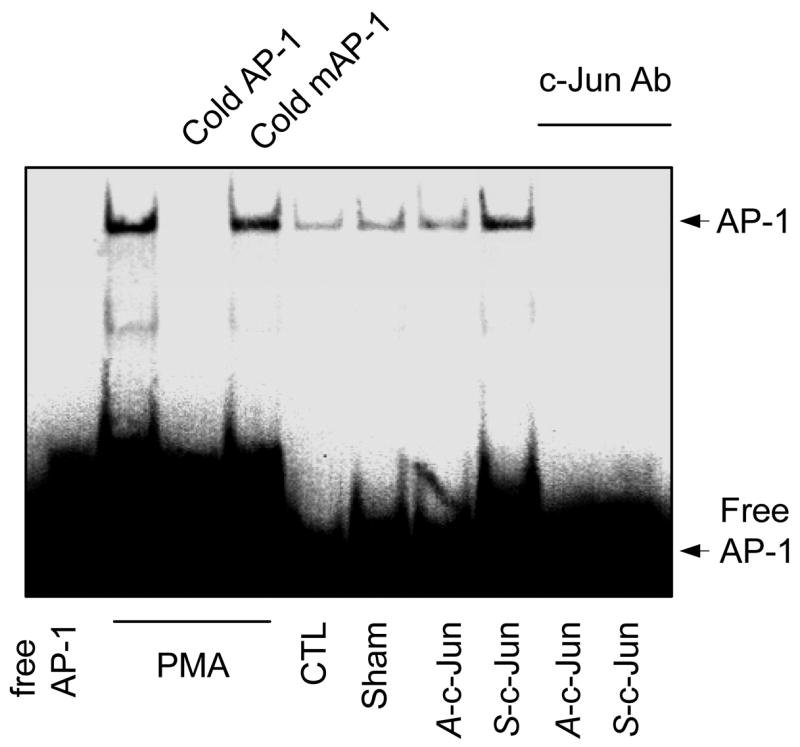
Subconfluent UAEC were infected with or without sham, A-c-Jun, or S-c-Jun adenoviruses (25 MOI) for 4hr. After 24hr incubation, nuclear protein samples were extracted. Nuclear protein extracts were also prepared from UAEC treated with PMA (100 nM) for 30min. The AP-1 DNA binding activity was measured by eletrophoretic mobility shift assay (EMSA) as described in Materials & Methods, using an oligonucleotide including the AP-1 site of sheep NOS3 promoter. Competition experiments were done with the same oligonucleotide but with mutations in the AP-1 site. -Jun is an antibody directed against the DNA binding domain of the c-Jun subunit of AP-1.
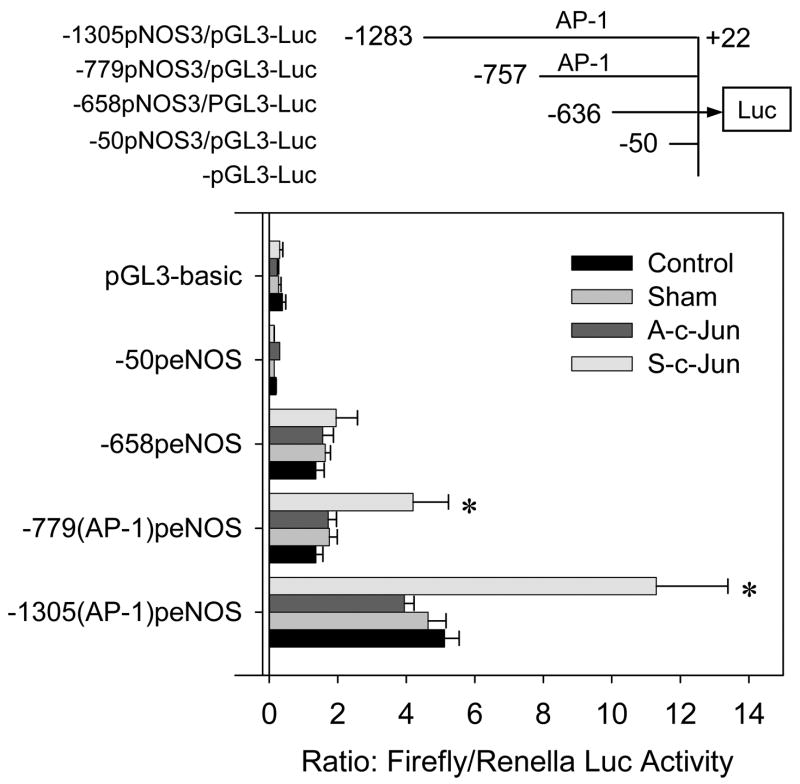
Effects of c-Jun overexpression on sheep NOS3 promoter activity
By using the 1305-bp 5′-flanking regulatory region of the sheep NOS3 gene isolated as above, we developed luciferase reporter constructs with the entire 1305bp sheep NOS3 promoter and deletions with or without the AP-1 sequence. After being transiently transfected into UAEC infected with or without sham, A-c-Jun, S-c-Jun adenoviruses, the activities of NOS3 promoter-driven luciferase reporter gene expression were assessed. As summarized in Fig. 9, compared to non-infected cells, transient NOS3 reporter gene expression of the 1305-bp sheep NOS3 promoter was increased significantly in cells infected with S-c-Jun, but not with sham or A-c-Jun adenoviruses. The deletion of the first 526-bp of the 1305-bp sheep NOS3 promoter, in which the AP-1 site was retained, resulted in a ~2.5-fold reduction in the 1305-bp sheep NOS3 promoter activity in cells with or without adenovirus infection. In addition, with this deletion promoter/reporter construct, the NOS3 promoter luciferase activity in S-c-jun adenovirus infected cells was nearly 1-fold greater than that in sham, A-c-jun adenovirus infected and non-infected control cells. Further deletion of the next 121-bp, in which the AP-1 site was deleted, did not result in additional decrease in basal promoter activity. However, when this deletion promoter/reporter construct was transfected into UAEC, no significant difference in the promoter activity was observed among cells infected with or without S-c-Jun, and sham or A-c-Jun adenoviruses as well as non-infected controls. Additional deletion of the next 586-bp of the sheep NOS3 promoter resulted in complete loss of luciferase reporter gene expression.
Fig. 9. Effects of c-Jun overexpression on sheep NOS3 promoter/luciferase reporter gene expression in transiently transfected into UAEC.
Subconfluent UAEC in 24-well plates were transiently transfected with pGL3-Luc, -1305pNOS3/pGL3-Luc (with an AP-1 site), -779pNOS3/pGL3-Luc (with an AP-1 site), -636pNOS3/pGL3-Luc (without an AP-1 site), and -50pNOS3/pGL3-Luc (without an AP-1 site) promoter-firefly luciferase gene constructs. The TK-Renilla luciferase vector (0.1μg/well) was co-transfected as an internal control. The cells were allowed to reach confluence in 24 hr and then infected with or without sham, A-c-Jun, or S-c-Jun adenoviruses (25 MOI) for an additional 24hr. The cells were then lysed and Firefly/Renilla luciferase activities in the extracts were measured by using the Promega Dual-Luciferase assay kit. Firefly luciferase activity was corrected by the renilla luciferase activity and expressed as a ratio. Results represent data (mean ± SEM) run in quadruple using different cell preparations derived from three pregnant ewes. *P<0.05 vs. controls.
To further evaluate the role of the AP-1 site in the transcriptional up-regulation of NOS3 by c-Jun overexpression, we then used site-directed mutagenesis to generate a mutated sheep NOS3 promoter luciferase reporter construct in which four of the seven nucleotides of the proximal AP-1 site in the wild-type sheep 1305-bp NOS3 promoter were substituted. The mutations of the AP-1 site did not change the basal luciferase activity of the wild-type sheep NOS3 promoter. The luciferase reporter gene expression in transient transfections of the constructs with either the wild-type or the AP-1 mutated sheep 1305-bp NOS3 promoters also did not differ among UAEC transfected with sham and A-c-jun adenoviruses or non-infected controls. However, the significantly increased luciferase reporter gene expression in UAEC infected with S-c-jun adenovirus only occurred in UAEC transiently transfected with the wild-type, but not with the AP-1 mutated sheep 1305-bp NOS3 promoter (Fig. 10).
Fig. 10. Effect of c-Jun overexpression on the activity of the human promoter mutated at its proximal AP-1 site.
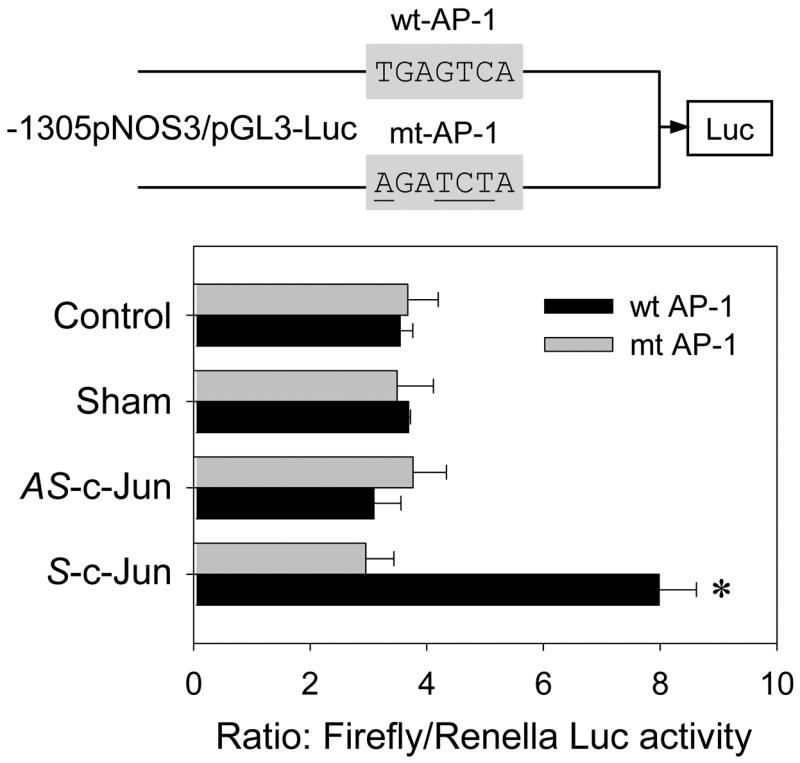
UAEC transiently transfected with the 1305-kb sheep NOS3 promoter-luciferase reporter, wild type (wt) or proximal AP-1 mutant (mt, mutated nucleotides were underlined), were infected with or without sham, A-c-jun, S-c-Jun adenoviruses (25 MOI). Luciferase activity was corrected and determined as in Fig. 8. Results represent data from three independent experiments (mean ± SEM). *P<0.05 vs. controls.
Effects of phorbol myristae acetate (PMA) on trans-activation of eNOS promoter and eNOS and c-Jun protein expression
Lastly, we determined the effects of a well-known AP-1 activator PMA on the trans-activation of sheep eNOS promoter and eNOS and c-Jun protein expressions in UAEC. As shown in Fig. 11A, transient transfection experiments showed that treatment with PMA (20 nM, 24 hr) significantly trans-activated the full-length sheep eNOS promoter but failed to do so when the promoter AP-1 site was mutated. When UAEC was treated with increasing concentrations of PMA (0, 1, 10, 25, 50, 100 nM) for 24 hr, eNOS and c-Jun protein expressions were both dose-dependently increased. It is of note that in comparison to untreated controls as little as 1 nM PMA stimulated eNOS and c-Jun protein expressions with similar potency to those up to 50 nM. However, PMA at 100 nM failed to stimulate eNOS and c-Jun protein expressions, possibly due to the fact that chronic treatment with high concentration PMA down-regulates protein kinase C (Chen et. al., 2001; Chen & Davis, 2003).
Fig. 11. Effect of phorbol myristate acetate (PMA) on the -activation of sheep promoter and c-Jun and NOS3 protein expression.
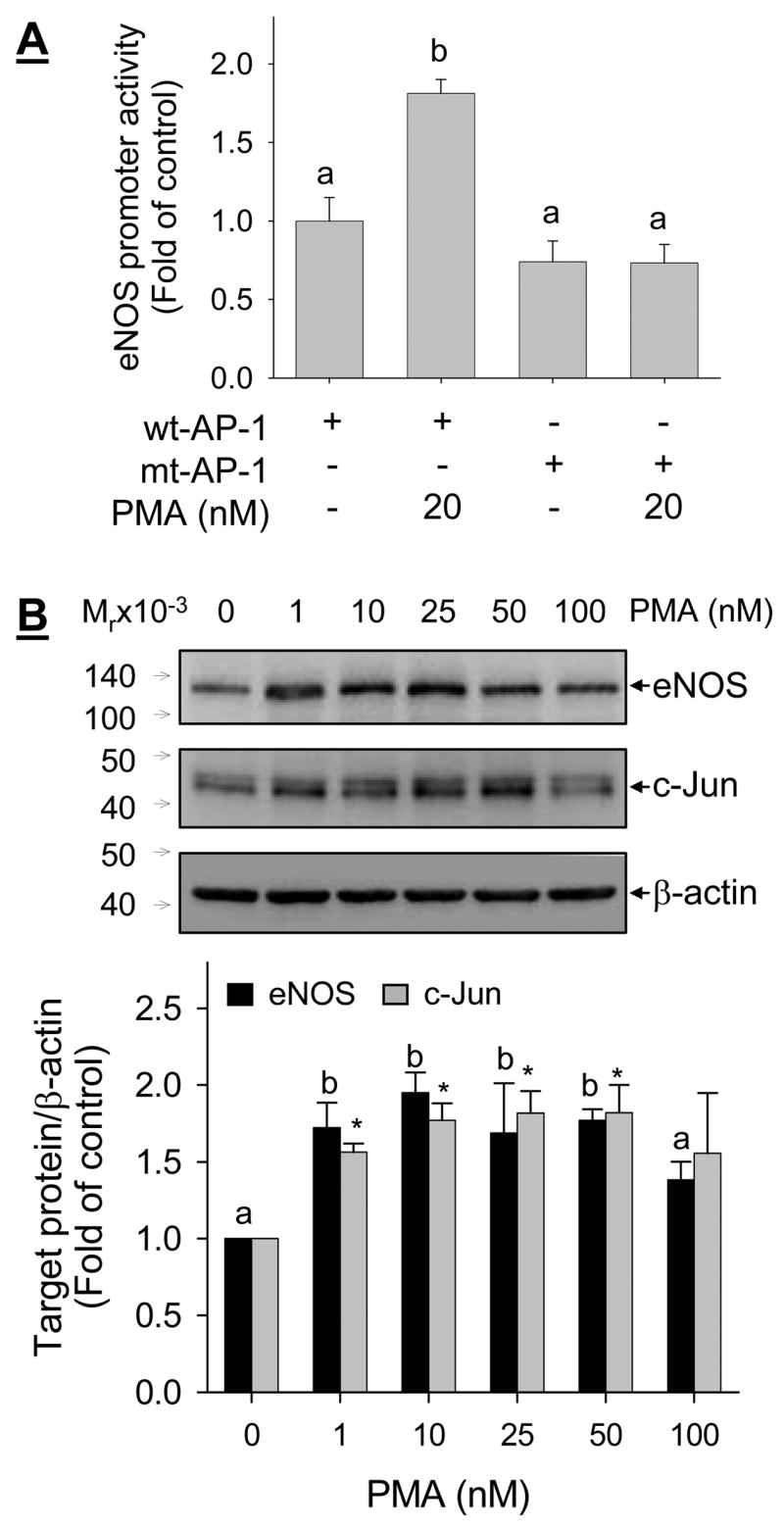
A: UAEC transiently transfected with the 1305-kb sheep NOS3 promoter-luciferase reporter, wild type (wt-AP-1) or proximal AP-1 mutant (mt-AP-1), as described in Fig. 10. After treated with 20 nM PMA for 24 hr, luciferase activity was determined as described in Fig. 9. B: UAEC were treated with increase concentrations of PMA for 24 hr, total protein lysates were prepared and protein levels of NOS3, c-Jun, and β-actin were measured by immunoblotting with their respective specific antibody. Results represent data (mean ± SEM) from four independent experiments using cells from different ewes. Bars with different superscripts differ significantly (p<0.05). * p<0.05 vs. controls in panel B.
DISSCUSSION
Nitric oxide produced locally by the uterine artery endothelium via NOS3 plays a crucial role in the regulation of uterine vasodilatation essential for the delivery of nutrients and oxygen supplies to the growing fetus during pregnancy (Sladek et al., 1997). The importance of the NOS3-NO system during pregnancy has recently been reinforced by the findings that uterine artery remodeling is impaired and litter size is reduced in NOS3-null compared to wild-type mice (van der Heijden et al., 2005). In vivo, there is solid evidence showing that uterine artery endothelial NOS3 protein is increased during pregnancy (Magness et al., 1997; Magness et al., 2001) and in the follicular phase of the estrous cycle or by estrogen replacement therapy in association with elevated circulating estrogen levels (Chen et al., 2006; Rosenfeld et al., 2003; Rupnow et al., 2001; Vagnoni et al., 1998). Available data also suggest that uterine artery endothelial NOS3 expression can be regulated at the level of transcription (Rosenfeld et al., 2003). Despite significant efforts have been paid to the regulation of NOS3 expression in the uterine and placental circulations, the mechanism(s) controlling uterine and placental artery endothelial NOS3 expression currently remains unknown.
Similar to the human (Karantzoulis-Fegaras et al., 1999) and mouse (Gnanapandithen et al., 1996) NOS3 promoters, the sheep NOS3 promoter isolated herein lacks a TATA-box, but possesses proximal elements such as Sp-1 and GATA motifs that features the NOS3 gene typically as a constitutively expressed gene (Forstermann et al., 1998). This is best exemplified by the fact that NOS3 mRNA and protein expression can be detected in various endothelial cells in vivo and in vitro under non-stimulated or stimulated conditions regardless of the origin of the cells. However, the NOS3 promoters of all species isolated so far, including sheep, also contain binding sites for multiple additional transcription factors participating in the complex regulation of NOS3 transcription in endothelial cells in a tissue/cell, and possibly species specific manner (Chan et al., 2004). Moreover, tissue and cell-specific transcriptional regulation of NOS3 expression is controlled by methylation (Chan et al., 2004) and histone acetylation (Fish et al., 2005; Gan et al., 2005) of the NOS3 promoter. In the present study, according to the sequence information of the 5′-flanking region of the sheep NOS3 gene, we hypothesized that the proximal consensus AP-1 (TGAGTCA) site positioned at the −682 to −676 upstream the ATG start codon in the sheep NOS3 gene plays an important role in the regulation of uterine artery endothelial NOS3 expression. We developed an adenoviral gene delivery system for overexpressing c-Jun, an AP-1 component, into sheep UAEC. Our data demonstrated in sheep UAEC that: 1) c-Jun overexpression is able to induce NOS3 promoter-driven luciferase reporter gene expression; 2) The stimulatory effects of c-Jun on NOS3 promoter activity is retained in a luciferase/reporter construct with progressive deletion upstream the proximal AP-1 site, but not in a construct with deletion downstream the proximal AP-1 site; 3) c-Jun overexpression is no longer able to stimulate the transient gene expression of a luciferase/reporter construct driven by the sheep 1305-bp NOS3 promoter in which the AP-1 site is mutated; 4) c-Jun overexpression increases the sheep NOS3 promoter c-Jun/AP-1 binding activity; 5) c-Jun increases NOS3 mRNA and protein expression. Thus, our data demonstrate that the c-Jun/AP-1 transcription factor regulates NOS3 expression at the level of transcription.
We also found that progressive deletions of the sheep NOS3 promoter results in significant reduction in its activity by using luciferase reporter assays. Deletion of the first 526 bases of the sheep 1305-bp NOS3 promoter resulted in a 2.5-fold reduction in the transient promoter reporter gene expression. Further deletion of the next 121 bases containing the AP-1 site did not result in additional decrease in the promoter activity. However, the deletion of two tightly clustered so called “positive regulatory domains” I and II proximal to the promoter (−104/−95 and −144/−115 relative to the transcription initiation) characterized in the human NOS3 promoter (Wariishi et al., 1995; Zhang et al., 1995) resulted in complete loss of the promoter activity. Thus, it is apparent that common mechanisms in the basal transcriptional control of NOS3 expression exist in the UAEC. Interestingly, we have found that a region between −613 to −268 upstream of the ATG start codon of sheep NOS3 gene possibly contains inhibitory cis-elements for NOS3 transcription because the luciferase/reporter gene expression of a deletion construct containing only −268 to +22 of the promoter was ~2-fold greater of that of a deletion construct containing −613 to +22 of the promoter (data not shown). To the best of our knowledge, this has not been reported previously, which is under further investigation in our laboratory.
The AP-1 transcription factors are Jun/Jun homodimer or Jun/Fos heterodimers (Karin et al., 1997). In this study, we have focused on c-Jun because UAEC expressed basal levels c-Jun, but not c-Fos, protein under the culture conditions. However, whether other members of the AP-1 transcription factors play a role in the regulation of NOS3 expression in UAEC also needs to be investigated. Furthermore, the AP-1 family oncogene products can be rapidly induced by a variety of physiological and pathophysiological stimuli important for cell function, including hormones such as estrogen (Shiozawa et al., 2004), growth factors (Inoue et al., 1995; Zheng et al., 1999), shear stress (Li et al., 2003), protein kinase C activators (Navarro-Antolin et al., 2000), tumor necrosis factor-α (Hanazawa et al., 1994), lipopolysaccharide (Schwartz et al., 1997; Wong et al., 2004), and angiotensin II (Clark et al., 1992). In the uterine and placental circulations, the role of AP-1 in estrogen (Chen et al., 2006; Vagnoni et al., 1998; Weiner et al., 1994) and angiogenic growth factors such as basic fibroblast growth factor (Zheng et al., 1999) as well as angiotensin II (Zheng et al., 2005) stimulation of endothelial NOS3 expression is currently unknown, which will be important to be investigated because of the key role(s) that these factors play in the regulation of uterine and placental vascular activity and reactivity during pregnancy.
It is also noteworthy that the regulation of endothelial NOS3 expression is complex. At the level of transcription, this is exemplified by the numerous cis-elements for binding of various transcription factors in the human NOS3 promoter. It is apparent that, similar to the human one (Wariishi et al., 1995; Zhang et al., 1995), the sheep NOS3 promoter also contains a consensus AP-1 site and seven half-estrogen receptor binding sites as well as many others (Fig. 1). Of note is that liganded steroid receptors can bind to the transcription factors, specificity protein 1 (Sp1) and Sp3, to be directed to GC rich regions of a given target gene promoter containing Sp1/Sp3 response elements (DeNardo et al., 2005). This mechanism may be implicated in estrogen regulation of NOS3 expression in UAEC since the so-called positive regulatory domains I and II proximal to the NOS3 promoter are GC-rich and contain multiple Sp1 sites (present study, (Fleming and Busse, 2003; Zhang et al., 1995)). Regardless, of specific interest to the uterine circulation, it would be interesting to test whether AP-1 plays a role in mediating estrogen stimulation of NOS3 expression by interacting with nuclear estrogen receptors, a pathway which has been demonstrated in other cell systems (Paech et al., 1997; Webb et al., 1999). Moreover, the regulation of NOS3 expression by a given physiological or pathophysiological extracellular stimulus involves ligand-receptor activation leading to specific downstream intracellular signaling towards coordinated interaction between nuclear transcription factors with one or more promoter regulatory cis-elements. Activation of extracellular-signal regulated protein kinases (ERK2/1) by physiological stimuli of uterine and placental endothelial cell functions such as estrogen (Chen et al, 2004) and angiogenic growth factors (Zheng et al, 1999) as well as enhanced activation of this pathway during pregnancy (Bird et al, 2003) is of critical interest since ERK2/1 is a well-known direct upstream kinase pathway for AP-1 activation and/or expression (Karin, 1997).
Once transcribed, the stability of NOS3 mRNA also is subjected to complex post-transcriptional regulation via the 3′-untranslational regulatory sequences interacting with RNA binding proteins (Fleming and Busse, 2003; McQuillan et al., 1994; Yoshizumi et al., 1993). Furthermore, NOS3 protein is very stable and with a half-life of 20 hr (Liu et al., 1995) and its enzymatic activity is controlled extensively by post-translational modifications of the protein (Michel and Feron, 1997; Shaul, 2002). Our primary focus in this study was on NOS3 transcription. Our results consistently support a concept that c-Jun/AP-1 plays an important role in regulating NOS3 transcription in endothelial cells. However, whether c-Jun overexpression has regulatory effects on other aspects of the NOS3-NO system, i.e., mRNA stability and post-transcriptional modifications awaits further investigation. In addition, we also have shown that total NOS activity is increased in c-Jun overexpressing UAEC. One possibility of the increased NOS activity is due to increased NOS3 protein expression as shown herein. However, protein expression and enzymatic activity are two different aspects of NOS3 regulation. For example, our data show different amplitudes of responses in NOS3 mRNA/protein expression and total NOS activity in c-Jun overexpressing cells. To this end, mechanism(s) involving calcium homeostasis might play a critical role in the regulation of NOS activity in UAEC by c-Jun/AP-1. This speculation is based on several lines of evidence: 1) NOS3 is a calcium-dependent enzyme (Fleming and Busse, 2003; Nathan and Xie, 1994); 2) in UAEC several recent reports have shown that intracellular calcium is important for pregnancy specific regulation of NO production by adenosine 5′-triphosphate (Yi and Bird, 2005; Yi et al., 2005); 3) Pregnancy-enhanced store-operated calcium channel function is associated with transient receptor potential channels (Gifford et al., 2006a), and calcium responses to adenosine 5′-triphosphate is due to greater capacitive calcium entry activity that might be originated from transient receptor potential channels (Gifford et al, 2006b); 4) We have recently found that c-Jun overexpression stimulated capacitive calcium entry activity in UAEC (Chen, et. al., unpublished data). However, additional in-depth studies are needed for deciphering the role of calcium homeostasis in the regulation of NOS activity by AP-1.
Supplementary Material
A: A 1283-bp 5′flanking region of NOS3 gene plus the beginning of the coding region (22-bp) was isolated and sequenced, then deposited to GenBank with an accession number AY684193. The nucleotide position number is relative to the ATG initiation codon (+1). The nucleotide sequences for putative cis-acting elements are boxed and labeled with different colors. Abbreviations for cis-elements are as follows: AP-1, activator protein 1; SRE-1, sterol regulatory element-1; AP-2, activator protein 2; SSRE, shear stress response element; NF-1, nuclear factor-1; and CRE, cAMP response element; ER half: estrogen half-palindromic motif; SRE: sterol regulatory element; APR: acute-phase response; MAZ: myc-associated zine-finger protein; YY: Ying-yang; PAX: paired box containing genes; and PEA3: Ets family transcription factor.
Acknowledgments
The present study was supported in part by NIH RO1 grants HL70562 and HL74947 (to D.B.C) and HL64703 (to JZ). E.M-G is partially supported by a Minority Supplemental Postdoctoral Fellowship from the National Heart, Lung and Blood Institute.
The authors are grateful to Dr. Yin Yu for providing adenoviral expression vectors, and Drs. Brace and Cheung for their assistance in animal handling.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bird IM, Sullivan JA, Di T, Cale JM, Zhang L, Zheng J, Magness RR. Pregnancy-dependent changes in cell signaling underlie changes in differential control of vasodilator production in uterine artery endothelial cells. Endocrinology. 2000;141:1107–1117. doi: 10.1210/endo.141.3.7367. [DOI] [PubMed] [Google Scholar]
- Bird IM, Zhang L, Magness RR. Possible mechanisms underlying pregnancy-induced changes in uterine artery endothelial function. Am J Physiol Regul Integr Comp Physiol. 2003;284:R245–258. doi: 10.1152/ajpregu.00108.2002. [DOI] [PubMed] [Google Scholar]
- Byers MJ, Zangl A, Phernetton TM, Lopez G, Chen DB, Magness RR. Endothelial vasodilator production by ovine uterine and systemic arteries: ovarian steroid and pregnancy control of ERalpha and ERbeta levels. J Physiol. 2005;565:85–99. doi: 10.1113/jphysiol.2005.085753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cale JM, Tsoi SC, Toppe M, Grummer MA, Ochiai M, Magness RR, Bird IM. Molecular cloning of ovine endothelial nitric oxide synthase and expression in COS-7 cells. J Soc Gynecol Investig. 2005;12:156–168. doi: 10.1016/j.jsgi.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Chan Y, Fish JE, D’Abreo C, Lin S, Robb GB, Teichert AM, Karantzoulis-Fegaras F, Keightley A, Steer BM, Marsden PA. The cell-specific expression of endothelial nitric-oxide synthase: a role for DNA methylation. J Biol Chem. 2004;279:35087–35100. doi: 10.1074/jbc.M405063200. [DOI] [PubMed] [Google Scholar]
- Chen DB, Fong HW, Davis JS. Induction of c-fos and c-jun messenger ribonucleic acid expression by prostaglandin F2alpha is mediated by a protein kinase C-dependent extracellular signal-regulated kinase mitogen-activated protein kinase pathway in bovine luteal cells. Endocrinology. 2001;142:887–895. doi: 10.1210/endo.142.2.7938. [DOI] [PubMed] [Google Scholar]
- Chen DB, Bird IM, Zheng J, Magness RR. Membrane estrogen receptor-dependent extracellular signal-regulated kinase pathway mediates acute activation of endothelial nitric oxide synthase by estrogen in uterine artery endothelial cells. Endocrinology. 2004;145:113–125. doi: 10.1210/en.2003-0547. [DOI] [PubMed] [Google Scholar]
- Chen DB, Davis JS. Epidermal growth factor stimulates c-fos and c-jun mRNA expression through Raf-1/MEK1/ERK2/1 dependent and independent pathways bovine luteal cells. Mol Cell Endocrinol. 2003;200:141–154. doi: 10.1016/s0303-7207(02)00379-9. [DOI] [PubMed] [Google Scholar]
- Chen DB, Jia S, King AG, Barker A, Li SM, Mata-Greenwood E, Zheng J, Magness RR. Global protein expression profiling underlines reciprocal regulation of caveolin 1 and endothelial nitric oxide synthase expression in ovariectomized sheep uterine artery by estrogen/progesterone replacement therapy. Biol Reprod. 2006;74:832–838. doi: 10.1095/biolreprod.105.049700. [DOI] [PubMed] [Google Scholar]
- Clark AJ, Balla T, Jones MR, Catt KJ. Stimulation of early gene expression by angiotensin II in bovine adrenal glomerulosa cells: roles of calcium and protein kinase C. Mol Endocrinol. 1992;6:1889–1898. doi: 10.1210/mend.6.11.1336125. [DOI] [PubMed] [Google Scholar]
- DeNardo DG, Kim HT, Hilsenbeck S, Cuba V, Tsimelzon A, Brown PH. Global gene expression analysis of estrogen receptor transcription factor cross talk in breast cancer: identification of estrogen-induced/activator protein-1-dependent genes. Mol Endocrinol. 2005;19:362–378. doi: 10.1210/me.2004-0267. [DOI] [PubMed] [Google Scholar]
- Fish JE, Matouk CC, Rachlis A, Lin S, Tai SC, D’Abreo C, Marsden PA. The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code. J Biol Chem. 2005;280:24824–24838. doi: 10.1074/jbc.M502115200. [DOI] [PubMed] [Google Scholar]
- Fleming I, Busse R. Molecular mechanisms involved in the regulation of the endothelial nitric oxide synthase. Am J Physiol Regul Integr Comp Physiol. 2003;284:R1–12. doi: 10.1152/ajpregu.00323.2002. [DOI] [PubMed] [Google Scholar]
- Forstermann U, Boissel JP, Kleinert H. Expressional control of the ‘constitutive’ isoforms of nitric oxide synthase (NOS I and NOS III) FASEB J. 1998;12:773–790. [PubMed] [Google Scholar]
- Gan Y, Shen YH, Wang J, Wang X, Utama B, Wang J, Wang XL. Role of histone deacetylation in cell-specific expression of endothelial nitric-oxide synthase. J Biol Chem. 2005;280:16467–16475. doi: 10.1074/jbc.M412960200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford SM, Yi FX, Bird IM. Pregnancy-enhanced store-operated Ca2+ channel function in uterine artery endothelial cells is associated with enhanced agonist-specific transient receptor potential channel 3-inositol 1,4,5-trisphosphate receptor 2 interaction. J Endocrinol. 2006a;190:385–395. doi: 10.1677/joe.1.06773. [DOI] [PubMed] [Google Scholar]
- Gifford SM, Yi FX, Bird IM. Pregnancy-enhanced Ca2+ responses to ATP in uterine artery endothelial cells is due to greater capacitative Ca2+ entry rather than altered receptor coupling. J Endocrinol. 2006b;190:373–384. doi: 10.1677/joe.1.06635. [DOI] [PubMed] [Google Scholar]
- Gnanapandithen K, Chen Z, Kau CL, Gorczynski RM, Marsden PA. Cloning and characterization of murine endothelial constitutive nitric oxide synthase. Biochim Biophys Acta. 1996;1308:103–106. doi: 10.1016/0167-4781(96)00098-x. [DOI] [PubMed] [Google Scholar]
- Hanazawa S, Takeshita A, Kitano S. Retinoic acid suppression of c-fos gene inhibits expression of tumor necrosis factor-alpha-induced monocyte chemoattractant JE/MCP-1 in clonal osteoblastic MC3T3-E1 cells. J Biol Chem. 1994;269:21379–21384. [PubMed] [Google Scholar]
- Inoue N, Venema RC, Sayegh HS, Ohara Y, Murphy TJ, Harrison DG. Molecular regulation of the bovine endothelial cell nitric oxide synthase by transforming growth factor-beta 1. Arterioscler Thromb Vasc Biol. 1995;15:1255–1261. doi: 10.1161/01.atv.15.8.1255. [DOI] [PubMed] [Google Scholar]
- Karantzoulis-Fegaras F, Antoniou H, Lai SL, Kulkarni G, D’Abreo C, Wong GK, Miller TL, Chan Y, Atkins J, Wang Y, Marsden PA. Characterization of the human endothelial nitric-oxide synthase promoter. J Biol Chem. 1999;274:3076–3093. doi: 10.1074/jbc.274.5.3076. [DOI] [PubMed] [Google Scholar]
- Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–246. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- Lang U, Baker RS, Braems G, Zygmunt M, Kunzel W, Clark KE. Uterine blood flow--a determinant of fetal growth. Eur J Obstet Gynecol Reprod Biol. 2003;110(Suppl 1):S55–61. doi: 10.1016/s0301-2115(03)00173-8. [DOI] [PubMed] [Google Scholar]
- Li Y, Zheng J, Bird IM, Magness RR. Effects of pulsatile shear stress on nitric oxide production and endothelial cell nitric oxide synthase expression by ovine fetoplacental artery endothelial cells. Biol Reprod. 2003;69:1053–1059. doi: 10.1095/biolreprod.102.013474. [DOI] [PubMed] [Google Scholar]
- Liao WX, Magness RR, Chen DB. Expression of estrogen receptors-alpha and -beta in the pregnant ovine uterine artery endothelial cells in vivo and in vitro. Biol Reprod. 2005;72:530–537. doi: 10.1095/biolreprod.104.035949. [DOI] [PubMed] [Google Scholar]
- Liao WX, Moore RK, Otsuka F, Shimasaki S. Effect of intracellular interactions on the processing and secretion of bone morphogenetic protein-15 (BMP-15) and growth and differentiation factor-9. Implication of the aberrant ovarian phenotype of BMP-15 mutant sheep. J Biol Chem. 2003;278:3713–3719. doi: 10.1074/jbc.M210598200. [DOI] [PubMed] [Google Scholar]
- Liu J, Garcia-Cardena G, Sessa WC. Biosynthesis and palmitoylation of endothelial nitric oxide synthase: mutagenesis of palmitoylation sites, cysteines-15 and/or -26, argues against depalmitoylation-induced translocation of the enzyme. Biochemistry. 1995;34:12333–12340. doi: 10.1021/bi00038a029. [DOI] [PubMed] [Google Scholar]
- Magness RR, Shaw CE, Phernetton TM, Zheng J, Bird IM. Endothelial vasodilator production by uterine and systemic arteries. II Pregnancy effects on NO synthase expression. Am J Physiol. 1997;272:H1730–1740. doi: 10.1152/ajpheart.1997.272.4.H1730. [DOI] [PubMed] [Google Scholar]
- Magness RR, Sullivan JA, Li Y, Phernetton TM, Bird IM. Endothelial vasodilator production by uterine and systemic arteries. VI Ovarian and pregnancy effects on eNOS and NO(x) Am J Physiol Heart Circ Physiol. 2001;280:H1692–1698. doi: 10.1152/ajpheart.2001.280.4.H1692. [DOI] [PubMed] [Google Scholar]
- Marsden PA, Heng HH, Scherer SW, Stewart RJ, Hall AV, Shi XM, Tsui LC, Schappert KT. Structure and chromosomal localization of the human constitutive endothelial nitric oxide synthase gene. J Biol Chem. 1993;268:17478–17488. [PubMed] [Google Scholar]
- McQuillan LP, Leung GK, Marsden PA, Kostyk SK, Kourembanas S. Hypoxia inhibits expression of eNOS via transcriptional and posttranscriptional mechanisms. Am J Physiol. 1994;267:H1921–1927. doi: 10.1152/ajpheart.1994.267.5.H1921. [DOI] [PubMed] [Google Scholar]
- Mershon JL, Baker RS, Clark KE. Estrogen increases iNOS expression in the ovine coronary artery. Am J Physiol Heart Circ Physiol. 2002;283:H1169–1180. doi: 10.1152/ajpheart.00397.2000. [DOI] [PubMed] [Google Scholar]
- Michel T, Feron O. Nitric oxide synthases: which, where, how, and why? J Clin Invest. 1997;100:2146–2152. doi: 10.1172/JCI119750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathan C, Xie QW. Nitric oxide synthases: roles, tolls, and controls. Cell. 1994;78:915–918. doi: 10.1016/0092-8674(94)90266-6. [DOI] [PubMed] [Google Scholar]
- Navarro-Antolin J, Rey-Campos J, Lamas S. Transcriptional induction of endothelial nitric oxide gene by cyclosporine A. A role for activator protein-1. J Biol Chem. 2000;275:3075–3080. doi: 10.1074/jbc.275.5.3075. [DOI] [PubMed] [Google Scholar]
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS. Differential ligand activation of estrogen receptors ERalpha and ERbeta at AP1 sites. Science. 1997;277:1508–1510. doi: 10.1126/science.277.5331.1508. [DOI] [PubMed] [Google Scholar]
- Palmer RM, Ashton DS, Moncada S. Vascular endothelial cells synthesize nitric oxide from L-arginine. Nature. 1988;333:664–666. doi: 10.1038/333664a0. [DOI] [PubMed] [Google Scholar]
- Rosenfeld CR, Chen C, Roy T, Liu X. Estrogen selectively up-regulates eNOS and nNOS in reproductive arteries by transcriptional mechanisms. J Soc Gynecol Investig. 2003;10:205–215. doi: 10.1016/s1071-5576(03)00049-2. [DOI] [PubMed] [Google Scholar]
- Rupnow HL, Phernetton TM, Shaw CE, Modrick ML, Bird IM, Magness RR. Endothelial vasodilator production by uterine and systemic arteries. VII Estrogen and progesterone effects on eNOS. Am J Physiol Heart Circ Physiol. 2001;280:H1699–1705. doi: 10.1152/ajpheart.2001.280.4.H1699. [DOI] [PubMed] [Google Scholar]
- Sakai M, Okuda A, Hatayama I, Sato K, Nishi S, Muramatsu M. Structure and expression of the rat c-jun messenger RNA: tissue distribution and increase during chemical hepatocarcinogenesis. Cancer Res. 1989;49:5633–5637. [PubMed] [Google Scholar]
- Schwartz D, Mendonca M, Schwartz I, Xia Y, Satriano J, Wilson CB, Blantz RC. Inhibition of constitutive nitric oxide synthase (NOS) by nitric oxide generated by inducible NOS after lipopolysaccharide administration provokes renal dysfunction in rats. J Clin Invest. 1997;100:439–448. doi: 10.1172/JCI119551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwemmer M, Bassenge E. Assembly and characterization of canine heart endothelial nitric oxide synthase cDNA and 5′-flanking sequence by homology (RT-)PCR cloning. Nitric Oxide. 1999;3:254–264. doi: 10.1006/niox.1999.0234. [DOI] [PubMed] [Google Scholar]
- Shaul PW. Regulation of endothelial nitric oxide synthase: location, location, location. Annu Rev Physiol. 2002;64:749–774. doi: 10.1146/annurev.physiol.64.081501.155952. [DOI] [PubMed] [Google Scholar]
- Shiozawa T, Miyamoto T, Kashima H, Nakayama K, Nikaido T, Konishi I. Estrogen-induced proliferation of normal endometrial glandular cells is initiated by transcriptional activation of cyclin D1 via binding of c-Jun to an AP-1 sequence. Oncogene. 2004;23:8603–8610. doi: 10.1038/sj.onc.1207849. [DOI] [PubMed] [Google Scholar]
- Sladek SM, Magness RR, Conrad KP. Nitric oxide and pregnancy. Am J Physiol. 1997;272:R441–463. doi: 10.1152/ajpregu.1997.272.2.R441. [DOI] [PubMed] [Google Scholar]
- Srinivasan S, Hatley ME, Bolick DT, Palmer LA, Edelstein D, Brownlee M, Hedrick CC. Hyperglycaemia-induced superoxide production decreases eNOS expression via AP-1 activation in aortic endothelial cells. Diabetologia. 2004;47:1727–1734. doi: 10.1007/s00125-004-1525-1. [DOI] [PubMed] [Google Scholar]
- Vagnoni KE, Magness RR. Estrogen and lipopolysaccharide stimulation of prostacyclin production and the levels of cyclooxygenase and nitric oxide synthase in ovine uterine arteries. Biol Reprod. 1998;59:1008–1015. doi: 10.1095/biolreprod59.4.1008. [DOI] [PubMed] [Google Scholar]
- Vagnoni KE, Shaw CE, Phernetton TM, Meglin BM, Bird IM, Magness RR. Endothelial vasodilator production by uterine and systemic arteries. III Ovarian and estrogen effects on NO synthase. Am J Physiol. 1998;275:H1845–1856. doi: 10.1152/ajpheart.1998.275.5.H1845. [DOI] [PubMed] [Google Scholar]
- van der Heijden OW, Essers YP, Fazzi G, Peeters LL, De Mey JG, van Eys GJ. Uterine artery remodeling and reproductive performance are impaired in endothelial nitric oxide synthase-deficient mice. Biol Reprod. 2005;72:1161–1168. doi: 10.1095/biolreprod.104.033985. [DOI] [PubMed] [Google Scholar]
- Venema RC, Nishida K, Alexander RW, Harrison DG, Murphy TJ. Organization of the bovine gene encoding the endothelial nitric oxide synthase. Biochim Biophys Acta. 1994;1218:413–420. doi: 10.1016/0167-4781(94)90195-3. [DOI] [PubMed] [Google Scholar]
- Wariishi S, Miyahara K, Toda K, Ogoshi S, Doi Y, Ohnishi S, Mitsui Y, Yui Y, Kawai C, Shizuta Y. A SP1 binding site in the GC-rich region is essential for a core promoter activity of the human endothelial nitric oxide synthase gene. Biochem Biophys Res Commun. 1995;216:729–735. doi: 10.1006/bbrc.1995.2682. [DOI] [PubMed] [Google Scholar]
- Webb P, Nguyen P, Valentine C, Lopez GN, Kwok GR, McInerney E, Katzenellenbogen BS, Enmark E, Gustafsson JA, Nilsson S, Kushner PJ. The estrogen receptor enhances AP-1 activity by two distinct mechanisms with different requirements for receptor transactivation functions. Mol Endocrinol. 1999;13:1672–1685. doi: 10.1210/mend.13.10.0357. [DOI] [PubMed] [Google Scholar]
- Wedgwood S, Mitchell CJ, Fineman JR, Black SM. Developmental differences in the shear stress-induced expression of endothelial NO synthase: changing role of AP-1. Am J Physiol Lung Cell Mol Physiol. 2003;284:L650–662. doi: 10.1152/ajplung.00252.2002. [DOI] [PubMed] [Google Scholar]
- Weiner CP, Lizasoain I, Baylis SA, Knowles RG, Charles IG, Moncada S. Induction of calcium-dependent nitric oxide synthases by sex hormones. Proc Natl Acad Sci U S A. 1994;91:5212–5216. doi: 10.1073/pnas.91.11.5212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong F, Hull C, Zhande R, Law J, Karsan A. Lipopolysaccharide initiates a TRAF6-mediated endothelial survival signal. Blood. 2004;103:4520–4526. doi: 10.1182/blood-2003-06-2118. [DOI] [PubMed] [Google Scholar]
- Yi FX, Bird IM. Pregnancy-specific modulatory role of mitochondria on adenosine 5′-triphosphate-induced cytosolic [Ca2+] signaling in uterine artery endothelial cells. Endocrinology. 2005;146:4844–50. doi: 10.1210/en.2005-0414. [DOI] [PubMed] [Google Scholar]
- Yi FX, Magness RR, Bird IM. Simultaneous imaging of [Ca2+]i and intracellular NO production in freshly isolated uterine artery endothelial cells: effects of ovarian cycle and pregnancy. Am J Physiol Regul Integr Comp Physiol. 2005;288:R140–8. doi: 10.1152/ajpregu.00302.2004. [DOI] [PubMed] [Google Scholar]
- Yoshizumi M, Perrella MA, Burnett JC, Jr, Lee ME. Tumor necrosis factor downregulates an endothelial nitric oxide synthase mRNA by shortening its half-life. Circ Res. 1993;73:205–209. doi: 10.1161/01.res.73.1.205. [DOI] [PubMed] [Google Scholar]
- Yu Y, Platoshyn O, Zhang J, Krick S, Zhao Y, Rubin LJ, Rothman A, Yuan JX. c-Jun decreases voltage-gated K(+) channel activity in pulmonary artery smooth muscle cells. Circulation. 2001;104:1557–1563. doi: 10.1161/hc3801.095662. [DOI] [PubMed] [Google Scholar]
- Zhang R, Min W, Sessa WC. Functional analysis of the human endothelial nitric oxide synthase promoter. Sp1 and GATA factors are necessary for basal transcription in endothelial cells. J Biol Chem. 1995;270:15320–15326. doi: 10.1074/jbc.270.25.15320. [DOI] [PubMed] [Google Scholar]
- Zheng J, Bird IM, Melsaether AN, Magness RR. Activation of the mitogen-activated protein kinase cascade is necessary but not sufficient for basic fibroblast growth factor- and epidermal growth factor-stimulated expression of endothelial nitric oxide synthase in ovine fetoplacental artery endothelial cells. Endocrinology. 1999;140:1399–1407. doi: 10.1210/endo.140.3.6542. [DOI] [PubMed] [Google Scholar]
- Zheng J, Wen Y, Chen DB, Bird IM, Magness RR. Angiotensin II elevates nitric oxide synthase 3 expression and nitric oxide production via a mitogen-activated protein kinase cascade in ovine fetoplacental artery endothelial cells. Biol Reprod. 2005;72:1421–1428. doi: 10.1095/biolreprod.104.039172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A: A 1283-bp 5′flanking region of NOS3 gene plus the beginning of the coding region (22-bp) was isolated and sequenced, then deposited to GenBank with an accession number AY684193. The nucleotide position number is relative to the ATG initiation codon (+1). The nucleotide sequences for putative cis-acting elements are boxed and labeled with different colors. Abbreviations for cis-elements are as follows: AP-1, activator protein 1; SRE-1, sterol regulatory element-1; AP-2, activator protein 2; SSRE, shear stress response element; NF-1, nuclear factor-1; and CRE, cAMP response element; ER half: estrogen half-palindromic motif; SRE: sterol regulatory element; APR: acute-phase response; MAZ: myc-associated zine-finger protein; YY: Ying-yang; PAX: paired box containing genes; and PEA3: Ets family transcription factor.