Abstract
Cancer is a complex disease that involves the accumulation of both genetic and epigenetic alterations of numerous genes. Data in the Genetic Alterations in Cancer database for gene mutations and allelic loss [loss of heterozygosity (LOH)] in human tumors (e.g. lung, oral, esophagus, stomach and colon/rectum) were reviewed. Results for the genes and pathways implicated in tumor development at these sites are presented. Mutation incidence, spectra and codon specificity are described for lung, larynx and oral tumors. LOH occurred more frequently than gene mutations in tumors from all sites examined. The cell cycle gene, TP53 (all sites), and cell signaling gene, APC (colorectal and gastric cancers), were the only genes with similar incidences of LOH and mutation. Alterations of one or more cell cycle and cell signaling genes were reported for tumors from each site. Site-specific activation was apparent in the cell signaling mitogen-activated protein kinase oncogenes (KRAS in lung, HRAS in oral cancers and BRAF in esophageal and colorectal cancers). Analysis of genetic changes in lung tumors showed that the incidence of mutations in the TP53 and KRAS genes and the incidence of LOH in the FHIT gene were significantly greater in smokers versus non-smokers (P < 0.01). In lung and oral cancers, the TP53 GC → TA transversion frequency increased with tobacco smoke exposure (P < 0.05). Furthermore, the TP53 mutational hot spots for lung and laryngeal cancers from smokers included codons 157, 245 and 273, whereas for oral tumors included codons 280 and 281.
Introduction
One of the cornerstones to understanding cancer is knowledge of the genes and mutations that initiate tumor development and give cells a selective growth advantage that allows tumor progression to occur. It is now well established that a clone of cancerous cells is unlikely to be the result of a single mutational event in a stem cell but rather a series of accumulated genetic and epigenetic events induced or inhibited by the interplay of DNA and environment (1–3). Single nucleotide polymorphisms are one mechanism whereby the genetic background has been implicated in defining an individual’s susceptibility to cancer; the Tp53 Arg/Pro polymorphism at codon 72 is one example (4–7), yet external environmental exposures also play an important role. These can give rise to a number of different genetic alterations, the most common of which include: chromosomal changes such as loss of heterozygosity (LOH), rearrangements, deletions, gains, polyploidy and translocations; gene mutations such as base substitutions, small insertions and deletions, allelic loss, amplification and rearrangements; and epigenetic events such as alteration in DNA methylation.
Perhaps the best-documented external factor associated with human cancer is that of exposure to tobacco smoke and lung cancer. Fifty years ago, Doll and Hill (8) first demonstrated a dose–response relationship between the number of cigarettes smoked and lung cancer death rate. Subsequent studies have established several molecular targets for tobacco smoke including the TP53, KRAS, FHIT, RB1 and HPRT genes (9–12). Recent studies continue to show that smoking is associated with cancer risk including not only lung cancer risk but also risk for oral, larynx, esophageal, gastric cancers and colorectal hyperplasia/polyps (13–15).
To help advance the understanding of how different genes contribute to tumorigenesis, databases have been designed to collect and report gene mutation data for many types of cancers. One of the best-known databases is the International Agency for Research on Cancer TP53 Mutation Database (http://www-p53.iarc.fr/). Others include the CDKN2A (https://biodesktop.uvm.edu/perl/p16) and androgen receptor gene databases (http://www.androgendb.mcgill.ca); the breast cancer gene (http://www.tumorgene.org/Breast/index.html) and the oral cancer gene (http://www.tumor-gene.org/Oral/oral.html) databases; and the molecular cytogenetics database (http://www.ncbi.nlm.nih.gov/sky/). These resources provide important data for single genes and/or cancers at single tumor sites, but in order to better understand the similarities and differences in tumorigenesis at different sites and define the types of alterations and accumulated genetic events that contribute to the tumor development, databases with information for multiple genes, cancer topographies (anatomical sites of the tumors), alterations and exposures are critical. Data from one such source, the Genetic Alterations in Cancer (GAC) database (http://dir-apps.niehs.nih.gov/gac/), are evaluated and summarized in this paper.
The GAC database was developed by National Institute of Environmental Health Sciences as a web-based system for collecting and summarizing data reported in the published literature for genetic alterations in various types of human cancers and laboratory rodent tumors (16). It provides a unique tool for assessing groups of related genes for mutations, allelic loss and homozygous deletions based on species and tumor site. Tumor morphologies from different topographical sites (regions of the body) and exposures to environmental agents can also be distinguished. Collating these types of data allows a broader view of tumor development and facilitates comparative analysis of multiple genes in genetic pathways to cancer rather than focusing on single genes.
Data in the GAC database for genetic alterations in human tumors were evaluated with an emphasis on results from smokers compared with non-smokers. Tumors sites were selected based on their potential association with tobacco smoke exposure (13–15). The results are summarized to demonstrate how disruption of genetic pathways, such as cell cycle control and cell signaling, by mutations and allelic loss in one or more genes can contribute to tumor development. This analysis is designed to provide further insight into the modes of action of carcinogenesis. We illustrate how changes in specific genes, and different patterns of gene alterations in various topographical sites, may serve as markers of exposure.
Materials and methods
Data selection
Mutation data were obtained from the GAC database (http://dir-apps.niehs.nih.gov/gac/) which was last updated in April 2007 (containing ~3175 records). A complete description of GAC, including data acquisition, was described previously (16).
Tumor groups were defined as ‘Spontaneous’ (no known exposure reported), ‘Tobacco (smoke)’ (samples from smokers, either current or previous cigarette/pipe) and ‘Unexposed (Tobacco)’ (samples from non-smokers reported to have never smoked). The datasets were unrestricted with regard to tumor origin (primary, metastatic or recurrent tumors) and morphology.
Data from human tumors evaluated for somatic mutations (missense, nonsense and silent point mutations, frameshift insertion and deletions, in-frame insertions and deletions) or LOH (single allele loss of intragenic markers or markers tightly linked to a gene) were selected for evaluation. The results are presented as separate datasets; bi-allelic alterations in single tumors are not shown. Genes included in the analysis are shown in Table I and are grouped in the cell cycle or cell signaling pathway in which they play a role (this is not necessarily their only pathway). Lists of references from which datasets were obtained are given as supplementary data (available at Carcinogenesis Online).
Table I.
Percentage of tumors with gene alterations in spontaneous human lung, oral, esophageal, stomach and colorectal cancers
| Lung
|
Oral
|
Esophagus
|
Stomach
|
Colon/rectum
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| % tumors with mutationsa | % tumors with LOH | % tumors with mutations | % tumors with LOH | % tumors with mutations | % tumors with LOH | % tumors with mutations | % tumors with LOH | % tumors with mutations | % tumors with LOH | |
| Cell cycle | ||||||||||
| BRCA1 | NA | NA | NA | NA | NA | 44 (42/96) | NA | 17 (11/64) | NA | 41 (201/494) |
| CDKN2A | 9 (45/494)b | NA | 7 (24/339)c | 60 (3/5) | 10 (69/700) | 59 (76/128) | 3 (9/354) | NA | 0 (0/77) | 19 (11/57) |
| CDKN2B | 0 (0/146) | 25 (2/8) | 0 (0/32) | NA | 2 (3/137) | NA | 0 (0/91) | NA | NA | NA |
| RB1 | 0 (0/21) | 47 (73/155) | NA | 12 (3/26) | 0 (0/49) | 40 (106/263) | NA | 31 (4/13) | 0 (0/33) | 14 (34/238) |
| TP53 | 42 (608/1457) | 49 (102/208) | 39 (261/661) | 48 (108/224) | 44 (1295/2928) | 57 (216/379) | 26 (749/2853) | 39 (148/375) | 40 (1368/3402) | 51 (606/1177) |
| Cell signaling: Wnt | ||||||||||
| APC | 1 (3/288) | 28 (20/72) | 4 (3/68) | 48 (11/23) | 2 (4/171) | 48 (99/207) | 18 (78/445) | 21 (81/383) | 36 (923/2563) | 33 (327/1004) |
| CTNNB1 | 0.8 (2/251) | NA | 0 (0/33) | NA | 0 (0/134) | NA | 4 (31/721) | NA | 6 (69/1199) | NA |
| Cell signaling: MAPK/ERK | ||||||||||
| BRAF | 2 (7/411) | NA | 0 (0/13) | NA | 11 (2/19) | NA | 1 (9/702) | NA | 14 (215/1559) | NA |
| HRAS | 1 (4/454) | 24 (21/89) | 11 (19/170) | 29 (7/24) | 0 (0/176) | 35 (6/17) | 2 (6/381) | 37 (7/19) | 1 (2/159) | 3 (1/35) |
| KRAS | 19 (506/2714) | NA | 4 (5/122) | NA | 2 (5/216) | NA | 6 (121/1949) | NA | 31 (2909/9504) | 0 (0/33) |
| NRAS | 1 (7/505) | NA | 0 (0/112) | NA | 0 (0/122) | NA | 2 (4/235) | NA | 6 (11/194) | NA |
| Cell signaling: apoptosis | ||||||||||
| DCC | NA | 41 (7/17) | NA | NA | NA | 26 (37/143) | NA | 33 (116/348) | NA | 48 (300/629) |
| FHIT | 2 (1/41) | 56 (199/353) | 0 (0/3) | NA | 34 (30/89) | 40 (61/153) | 5 (6/121) | 32 (79/248) | NA | 40 (58/146) |
NA, not available; ERK, extracellular signal-regulated kinase.
Mutations were somatic missense, nonsense and silent point mutations, frameshift deletions and insertions, in-frame deletions and insertions.
Values in parentheses show number of tumors with mutation/total number of tumors studied.
Values in parentheses show number of tumors with LOH/total number of informative samples.
Statistical analysis
Gene mutation and LOH data for human lung, oral, esophageal, stomach and colon/rectum tumors were retrieved from the GAC database of peer-reviewed literature. To analyze the incidence of alterations, data were organized by tumor site and agent group [spontaneous, tobacco smoke, unexposed (tobacco)]. Data from detailed data lists showing the results for each tumor sample from all studies retrieved were generated by the program, captured and prepared for broad-based statistical analysis. This included data from a total of 181 peer-reviewed studies of lung tumors, 83 studies of oral tumors, 133 studies of esophageal tumors, 163 studies of stomach tumors and 233 studies of colon/rectum tumors from smokers, non-smokers or individuals for which smoking status was not reported (references to all studies are in the supplementary data available at Carcinogenesis Online).
The mutation incidence was compared for pairs of data groups (e.g. data for TP53 or KRAS or APC, etc. from lung versus oral tumors, lung versus esophageal tumors, oral versus stomach tumors, etc. or lung tumors from smokers versus non-smokers) using a standard two-sample Z-statistic for proportions. However, since sample proportions were derived from multiple studies and variables between studies (e.g. age, gender, exposure, and other environmental factors) were not specifically factored in, extra-variation between studies was taken into account by using a non-parametric estimator for extra binomial variability (17). The P values for the tests were derived using a non-parametric bootstrap methodology (18). TP53 mutational spectra were compared for smokers and non-smokers in lung and oral cancers by the Chi-square test.
Results
Altered genetic pathways
Data for the incidence of somatic point mutations, in-frame and frameshift insertions and deletions and LOH at gene-specific markers in human lung, oral, esophageal, gastric and colorectal cancers are summarized in Table I. The data are from studies of spontaneous tumors (exposure to potential carcinogens was not reported) and are organized by tumor site and genetic pathway. Gene mutations were universally studied more often than LOH for all of the genes and tumor sites, yet LOH generally occurred at higher frequencies.
Cell cycle genes
Disruption of the cell cycle was a common event in all cancers studied (Table I). This was primarily the result of alterations in the TP53 gene, which occurred through both LOH and point mutations in approximately 40–50% of tumors studied. In other cell cycle genes, allelic loss was typically found more often than point mutations (see the CDKN2A gene for oral, esophageal and colon/rectum tumors and RB1 gene for lung, esophagus and colorectal cancers).
Allelic loss of BRCA1, CDKN2A, RB1 and TP53 was detected in colorectal cancer but mutations were only reported for TP53. LOH was also reported for these same four genes in esophageal tumors, but at a higher frequency than in colorectal cancer. Mutations were seen in TP53 as well as in the CDK4 inhibitor genes, CDKN2A and CDKN2B. In gastric tumors, genes that were studied for either allelic loss or mutations demonstrated low incidences of mutations in CDKN2A and CDKN2B. Moreover, the incidence of TP53 mutations was significantly lower at this site than at all the other topographic sites studied (P < 0.01). Fewer studies examined the cell cycle genes in oral cancer, but of those that did, CDKN2A mutations were detected in 7% of the cases, CDKN2B mutations were not found and LOH in RB1 occurred in 12% of cases. TP53 represented the majority of the data for this group with mutations in 39% and LOH in 48% of the tumors. Lung tumors showed a high incidence of TP53 and RB1 allelic loss and some evidence of CDKN2B loss but mutations only in TP53 (42%) and CDKN2A (9%).
Cell signaling: Wnt
Activation of the Wnt pathway in tumor development is most often studied through the APC and CTNNB1 genes; however, at the topographical sites included in Table I, there was little evidence of CTNNB1 mutations reported in the studies reviewed. In lung and oral tumors, only two of >250 tumors analyzed had mutations and in digestive tract cancers, the incidence of mutation was ~5% for >2000 tumors. Studies of LOH at these tumor sites were not available.
Potential loss of APC function was indicated by the presence of both mutations and LOH in colon/rectum (36% mutations and 33% LOH) and gastric (18% mutations and 21% LOH) cancers. In contrast, for lung, oral and esophageal tumors APC alterations were reported most frequently as LOH rather than mutations. The incidences of allelic loss were 28% (lung) and 48% (oral and esophagus) and were at a level comparable with that in colorectal tumors but gene mutations occurred in < 5% of the tumors analyzed.
Cell signaling: mitogen-activated protein kinase/extracellular signal-regulated kinase
Activation of the mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase pathway oncogenes through mutation or allelic loss occurred most frequently in colorectal cancer, primarily through mutations of the KRAS (31%) and BRAF (14%) genes (Table I). In gastric cancers, the MAPK pathway was rarely activated by gene mutations but evidence from a low number of tumors suggests that HRAS allelic loss may occur. Similar allelic losses were detected in esophageal, oral and lung cancers but further studies are needed to verify this. Likewise, evidence based on a low number of tumors suggests that the BRAF gene may be activated more frequently in esophageal tumors than the RAS genes, which were only rarely activated.
The incidence of KRAS mutations was significantly higher (P < 0.01) in spontaneous lung tumors (19%) compared with oral tumors (4%). Overall, KRAS mutations occurred at a significantly higher incidence in colon/rectum tumors (31%) than at any other site examined. HRAS mutations were more prevalent in oral tumors than lung or digestive tract tumors, reaching significance in esophagus (P < 0.05) and stomach (P < 0.05); this has been linked to tobacco exposure, primarily chewing tobacco. (19). BRAF and NRAS alterations were uncommon in lung, oral and gastric tumors.
Cell signaling: apoptosis
Other genes implicated in tumorigenesis include DCC and FHIT; both are thought to be tumor suppressor genes. DCC functions as a dependence receptor required for apoptosis induction (20). FHIT also induces apoptosis (21). In digestive tract tumors (esophagus, stomach and colon/rectum), allelic losses of DCC or FHIT were detected in 26–48% of all cases (Table I). In lung tumors, loss of a FHIT allele was detected in 56% of cases and evidence based on a low number of tumors suggested that DCC allele loss was also frequent (41%). Results from one study of lung tumors indicated that point mutations in FHIT are rare (22); mutations were also infrequent in studies of stomach tumors (6 of 121 tumors) but not esophageal cancer (34% of 89 tumors had mutations).
Gene pathways in lung cancer of smokers and non-smokers
One of the strongest etiological factors for the development of tumors in lung and head and neck cancers is tobacco smoke. Although tumors shown in Table I were considered spontaneous in nature, the group undoubtedly comprised smokers and non-smokers. Exposures to polycyclic aromatic hydrocarbons including benzo[a]pyrene, aromatic amines, aldehydes and tobacco-specific nitrosamines from tobacco smoke cause DNA adduct formation and elicit DNA alterations (23–25). To analyze the extent of these alterations, we have compared incidences of mutations and LOH in lung tumors of smokers and non-smokers (Table II). While the list of genes studied was not exhaustive, both smokers and non-smokers demonstrated alterations in the same genes but with some significant differences in the incidence of alterations. The most frequently altered cell cycle gene was TP53 (Table II) where a statistically significant increase in the incidence of mutations was observed between smokers (39%) and non-smokers (26%; P < 0.01). Allele loss occurred in ~40% of cases but this did not vary with tobacco exposure. Mutations in CDKN2A were rare and although the incidence of LOH was higher it did not differ significantly between smokers and non-smokers. In the Wnt pathway, evidence based on a low number of tumors suggests that APC loss occurred less frequently in smokers than non-smokers. The number of KRAS mutations was significantly higher in smokers (20%) compared with non-smokers (3%; P < 0.001), yet no mutations were detectable in HRAS or NRAS genes (Table II). Data for cell signaling pathways involving FHIT and DCC genes were restricted to allelic loss and showed FHIT LOH occurred significantly more frequently in tumors from smokers compared with non-smokers (P < 0.01). DCC loss was evident in 16% of smokers but did not occur in non-smokers.
Table II.
Percentage of lung tumors from smokers and non-smokers with gene alterations
| Tobacco smokea |
Unexposedb |
|||
|---|---|---|---|---|
| % tumors with mutations | % tumors with LOH | % tumors with mutations | % tumors with LOH | |
| Cell cycle | ||||
| CDKN2A | 4 (6/136) | 33 (8/24) | 0 (0/47) | 29 (5/17) |
| CDKN2B | NA | NA | NA | NA |
| RB1 | NA | 47 (47/99) | NA | 19 (10/53) |
| TP53 | 39c (627/1591) | 38 (47/125) | 26 (105/410) | 40 (14/35) |
| Cell signaling: Wnt | ||||
| APC | NA | 43 (31/72) | NA | 67 (4/6) |
| Cell signaling: MAPK/ERK | ||||
| BRAF | NA | NA | NA | NA |
| HRAS | 0 (0/122) | NA | 0 (0/42) | NA |
| KRAS | 20c (616/3114) | NA | 3 (22/696) | NA |
| NRAS | 0 (0/193) | NA | 0 (0/69) | NA |
| Cell signaling: apoptosis | ||||
| DCC | NA | 16 (13/81) | NA | 0 (0/9) |
| FHIT | NA | 57c (262/460) | NA | 27 (34/126) |
ERK, extracellular signal-regulated kinase.
Tumors from current or previous tobacco smokers.
Tumors from non-smokers (never smokers).
Per cent tumors with alteration statistically higher in smokers compared with unexposed, non-smokers (P < 0.01).
TP53 spectra in tumors: smokers and non-smokers
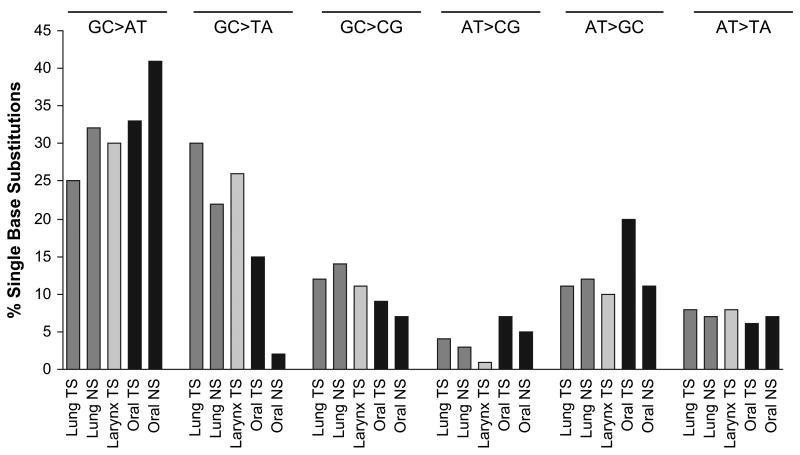
Evidence shows that the mutational specificity of tobacco smoke occurs at the level of the base rather than codon with GC → TA transversions indicative of exposure (26,27). To analyze the effect of tobacco smoke exposure on the incidence of single base substitutions at primary sites of smoke deposition: lung, larynx and oral cavity, we have compared TP53 mutational spectra from smokers and non-smokers (Figure 1). Laryngeal tumors from non-smokers were not included in these analyses due to insufficient data. When all single base substitutions were considered, a higher frequency of GC → AT and GC → TA substitutions occurred in lung, larynx and oral tumors compared with other substitutions. Substitutions of AT bases and GC → CG transversions occurred infrequently (< 20% of cases) and did not vary significantly with tobacco smoke exposure. Figure 1 demonstrates the percentage of single base substitutions that occurred in different tumor groups. No difference was observed between the percentage of GC → AT transitions and GC → TA transversions in lung (25% versus 30%) and laryngeal (30% versus 26%) tumors of smokers in contrast to oral tumors where GC → AT substitutions in smokers (33%) were significantly higher than GC → TA transversions (15%; P < 0.001). Similarly, in oral tumors from non-smokers the number of GC → AT transitions (41%) was significantly higher than the number of GC → TA transversions (2%; P < 0.001).
Fig. 1.
TP53 mutational spectra for lung, oral and laryngeal cancers showing the total percentage of single base pair substitutions that were GC → AT, GC → TA, GC → CG, AT → CG, AT → GC and AT → TA for the entire gene. TS indicates tumors arose in current or previous tobacco smokers; NS indicates tumors arose in non-smokers. Black bars designate oral cancers; dark gray designate lung and light gray designate larynx cancers.
When the frequency of substitutions was compared between smokers and non-smokers, it was evident that exposure to tobacco smoke significantly increased the number of GC → TA transversions. In lung tumors, the percentage of GC → TA mutations in non-smokers was 22%, whereas in smokers it was 30% (P < 0.05) with the increase occurring predominantly at CpG sites (data not shown). Oral cancers displayed a similar increase in GC → TA substitutions after exposure to tobacco smoke (2% in non-smokers compared with 15% in smokers; P < 0.05) and these occurred at both CpG and non-CpG sites. This increase was accompanied by a decrease in the frequency of GC → AT transitions following exposure to tobacco. In neither oral nor lung cancers did the changes in frequency of GC → AT substitutions between smokers and non-smokers reach significance. Substitutions of other bases did not differ significantly with tobacco smoke exposure (GC → CG, AT → CG, AT → GC and AT → TA).
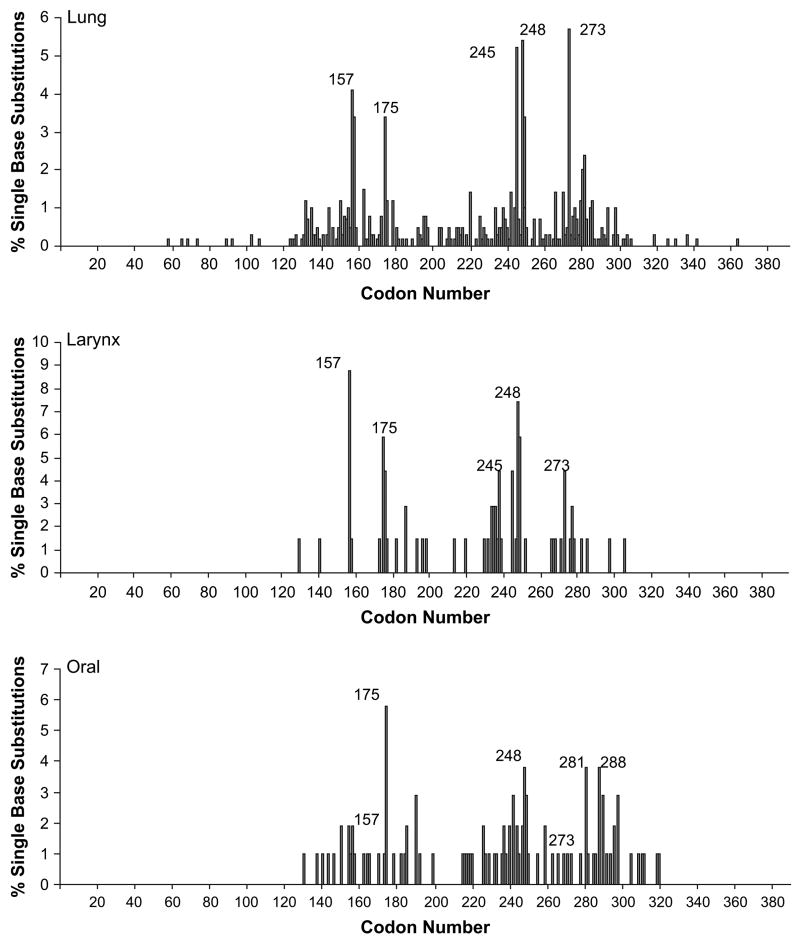
Although the mutational specificity of tobacco smoke occurs at the nucleotide level rather than the codon level, in vitro evidence demonstrates that benzo[a]pyrene binds with greatest strength to codons 157, 248 and 273 (28–30). We have analyzed these codons for the presence of mutations. Positional spectra for tumors from smokers clearly reveal the presence of mutations at these codons in lung and larynx tumors (Figure 2). In oral tumors, mutations in these codons occurred most frequently at codon 248; codons 157 and 273 mutations were less numerous. Mutations at codon 175 were common in all cancers and also codon 245 in lung and larynx tumors. The latter codons are considered common mutational hot spots present in many types of cancer and are not necessarily associated with tobacco exposure (31).
Fig. 2.
TP53 positional spectra showing the percentage of single base substitutions occurring at each codon in lung, larynx and oral tumors from smokers.
To determine whether the base substitutions in the most frequently mutated codons in smokers (as reported in the literature; 28, 29, 30, 31) showed specificity for tobacco smoke exposure, the percentage of GC → TA transversions occurring at codons 157, 175, 245, 248 and 273 was determined (Table III). In tumors other than lung cancer, this type of substitution is an infrequent occurrence (31); however, in lung tumors from smokers all the frequently mutated codons demonstrated GC → TA transversions (25–88%) indicating a possible role for tobacco smoke in their formation. In comparison, tumors from non-smokers rarely exhibited this substitution (Table III). Codons 157, 245 and 273 harbored GC → TA transversions in larynx cancers but of these, only codon 157 was a mutational hot spot in smokers. Codons 175 and 248 did not display GC → TA changes, instead GC → AT and GC → CG were observed. In oral cancers from smokers, codon 157 displayed 100% GC → TA transversions, yet this was not a frequently mutated codon. Mutational hot spots were evident at codons 175, 248, 281 and 288, yet none of these codons displayed GC → TA changes. At other codons, the percentage of GC → TA substitutions was 66% and 100% at codons 191 and 298, respectively (data not shown). In non-smokers, mutations were not observed at codons 157 and 175 and none of the substitutions at codons 245, 248 and 273 were GC → TA.
Table III.
Percentage of TP53 mutations at codons 157, 175, 245, 248 and 273 that were GC → TA transversions
| % GC → TA transversion mutations
|
|||||
|---|---|---|---|---|---|
| Lung TS | Lung NS | Larynx TS | Oral TS | Oral NS | |
| Codon 157 | 88 | 0 | 50 | 100 | NA |
| Codon 175 | 30 | 0 | 0 | 0 | NA |
| Codon 245 | 61 | 0 | 67 | 0 | 0 |
| Codon 248 | 25 | 0 | 0 | 0 | 0 |
| Codon 273 | 50 | 17 | 33 | 0 | 0 |
TS, tobacco smokers (current or previous); NS, non-smokers (never); NA, no mutation data available.
Discussion
In this paper, we have presented an analysis of published data collected from a large number of studies and analyzed using the GAC database. In performing this analysis, we have combined data from individual studies, some of which were small, to look for overall trends in gene alterations in tumors while minimizing the effect that single study anomalies have on the conclusions.
Current understanding of the molecular pathways of cancer development describes a multitude of genetic and epigenetic changes that accumulate as tumors develop (32). To understand the roles played by different genes in this process, we have characterized some of the genetic changes that occur in tumors from different topographical sites. Several mechanisms are known to activate or inactivate genes; we have presented data for two that are commonly studied: mutation and gene LOH. Other types of alterations are known to affect genes including homozygous deletion, methylation and chromosome LOH; these undoubtedly also play a vital role in tumor development but have not been evaluated here.
Greenman et al. (33) recently analyzed 518 protein kinase genes from 210 diverse cancers for mutations. The authors concluded that the majority of the somatic mutations detected in these genes were ‘passenger’ mutations that did not directly effect tumor development. Similarly, Sjoblom et al. (34) sequenced 13 023 different genes in breast and colorectal cancers and showed that on average ~90 mutations per tumor were observed and that 11 genes per tumor were mutated at a significant frequency. However, these studies did not take into account other types of genetic alteration such as LOH that also play a significant role in tumor formation. The data we have presented in this paper demonstrates that in oral tumors and tumors of the digestive tract and lung, genes are more frequently affected by allele loss than by mutation. Gene LOH was reported in each tumor group analyzed for allelic loss and the incidence was >30% in over two-thirds of the studies. These data probably represent an underestimate of the total frequency of allelic loss because analysis was restricted to intragenic markers and markers tightly linked to a gene. Chromosome markers located in the vicinity of a gene or gene locus that could conceivably extend into the gene were not considered in these analyses of gene LOH.
When gene mutations are considered in parallel with allele loss, it is evident that TP53 was one of the few genes inactivated at similar frequencies by allele loss and mutation; this occurred at all cancer sites examined. The only other example of this occurring was in the APC gene and this was restricted to gastric and colorectal cancers. Although the datasets defining the mutation and LOH data are distinct and cannot be used to infer bi-allelic inactivation, it is tempting to conclude that this data show TP53 and in some instances APC fulfill Knudson’s theory of gene inactivation in cancer development. Although several other tumor suppressor genes have been studied, they typically demonstrated very low levels of gene mutations in association with higher levels of LOH. Whether these genes perform a role in cancer development and what that role is remains unclear but possibly gene inactivation occurs by an alternative mechanism: gene methylation or germ line mutation. Alternatively, single allele inactivation may exert a dominant oncogenic effect or it may simply confer a predisposition to cancer. The number of genes and topographical sites where this occurred might suggest this as an important avenue of future research.
Data for alterations of oncogenes were less numerous and largely restricted to mutation studies of the RAS and BRAF genes of the MAPK/extracellular signal-regulated kinase pathway. No conclusions could be drawn concerning the role LOH plays in activation of these genes because of the lack of information but mutations appeared to play site-specific roles in the development of lung (KRAS), oral (HRAS), esophageal (BRAF) and colorectal (BRAF and KRAS) cancers. In the case of lung and oral cancers, it is possible that these were the result of site-specific exposures to environmental carcinogens (tobacco smoke or chewing tobacco). Still with the addition of allelic loss data for these genes, the specificity of gene activation for specific topographies may be negated.
When considered by topography, it is evident that alterations in cell signaling play a role in cancer formation: in colorectal and gastric cancer through genes of the Wnt pathway and the cell cycle genes, particularly TP53, at all sites of the digestive tract, oral and lung cancers. The potential for cell cycle disruption was particularly evident in esophageal cancers where LOH was detectable in a number of different genes. Constitutive transcriptional activation of genes involved in signal transduction in the MAPK/extracellular signal-regulated kinase pathway occurred most frequently in colorectal and lung cancers. All topographies but particularly lung cancers demonstrated alterations in FHIT and DCC, suggesting that inhibition of apoptosis was common to many tumors.
Some of the most well documented gene alterations in these data-sets were KRAS mutations in lung cancers. The reason for this abundance of data is that it represents an instance where a common environmental exposure, tobacco smoke, evokes a specific genetic response (35). When data for tumors with known exposure to tobacco smoke are compared with those that were unexposed, the incidence of KRAS and TP53 mutations was significantly higher in smokers. Likewise, the incidence of allelic loss of the FHIT gene in smokers was significantly higher than in non-smokers. Other gene alterations have not been reported in sufficient numbers to draw conclusions about the effect of smoking on their incidence. This analysis does demonstrate that the range of genes activated in smokers and non-smokers was identical, suggesting that one mechanism by which tobacco can give rise to lung cancer is through an increased frequency of genetic alterations. Of course, these analyses are limited by the scope of the published literature; studies tend to focus on the same, well-characterized genes resulting in an absence of information for many genes and genetic pathways. In future, studies of a broader spectrum of genes would help to ascertain whether tumorigenesis in smoking-related lung cancers is the result of alterations in genetic pathways unique to tobacco smoke exposure and distinct from tumors with no exposure to tobacco.
There are >60 carcinogenic components of tobacco smoke which include polycyclic aromatic hydrocarbons, aromatic amines, aldehydes and nitrosamines (15); direct exposure to these carcinogens occurs not only in lung but also in the upper aerodigestive tract, particularly the mouth and larynx. In the lung, it has been shown that preferential binding of benzo[a]pyrene and acrolein to guanine residues leads to an increased incidence of GC → TA transversions (25,36). From this it follows that GC → TA transversions do not frequently occur at sites not directly exposed to tobacco smoke (37,27). Indeed, our analysis of oral tumors from non-smokers demonstrated this; GC → TA substitutions were present as only 2% of the mutations. Surprisingly, this was not the case for lung cancers from non-smokers where 22% of alterations were GC → TA transversions. It is possible that this number was an overestimation of the true rate due to the inclusion of a potentially anomalous study. Gao et al. (38) reported a 66% mutation rate in lung tumors of non-smokers compared with < 36% for all other studies. Each tumor in this study demonstrated multiple mutations such that 10 of 15 mutated tumors had 48 mutations comprising 31% of the total mutations for the group. This undoubtedly influenced the results for this group, yet even if this study is excluded, the percentage of mutations that were GC → TA in lung tumors (16% excluding the Gao study) is significantly higher than oral tumors (2%). Why this should occur is not clear but it is possible that lung exposure to polycyclic aromatic hydrocarbons, aromatic amines and nitrosamines derives from alternative sources that are not pertinent to oral tumorigenesis. One of these might be side-stream tobacco smoke for passive/involuntary smokers. Qualitatively, sidestream smoke has the same chemical constituents as mainstream smoke and could result in exposure in non-smokers (39). Another possible exposure that might present an environment risk to non-smokers is the complex chemical mixtures of diesel fuel and vehicle exhaust fumes (40,41). Despite the differences in the lung and oral non-smoking groups, our data show an increase in the percentage of GC → TA transversions in tobacco smokers; this was most apparent in oral tumors but was also evident in lung tumors. The increase in GC → TA transversions was associated with an accompanying decrease in the percentage of GC → AT transitions; this was seen in both lung and oral tumors and was mirrored in laryngeal tumors of smokers. The fact that this decrease occurred particularly at CpG sites supports the view that it was the consequence of a chemical exposure (42).
Efforts made to analyze some of the individual chemical components of tobacco smoke in lung tumors of laboratory rodents have supported these findings; GC → TA transversions were prevalent in lung tumors of mice exposed to benzo[a]pyrene (16,43–45). Interestingly, 4-(N-nitrosomethylamino)-1-(3-pyridyl)-1-butanone also found in a variety of tobacco products (chewing tobacco, snuff, cigarettes and cigars) primarily causes GC → AT transitions in exposed mice (16,46–49). Although exposure to 4-(N-nitrosomethylamino)-1-(3-pyridyl)-1-butanone undoubtedly occurs in smokers, our analysis of the human data in the literature did not demonstrate any increase in the percentage of GC → AT transitions in lung or oral cancers.
Unlike other carcinogens such as aflatoxin B1, the mutagenic specificity of tobacco smoke exposure is defined at the base level by G → T transversions rather than at the codon level (50–52). Still comparison of positional spectra and incidences of GC → TA transversions for lung, larynx and oral tumors from smokers define several codons as hot spots of mutations in TP53, all of which occur in the DNA-binding domain of the protein. For both lung and larynx, these were codons 157, 245, 248 and 273; this agrees with in vitro studies that show benzo[a]pyrene derivatives bind with greatest strength to codons 157, 248 and 273 (28,51,52). In contrast, mutational hot spots in oral tumors were restricted to codons 175 and 248 when the types of base change occurring at these codons was considered codon 248 showed 0–25% incidence of G → T substitutions. This would suggest that the benzo[a]pyrene component of tobacco smoke did not play a prominent role in mutation at this site.
Codons 175 and 245 are commonly mutated in different types of cancer and are not usually associated with specific exposures. Despite this, lung and larynx (codon 245 only) tumors frequently demonstrate G → T transversions at these sites, suggesting that tobacco smoke may play a role in their mutation in some instances. In oral cancer, this did not occur; codon 175 and 245 mutations were not G → T transversions. Of course, this does not preclude tobacco smoke from playing a role in their mutation but it is unlikely that benzo[a]pyrene is involved; instead 4-(N-nitrosomethylamino)-1-(3-pyridyl)-1-butanone may play a role. Moreover, the sites at which the highest incidences of G → T substitutions occurred were not mutational hot spots. It is unclear why the differences in mutational hot spots exist for these topographies; there was a difference in tumor morphology between oral (all squamous cell carcinomas) and lung (non-small cell and small cell carcinomas) cancer but this was unlikely to be one of the factors because laryngeal tumors were all squamous cell carcinomas. Perhaps, they are a reflection of prominent roles played by other factors such as human papilloma virus (HPV) or alcohol in the development of oral tumors. Further studies are needed to resolve these possibilities.
In conclusion, this paper has highlighted how study of peer-reviewed literature using the GAC database (http://dir-apps.niehs.nih.gov/gac/) can bring about a new understanding of the carcinogenic processes. We show that generally the same genes are part of multiple pathways to cancer in all target sites examined (lung, oral, esophagus, stomach, colon/rectum) with changes commonly occurring in TP53, Ras family and CDK family genes. What distinguished gene changes at a one particular target site versus another was the per cent incidence of mutation or LOH for a particular gene. Moreover, environmental exposure defined a further level of specificity with the spectra of substitutions being characteristic of exposure. Compiling information on gene changes in cancer identifies target gene changes that can be used to develop biomarkers for the cancer disease process and strategies for disease prevention.
Supplementary Material
Supplementary material
Supplementary data can be found at http://carcin.oxfordjournals.org/
Acknowledgments
Funding
The Genetic Alterations in Cancer System (https://dir-apps.niehs.nih.gov/gac/) and cancer analyses supported by the Intramural Research Program of the National Institute of Environmental Health Sciences; National Institutes of Health (Contract No. N43-ES-15477).
We thank Dr G. Kissling and K. Witt for their review of this paper and helpful suggestions; also A. Rashid for development of the program routines that operate the GAC system.
Abbreviations
- GAC
Genetic Alterations in Cancer
- LOH
loss of heterozygosity
- MAPK
mitogen-activated protein kinase
Footnotes
Conflict of Interest Statement: None declared.
Contributor Information
I.A. Lea, Integrated Laboratory Systems, Inc., Research Triangle Park, NC 27709, USA
M.A. Jackson, Integrated Laboratory Systems, Inc., Research Triangle Park, NC 27709, USA
X. Li, Integrated Laboratory Systems, Inc., Research Triangle Park, NC 27709, USA
S. Bailey, Integrated Laboratory Systems, Inc., Research Triangle Park, NC 27709, USA
S.D. Peddada, National Institutes of Environmental Health Sciences, Research Triangle Park, NC 27709, USA
J.K. Dunnick, National Institutes of Environmental Health Sciences, Research Triangle Park, NC 27709, USA
References
- 1.Armitage P, et al. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br J Cancer. 2004;91:1983–1989. doi: 10.1038/sj.bjc.6602297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doll R. The age distribution of cancer and a multistage theory of carcinogenesis. Int J Epidemiol. 2004;33:1183–1184. doi: 10.1093/ije/dyh359. [DOI] [PubMed] [Google Scholar]
- 3.Shields PG, et al. Cancer risk and low-penetrance susceptibility genes in gene-environment interactions. J Clin Oncol. 2000;18:2309–2315. doi: 10.1200/JCO.2000.18.11.2309. [DOI] [PubMed] [Google Scholar]
- 4.Hu Y, et al. The p53 codon 72 proline allele is associated with p53 gene mutations in non-small cell lung cancer. Clin Cancer Res. 2005;11:2502–2509. doi: 10.1158/1078-0432.CCR-04-1913. [DOI] [PubMed] [Google Scholar]
- 5.Agorastos T, et al. p53 codon 72 polymorphism and risk of intra-epithelial and invasive cervical neoplasia in Greek women. Eur J Cancer Prev. 2000;9:113–118. doi: 10.1097/00008469-200004000-00007. [DOI] [PubMed] [Google Scholar]
- 6.Clapp RW, et al. Environmental and occupational causes of cancer re-visited. J Public Health Policy. 2006;27:61–76. doi: 10.1057/palgrave.jphp.3200055. [DOI] [PubMed] [Google Scholar]
- 7.Danaei G, et al. Causes of cancer in the world: comparative risk assessment of nine behavioral and environmental risk factors. Lancet. 2005;366:1784–1793. doi: 10.1016/S0140-6736(05)67725-2. [DOI] [PubMed] [Google Scholar]
- 8.Doll R, et al. The mortality of doctors in relation to their smoking habits: a preliminary report. Br Med J. 1954;228:1451–1455. doi: 10.1136/bmj.1.4877.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gealy R, et al. Comparison of mutations in the p53 and K-ras genes in lung carcinomas from smoking and nonsmoking women. Cancer Epidemiol Biomarkers Prev. 1999;8:297–302. [PubMed] [Google Scholar]
- 10.Husgafvel-Pursiainen K, et al. Cigarette smoking and p53 mutations in lung cancer and bladder cancer. Environ Health Perspect. 1996;104:553–556. doi: 10.1289/ehp.96104s3553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wistuba II, et al. Molecular genetics of small cell lung carcinomas. Semin Oncol. 2001;28(suppl 4):3–13. [PubMed] [Google Scholar]
- 12.Maisson PP, et al. The molecular basis of lung cancer: molecular abnormalities and therapeutic implications. Respir Res. 2003;4:12. doi: 10.1186/1465-9921-4-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lubin JH, et al. Cigarette smoking and cancer risk: modeling total exposure and intensity. Am J Epidemiol. 2007;166:479–489. doi: 10.1093/aje/kwm089. [DOI] [PubMed] [Google Scholar]
- 14.Ji BT, et al. Tobacco smoking and colorectal hyperplastic and adenomatous polyps. Cancer Epidemiol Biomarkers Prev. 2006;15:897–901. doi: 10.1158/1055-9965.EPI-05-0883. [DOI] [PubMed] [Google Scholar]
- 15.International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. Vol. 83. IARC; Lyon: 2004. Tobacco smoke and involuntary smoking. [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson MA, et al. Genetic alterations in cancer knowledge system: analysis of gene mutation in mouse and human liver and lung tumors. Toxicol Sci. 2006;90:400–418. doi: 10.1093/toxsci/kfj101. [DOI] [PubMed] [Google Scholar]
- 17.McCullagh P, et al. Generalized Linear Models. Chapman and Hall; New York: 1997. [Google Scholar]
- 18.Efron B, et al. An Introduction to Bootstrap. Chapman and Hall; New York: 1993. [Google Scholar]
- 19.Saranath D, et al. High frequency mutation in codons 12 and 61 of H-ras oncogene in chewing tobacco-related human oral carcinoma in India. Br J Cancer. 1991;63:573–578. doi: 10.1038/bjc.1991.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Forcet C, et al. The dependence receptor DCC (deleted in colorectal cancer) defines an alternative mechanism for caspase activation. Proc Natl Acad Sci USA. 2001;9:3416–3421. doi: 10.1073/pnas.051378298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roz L, et al. The apoptotic pathway by the Fhit protein I lung cancer cell lines is not affected by Bcl-2 or Bcl-x(L) overexpression. Oncogene. 2004;23:9102–9110. doi: 10.1038/sj.onc.1208142. [DOI] [PubMed] [Google Scholar]
- 22.Fong KM, et al. FHITand FRA3B 3p14.2 allele loss are common in lung cancer and preneoplastic bronchial lesions and are associated with cancer-related FHIT cDNA splicing aberrations. Cancer Res. 1997;57:2256–2267. [PubMed] [Google Scholar]
- 23.Hecht SS. Tobacco smoke carcinogens and lung cancer. J Natl Cancer Inst. 1999;91:1194–1210. doi: 10.1093/jnci/91.14.1194. [DOI] [PubMed] [Google Scholar]
- 24.Wei Q, et al. Benzo(a)pyrene diol epoxide-induced chromosomal aberrations and risk of lung cancer. Cancer Res. 1996;56:3975–3979. [PubMed] [Google Scholar]
- 25.Feng Z, et al. Acrolein is a major cigarette-related lung cancer agent: preferential binding at p53 mutational hotspots and inhibition of DNA repair. Proc Natl Acad Sci USA. 2006;17:15404–15409. doi: 10.1073/pnas.0607031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greenblatt MS, et al. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 27.Hussain SP, et al. p53 mutation spectrum and load: the generation of hypotheses linking the exposure of endogenous and exogenous carcinogens to human cancer. Mutat Res. 1999;428:23–32. doi: 10.1016/s1383-5742(99)00028-9. [DOI] [PubMed] [Google Scholar]
- 28.Denissenko MF, et al. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 29.Hernandez-Boussard TM, et al. A specific spectrum of p53 mutations in lung cancer from smokers: review of mutations compiled in the IARC p53 database. Environ Health Perspect. 1998;106:385–391. doi: 10.1289/ehp.98106385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith LE, et al. Targeting of lung cancer mutational hotspots by polycyclic aromatic hydrocarbons. J Natl Cancer Inst. 2000;92:803–811. doi: 10.1093/jnci/92.10.803. [DOI] [PubMed] [Google Scholar]
- 31.Hainaut P, et al. TP53 mutation spectrum in lung cancer and mutagenic signature of components of tobacco smoke: lessons from the IARC TP53 mutation database. Mutagenesis. 2001;16:551–553. doi: 10.1093/mutage/16.6.551. [DOI] [PubMed] [Google Scholar]
- 32.Osada H, et al. Genetic alterations of multiple tumor suppressors and oncogenes in the carcinogenesis and progression of lung cancer. Oncogene. 2002;21:7421–7434. doi: 10.1038/sj.onc.1205802. [DOI] [PubMed] [Google Scholar]
- 33.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sjoblom T, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 35.Westra WH, et al. K-ras oncogene activation in lung adenocarcinomas from former smokers. Evidence that K-ras mutations are an early and irreversible event in the development of adenocarcinomas of the lung. Cancer. 1993;72:432–438. doi: 10.1002/1097-0142(19930715)72:2<432::aid-cncr2820720219>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 36.Soussi T. The p53 tumor suppressor gene: a model for molecular epidemiology of human cancer. Mol Med Today. 1996;2:32–37. doi: 10.1016/1357-4310(96)88756-9. [DOI] [PubMed] [Google Scholar]
- 37.Bennett WP, et al. Molecular epidemiology of human cancer risk: gene-environment interactions and mutation spectrum in human lung cancer. J Pathol. 1999;187:8–18. doi: 10.1002/(SICI)1096-9896(199901)187:1<8::AID-PATH232>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 38.Gao HG, et al. Distribution of p53 and K-ras mutations in human lung cancer tissues. Carcinogenesis. 1997;18:473–478. doi: 10.1093/carcin/18.3.473. [DOI] [PubMed] [Google Scholar]
- 39.Husgafvel-Pursiainen K. Genotoxicity of environmental tobacco smoke: a review. Mutat Res. 2004;567:427–445. doi: 10.1016/j.mrrev.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 40.Bruske-Hohlfeld I, et al. Lung cancer risk in male workers occupationally exposed to diesel motor emissions in Germany. Am J Ind Med. 1999;36:405–414. doi: 10.1002/(sici)1097-0274(199910)36:4<405::aid-ajim1>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 41.Lipsett M, et al. Occupational exposure to diesel exhaust and lung cancer: a meta-analysis. Am J Public Health. 1999;89:1009–1017. doi: 10.2105/ajph.89.7.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones PA, et al. From gene to carcinogen: a rapidly evolving field in molecular epidemiology. Cancer Res. 1991;51:3617–3620. [PubMed] [Google Scholar]
- 43.Chen B, et al. Allele-specific activation and expression of the K-ras gene in hybrid mouse lung tumors induced by chemical carcinogens. Car-cinogenesis. 1994;15:2031–2035. doi: 10.1093/carcin/15.9.2031. [DOI] [PubMed] [Google Scholar]
- 44.Gray DL, et al. The effects of a binary mixture of benzo(a)pyrene and 7H-dibenzo(c,g)carbazole on lung tumors and K-ras oncogene mutations in strain A/J mice. Exp Lung Res. 2001;27:245–253. doi: 10.1080/019021401300054000. [DOI] [PubMed] [Google Scholar]
- 45.Mass MJ, et al. Ki-ras oncogene mutations in tumors and DNA adducts formed by benz[j]aceanthrylene and benzo[a]pyrene in the lungs of strain A/J mice. Mol Carcinog. 1993;8:186–192. doi: 10.1002/mc.2940080309. [DOI] [PubMed] [Google Scholar]
- 46.Matzinger SA, et al. K-ras mutations in lung tumors from A/J and A/J × TSG-p53 F1 mice treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and phenethyl isothiocyanate. Carcinogenesis. 1995;16:2487–2492. doi: 10.1093/carcin/16.10.2487. [DOI] [PubMed] [Google Scholar]
- 47.Chen B, et al. Dose-dependent ras mutation spectra in N-nitroso-diethylamine induced mouse liver tumors and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone induced mouse lung tumors. Carcinogenesis. 1993;14:1603–1608. doi: 10.1093/carcin/14.8.1603. [DOI] [PubMed] [Google Scholar]
- 48.Kawano R, et al. Effects of K-ras gene mutations in the development of lung lesions induced by 4-(N-methyl-n-nitrosamino)-1-(3-pyridyl)-1-butanone in A/J mice. Jpn J Cancer Res. 1996;87:44–50. doi: 10.1111/j.1349-7006.1996.tb00198.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ronai ZA, et al. G to A transitions and G to T transversions in codon 12 of the Ki-ras oncogene isolated from mouse lung tumors induced by 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and related DNA methylating and pyridyloxobutylating agents. Carcinogenesis. 1993;14:2419–2422. doi: 10.1093/carcin/14.11.2419. [DOI] [PubMed] [Google Scholar]
- 50.Denissenko MF, et al. Slow repair of bulky DNA adducts along the nontranscribed strand of the human p53 gene may explain the strand bias of transversion mutations in cancers. Oncogene. 1998;16:1241–1247. doi: 10.1038/sj.onc.1201647. [DOI] [PubMed] [Google Scholar]
- 51.Hainaut P, et al. Patterns of G>T transversions in lung cancers reflect the primary mutagenic signature of DNA-damage by tobacco smoke. Carcinogenesis. 2001;22:367–374. doi: 10.1093/carcin/22.3.367. [DOI] [PubMed] [Google Scholar]
- 52.Le Calvez F, et al. TP53 and KRAS mutation load and types in lung cancers in relation to tobacco smoke: distinct patterns in never, former and current smokers. Cancer Res. 2005;64:50767–55083. doi: 10.1158/0008-5472.CAN-05-0551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary data can be found at http://carcin.oxfordjournals.org/