Abstract
The Raf-1 serine/threonine kinase is a key protein involved in the transmission of many growth and developmental signals. In this report, we show that autoinhibition mediated by the noncatalytic, N-terminal regulatory region of Raf-1 is an important mechanism regulating Raf-1 function. The inhibition of the regulatory region occurs, at least in part, through binding interactions involving the cysteine-rich domain. Events that disrupt this autoinhibition, such as mutation of the cysteine-rich domain or a mutation mimicking an activating phosphorylation event (Y340D), alleviate the repression of the regulatory region and increase Raf-1 activity. Based on the striking similarites between the autoregulation of the serine/threonine kinases protein kinase C, Byr2, and Raf-1, we propose that relief of autorepression and activation at the plasma membrane is an evolutionarily conserved mechanism of kinase regulation.
Keywords: signal transduction
The Raf-1 serine/threonine kinase is a central component in many signaling pathways, functioning to link activated tyrosine kinases and Ras to mitogen and extracellular regulated kinase 1 (MEK1) and mitogen-activated protein kinase (MAPK) (1–3). Because of the essential nature of Raf-1 in signal transduction, considerable effort has been placed on elucidating the mechanisms involved in regulating its activity. Although significant advances have been made, the regulation of Raf-1 is a highly complex process that, even now, is not fully understood (reviewed in ref. 3). One of the earliest findings that provided insight into Raf-1 regulation was the observation that many oncogenic Raf-1 proteins do not contain the noncatalytic N-terminal region (4–12). Deletion analysis subsequently confirmed that truncation of Raf-1 by removal of its N terminus generates a constitutively active and transforming protein (13, 14). Based on these findings, it has been suggested that the N-terminal region of Raf-1 functions to inhibit and regulate the activity of the C-terminal catalytic domain.
The N-terminal regulatory region of Raf-1 contains two of the three conserved sequences present in all Raf molecules, CR1 and CR2 (15, 16). CR1 encompasses residues 62–194 and contains a Ras-binding domain (RBD; residues 51–131, refs. 17–19) and a cysteine-rich domain (CRD; residues 139–184, ref. 20). CR2 is a 14-amino acid sequence that is rich in serine and threonine residues. Ser-259, found within CR2, is an in vivo phosphorylation site that mediates the binding of Raf-1 to the 14–3-3 family of proteins (21–23). The last conserved domain, CR3, is located within the C-terminal region of Raf-1 (15, 16). CR3 constitutes the protein kinase domain and contains another phosphorylation-dependent 14–3-3-binding site (Ser-621, refs. 22 and 23), as well as activational sites of in vivo phosphorylation (Tyr-340, -341, ref. 24, and Ser-338, -339, ref. 25).
If the N-terminal regulatory region does inhibit Raf-1 activity, then the irreversible nature of deleting this region would generate Raf-1 molecules that are unregulated and constitutively active (as has been shown in refs. 4–14). However, under normal signaling conditions, repression by the regulatory region would be expected to be relieved by reversible modifications, such that the activity of the catalytic domain could be precisely regulated. The current model for Raf-1 activation (reviewed in ref. 3) proposes that in response to Ras activation, Raf-1 translocates to the membrane and binds to GTP-loaded Ras. The binding of the Raf RBD to the effector domain of Ras then allows the CRD to contact Ras and/or membrane phospholipids. These interactions and other modifications occurring at the membrane, including phosphorylation of Tyr-340, -341, and Ser-338, -339, result in Raf-1 activation. Thus, based on the autoinhibition model, many of the events known to be required for Raf-1 activation may help to relieve the repression of the regulatory region.
In this report we have taken a molecular and biochemical approach to determine whether autoinhibition plays a role in regulating Raf-1 activity. By expressing the regulatory and catalytic regions of Raf-1 as separate proteins, we find that the isolated regulatory region can suppress in trans the biological and enzymatic activity of the catalytic domain, indicating that the regulatory region does indeed function as a repressor of Raf-1 activity.
MATERIALS AND METHODS
Construction of Raf-1, Reg/Raf, and Cat/Raf Plasmids.
The Reg/Raf construct was generated by PCR amplification of a DNA fragment corresponding to amino acid residues 1–330 of Raf-1. The PCR product was then inserted into the pA vector immediately downstream of sequences encoding the FLAG epitope tag. Reg/Raf mutant constructs were obtained by site-directed mutagenesis by using the appropriate oligonucleotides to introduce the desired base changes. The specific base changes in all mutant constructs were confirmed by sequence analysis. The full-length Raf-1 (residues 1–648) mutant constructs and the Cat/Raf (residues 306–648) constructs, which did not contain an epitope tag, have been described (21, 24, 26, 27).
Oocyte Injection and Analysis.
Xenopus laevis oocytes were isolated and defolliculated as previously described (26). Defolliculated oocytes were injected with buffer or with 30 ng of in vitro-transcribed RNA encoding the Reg/Raf proteins. Four to eight hours later, after the Reg/Raf protein had been synthesized, the oocytes were injected with RNA encoding Cat/Raf (15 ng), Ha-RasV12 (15 ng), Tpr-Met (30 ng), or v-Mos (30 ng) or were treated with 5 μg/ml progesterone. Oocytes were scored for germinal vesicle breakdown (GVBD). This observation was verified by manual dissection of oocytes after fixation in 8% trichloroacetic acid.
Preparation of Cell Lysates, Immunoprecipitation, and in Vitro Protein Kinase Assays.
Oocytes were lysed by trituration with a pipette tip in either Nonidet P-40 (NP-40) lysis buffer [20 mM Tris, pH 8.0/137 mM NaCl/10% glycerol/1% NP-40/2 mM EDTA/1 mM phenylmethylsulfonyl fluoride (PMSF)/aprotinin (0.15 unit/ml)/20 μM leupeptin/5 mM sodium vanadate] or RIPA buffer [20 mM Tris, pH 8.0/137 mM NaCl/10% glycerol/1% NP-40/0.5% sodium deoxycholate/0.1% SDS/2 mM EDTA/1 mM PMSF/aprotinin (0.15 unit/ml)/20 μM leupeptin/5 mM sodium vanadate] (10 μl per oocyte). Insoluble material was pelleted by centrifugation at 14,000 × g for 10 min at 4°C and cell lysates were equalized for protein expression by immunoblot analysis. Immunoprecipitation assays were performed by incubating cell lysates with the appropriate antibody for 4–6 hr at 4°C. The antigen–antibody complexes were collected with protein-G Sepharose beads (Pharmacia). The immunoprecipitates were then washed four times with ice-cold NP-40 lysis buffer and either analyzed by SDS/PAGE or by in vitro protein kinase assays (26).
RESULTS
The N-Terminal Regulatory Region of Raf-1 Inhibits the Biological and Enzymatic Activity of the C-Terminal Catalytic Domain.
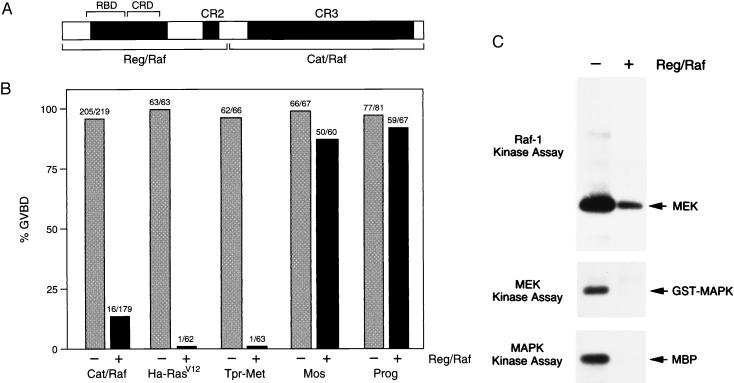
To determine whether the Raf-1 regulatory region functions to repress the activity of the catalytic domain, we first generated separate proteins encoding each region of Raf-1 (Reg/Raf and Cat/Raf; Fig. 1A). We then examined the effect of Reg/Raf on the biological activity of Cat/Raf in trans, by using the Xenopus oocyte meiotic maturation assay. The isolated catalytic domain of Raf-1 has previously been shown to be a constitutively active protein that has transforming potential (14) and can induce oocyte maturation (as evidenced by GVBD and activation of cdc2; ref. 26). Therefore, as expected, expression of Cat/Raf efficiently induced maturation in 93% of the oocytes (Fig. 1B). However, when Cat/Raf was expressed in the presence of Reg/Raf, only 9% of the oocytes underwent GVBD (Fig. 1B), suggesting an inhibitory effect of Reg/Raf on Cat/Raf activity. To examine whether Reg/Raf is a nonspecific inhibitor of oocyte maturation, we assayed the effect of Reg/Raf on other inducers of oocyte maturation. Reg/Raf had little or no effect on Ras-independent maturation induced by v-Mos expression or progesterone treatment (Fig. 1B), indicating that Reg/Raf is not a general inhibitor of oocyte maturation. However, Reg/Raf blocked Ras-dependent GVBD, as demonstrated by the inhibition of maturation induced by Ha-RasV12 and the activated receptor tyrosine kinase Tpr-Met (Fig. 1B). Because Reg/Raf contains the Raf-1 RBD, this inhibitory effect most likely reflects the ability of Reg/Raf to bind to activated Ras and prevent its interaction with downstream effectors, such as endogenous Xenopus Raf (28).
Figure 1.
Reg/Raf inhibits the biological and enzymatic activity of Cat/Raf. (A) Schematic depiction of the Reg/Raf and Cat/Raf proteins. (B) Effect of Reg/Raf on Xenopus oocyte meiotic maturation. Oocytes were injected with buffer (−) or RNA encoding the Reg/Raf protein (+). Four to eight hours later, the oocytes were injected with RNA encoding Cat/Raf-1, Ha-RasV12, Tpr-Met, or v-Mos or were treated with progesterone (Prog). GVBD was then scored. Numbers shown represent a compilation of at least five (Cat/Raf) or two (Ha-RasV12, Tpr-Met, v-Mos, Prog) independent experiments where equivalent amounts of Reg/Raf proteins were expressed. (C) Reg/Raf inhibits the enzymatic activity of Cat/Raf and the activation of MEK1 and MAPK. Cat/Raf (Top), MEK1 (Middle), and MAPK (Bottom) proteins were immunoprecipitated from Xenopus oocytes expressing Cat/Raf alone (−) or coexpressing Cat/Raf and Reg/Raf (+), and in vitro protein kinase assays were performed. The positions of MEK1, glutathione S-transferase (GST)-MAPK, or myelin basic protein (MBP), used as exogenous substrates in these assays, are indicated.
To investigate the nature of the block caused by Reg/Raf, we next examined the enzymatic activity of Cat/Raf in the presence and absence of Reg/Raf. Cat/Raf proteins were immunoprecipitated from oocyte lysates and immune complex kinase assays were performed by using MEK1 as an exogenous substrate. As shown in Fig. 1C, the kinase activity of Cat/Raf was significantly reduced (>10-fold) in oocytes coexpressing Reg/Raf. In addition, when we examined the enzymatic activities of MEK1 and MAPK (the endogenous Xenopus kinases functioning downstream of Cat/Raf), we found that although both MEK1 and MAPK were activated in oocytes expressing Cat/Raf alone, their activation was completely blocked in oocytes coexpressing Reg/Raf and Cat/Raf (Fig. 1C). Taken together, these findings indicate that Reg/Raf can inhibit the biological and enzymatic activity of Cat/Raf and can prevent Cat/Raf from transmitting a signal to MEK1 and MAPK.
Association of the Reg/Raf and Cat/Raf Proteins.
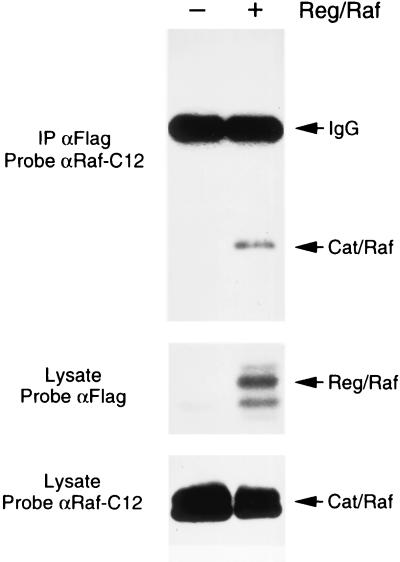
A potential model for the Reg/Raf inhibitory effect is that Reg/Raf interacts with Cat/Raf and prevents either the activation of Cat/Raf or the binding of Cat/Raf to its substrates. To test this model, we examined whether an interaction between Reg/Raf and Cat/Raf could be detected. Cat/Raf proteins were expressed alone or with FLAG-tagged Reg/Raf in oocytes. FLAG immunoprecipitates were then prepared and examined for the presence of Cat/Raf. By immunoblot analysis, Cat/Raf was detected in the FLAG immunoprecipitates, but only when Reg/Raf was present (Fig. 2). In similar experiments, Cat/Raf was unable to coimmunoprecipitate with FLAG-tagged full-length Raf-1 or with the amino-terminal domain of kinase suppressor of Ras (KSR; data not shown). Thus, these results indicate that Reg/Raf and Cat/Raf do specifically associate, and provide evidence that the Reg/Raf repression may be mediated, at least in part, by a physical interaction with Cat/Raf.
Figure 2.
Association of the Reg/Raf and Cat/Raf proteins. FLAG immunoprecipitates were prepared from Xenopus oocytes expressing Cat/Raf alone (−) or coexpressing Cat/Raf and FLAG-tagged Reg/Raf (+). The immunoprecipitates were resolved by electrophoresis on an SDS/8% polyacrylamide gel and examined for the presence of Cat/Raf by immunoblotting with antibodies directed against the C-terminus of Raf-1 (Top). Total oocyte lysates were analyzed by immunoblotting to evaluate the expression level of FLAG-tagged Reg/Raf (Middle), and Cat/Raf (Bottom).
Mutations in the Raf-1 CRD Specifically Disrupt Reg/Raf Inhibition of Cat/Raf.
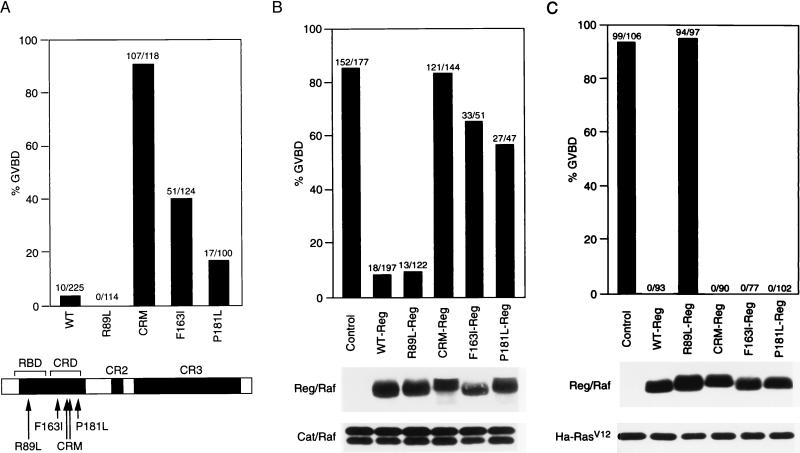
Previous studies have shown that mutation of certain amino acid residues in the N-terminal regulatory region can significantly alter the biological activity of Raf-1. One such mutation, R89L, is located in the RBD and prevents the binding and activation of Raf-1 by Ras (17, 28, 29). In contrast, mutations located in the Raf-1 CRD have been found to increase both the biological and enzymatic activity of full-length Raf-1 proteins (21, 27). These CRD mutations include a substitution of two serines for cysteines at residues 165 and 168 (CRM), substitution of isoleucine for phenylalanine at residue 163 (F163I), and substitution of leucine for proline at residue 181 (P181L). The CRM mutation disrupts a putative cysteine finger motif within the CRD and the F163I and P181L mutations are activating substitutions that were first identified in Drosophila to suppress the effect of a Ras-binding site mutation (30). As expected, expression of full-length wild-type (WT)- and R89L-Raf-1 proteins did not significantly induce oocyte maturation, with WT- and R89L-Raf-1 inducing GVBD in 4% and 0% of the oocytes, respectively (Fig. 3A). However, full-length Raf-1 proteins containing mutations in the CRD were able to promote maturation, with CRM-, F163I-, and P181L-Raf-1 proteins inducing GVBD in 91%, 41%, and 17% of the oocytes, respectively (Fig. 3A). To determine whether the RBD and CRD mutations would alter the repressor activity of the regulatory region, we generated Reg/Raf proteins containing these mutations and examined their ability to inhibit Cat/Raf-mediated maturation. As a control, the mutant Reg/Raf proteins were also assayed for their ability to block Ha-RasV12-induced GVBD. In comparison to WT Reg/Raf, we found that Reg/Raf proteins containing the R89L mutation were unable to block Ha-RasV12-mediated maturation (Fig. 3C), but were still fully competent to inhibit maturation induced by Cat/Raf (Fig. 3B). In contrast, Reg/Raf proteins containing the CRD mutations exhibited a decreased ability to suppress Cat/Raf-induced maturation (Fig. 3B), but were still fully competent to block maturation mediated by Ha-RasV12 (Fig. 3C). Interestingly, the reduction in the repressor activity of Reg/Raf correlated with the increased biological activity of the full-length Raf-1 proteins, in that the CRD mutation having the greatest activational effect on full-length Raf-1 also resulted in the greatest reduction in Reg/Raf inhibition. These findings indicate that an intact CRD is critical for the full autorepressor activity of Reg/Raf and demonstrate further that the domain of the regulatory region involved in suppressing Cat-Raf activity is distinct from that required to block Ras-dependent signaling.
Figure 3.
Regulatory region mutations alter the inhibitory effect of Reg/Raf. (A) Effects of regulatory region mutations on the activity of full-length Raf-1 proteins. Oocytes were injected with RNA (30 ng) encoding full-length WT-, R89L-, CRM-, F163I-, or P181L-Raf-1 and scored for GVBD. (B) Effects of regulatory region mutations on Reg/Raf inhibition of Cat/Raf. Oocytes were first injected with buffer (Control) or RNA encoding WT-, R89L-, CRM-, F163I-, or P181L-Reg/Raf. Four to eight hours later, oocytes were injected with RNA encoding Cat/Raf and then scored for GVBD. Oocyte lysates were analyzed by immunoblotting to evaluate the expression level of FLAG-tagged Reg/Raf (Middle) and Cat/Raf-1 (Bottom). (C) Effects of regulatory region mutations on the inhibition of Ha-RasV12 by Reg/Raf. Oocytes were injected with buffer (Control) or RNA encoding WT-, R89L-, CRM-, F163I-, or P181L-Reg/Raf. Four to eight hours later, oocytes were injected with RNA encoding Ha-RasV12 and then scored for GVBD. Oocyte lysates were analyzed by immunoblotting to evaluate the expression level of FLAG-tagged Reg/Raf (Middle) and Ha-RasV12 (Bottom). The numbers shown in A, B, and C represent a compilation of three independent experiments; the protein analysis shown in B and C is from one typical experiment.
Binding of 14–3-3 to Mutant Reg/Raf Proteins.
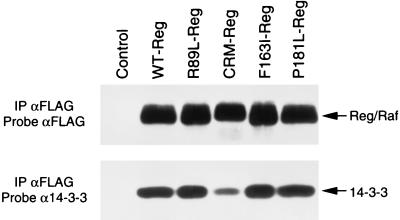
The Raf-1 CRD is an atypical cysteine-rich C1 domain that does not bind diacylglycerol or phorbol esters (31). However, the Raf-1 CRD has been reported to interact with phosphatidylserine and Ras (20, 32–36). In addition, although 14–3-3 has been best characterized to be a phosphoserine-binding protein that associates with Raf-1 at Ser-259 and Ser-621 (21, 23, 37), an interaction between the Raf-1 CRD and 14–3-3 has also been reported (21, 38). The association of 14–3-3 with the Raf-1 CRD has been suggested to inhibit Raf-1 activity, because mutations that reduce the binding of 14–3-3 to an isolated CRD-containing protein increase the activity of full-length Raf-1 (38). To determine whether the CRM, F163I, and P181L mutations reduce the inhibitory activity of Reg/Raf by interfering with 14–3-3 binding, we examined the mutant Reg/Raf proteins for their ability to associate with 14–3-3. By immunoblot analysis, 14–3-3 was detected in immunoprecipitates from all Reg/Raf proteins (Fig. 4). A reduction in 14–3-3 binding was observed with CRM-Reg/Raf; however, F163I- and P181L-Reg/Raf interacted with 14–3-3 as efficiently as did WT- and R89L-Reg/Raf. Therefore, the Reg/Raf inhibitory activity does not appear to correlate with 14–3-3 binding.
Figure 4.
Binding of 14–3-3 to WT and mutant Reg/Raf proteins. FLAG-tagged Reg/Raf proteins were immunoprecipitated from oocytes lysed in RIPA buffer. The immunoprecipitates were washed extensively and then incubated for 2 hr with extracts from 293 cells (as a source of exogenous 14–3-3). The immunoprecipitates were examined by immunoblot analysis by using FLAG (Upper) and 14–3-3 (Lower) antibodies.
Activating Mutations in Cat/Raf Prevent Reg/Raf Inhibition.
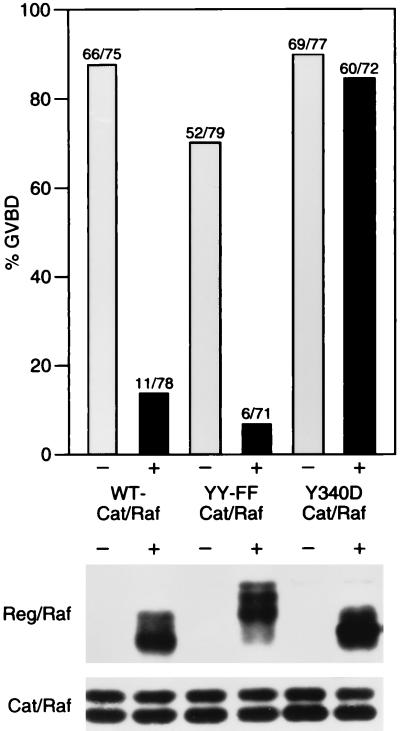
Tyrosine residues 340 and 341, located in the Raf-1 catalytic domain, have been identified to be activating sites of in vivo phosphorylation (24, 39). Mutation of these tyrosines to negatively charged aspartic acid residues constitutively activates the enzymatic and biological activity of full-length Raf-1, presumably by mimicking constitutive phosphorylation of these sites (24, 39). In contrast, replacing these tyrosines with uncharged phenylalanine residues generates a full-length Raf-1 protein that cannot be activated (24). To determine whether mutation of tyrosine 340 and 341 has any effect on the ability of Reg/Raf to suppress Cat/Raf activity, we generated Y340D- and YY340,341FF-Cat/Raf proteins and examined their biological activity in the presence and absence of WT-Reg/Raf. As previously reported, expression of either Y340D- or YY340,341FF-Cat/Raf was able to promote oocyte maturation (24). GVBD mediated by YY340,341FF-Cat/Raf was suppressed by Reg/Raf; however, maturation mediated by Y340D-Cat/Raf was not (Fig. 5). In addition, we found that Y340D-Cat/Raf was unable to coimmunoprecipitate with Reg/Raf (data not shown). These results demonstrate that Y340D-Cat/Raf cannot be repressed by Reg/Raf and indicate further that modification of residues in the Raf-1 catalytic domain may activate the full-length Raf-1 protein by relieving the inhibitory effect of the regulatory region.
Figure 5.
Activating mutations in Cat/Raf prevent Reg/Raf suppression. Oocytes were injected with buffer (−) or RNA encoding Reg/Raf (+). Four to eight hours later, oocytes were injected with RNA encoding WT-, YY340,341FF-, or Y340D-Cat/Raf and then scored for GVBD. Oocyte lysates were analyzed by immunoblotting to evaluate the expression level of FLAG-tagged Reg/Raf (Middle) and Cat/Raf (Bottom). Numbers shown represent a compilation of three independent experiments where equivalent amounts of Reg/Raf and Cat/Raf proteins were expressed; the protein analysis is from one typical experiment.
DISCUSSION
The Raf-1 serine/threonine kinase is an important signal-transducing molecule that functions in many growth and developmental pathways. In response to signaling events, the activation of Raf-1 is an intricate multistep process involving a change in subcellular localization, protein interactions, and phosphorylation events. In this report, we have taken a molecular and biochemical approach to determine whether autoinhibition also plays a role in regulating Raf-1 activity.
By using meiotic maturation of Xenopus oocytes as an assay system, we found that a protein encoding the Raf-1 regulatory region (Reg/Raf) can repress in trans the activity of a protein encoding the Raf-1 catalytic domain (Cat/Raf). Reg/Raf suppressed Cat/Raf-induced oocyte maturation, inhibited Cat/Raf enzymatic activity, and prevented Cat/Raf from activating the downstream kinases MEK1 and MAPK. Reg/Raf is not a general inhibitor of oocyte maturation, because the presence of this protein did not block v-Mos or progesterone-induced GVBD. Reg/Raf does contain the RBD and, therefore, was able to block Ras-dependent oocyte maturation. This finding is consistent with previous studies showing that the isolated regulatory region can inhibit Ras-mediated signaling in mammalian cells (40–42). However, our mutational analysis of the regulatory region revealed that the inhibition of Ras signaling and Cat/Raf signaling were separate events. Mutation of a critical arginine residue required for Ras binding (R89L, refs. 28 and 29) completely eliminated Reg/Raf inhibition of Ras-dependent signaling, but had no effect on the repression of Cat/Raf activity, demonstrating that the effect observed with Reg/Raf could not simply be attributed to a block of endogenous Xenopus Ras activity. Furthermore, mutations in the Raf-1 CRD significantly altered the repressor activity exhibited toward Cat/Raf, yet had no effect on Reg/Raf inhibition of Ras signaling. Thus, although the RBD is the domain of the regulatory region required for blocking Ras activity, the CRD is the domain critical for the autorepressor activity. The fact that two of the CRD mutations (F163I and P181L) were first identified as activating mutations in the Drosophila Raf-1 protein suggests that the role of the CRD and the autorepressor activity of the regulatory domain are conserved.
In addressing the mechanism by which the regulatory region inhibits the activity of the catalytic domain, we found that Cat/Raf was able to specifically coimmunoprecipitate with Reg/Raf, suggesting that a physical interaction does occur. This association could be a direct intramolecular interaction or could be an indirect interaction mediated by a third molecule. The Raf-1 CRD, which is required for the autoinhibitory activity, has been reported to associate with 14–3-3, and binding of 14–3-3 to the CRD has been suggested to negatively regulate Raf-1 activity (38). However, by using our panel of Reg/Raf mutant proteins, we did not observe a correlation between 14–3-3 binding and Reg/Raf inhibition of Cat/Raf activity. Nevertheless, because the CRM mutation, which had the greatest effect on Reg/Raf inhibitory activity, exhibited reduced 14–3-3 binding, we cannot rule out the possibility that in the context of the full-length Raf-1 protein, 14–3-3 binding might help stabilize the autoinhibited “inactive” Raf-1 conformation.
If Raf-1 autoinhibition is mediated by intramolecular interactions, then it would be expected that activating mutations in either the regulatory region or the catalytic domain might increase Raf-1 activity by disrupting these interactions. Our data suggest that this is indeed the case. A mutation in the catalytic domain, which mimics an activating phosphorylation event at Tyr-340, and activating mutations in the CRD were found to greatly reduce or to eliminate Reg/Raf inhibition of Cat/Raf activity. Analysis of proteins containing the CRD mutations also revealed a correlation between the reduction in Reg/Raf repression and the increase in full-length Raf-1 activity. Thus, in the context of the full-length Raf-1 molecule, it appears that any event disrupting the autoinhibition—such as mutation, protein interactions, and phosphorylation—may increase the enzymatic and biological activity of Raf-1 by relieving the repression of the regulatory region.
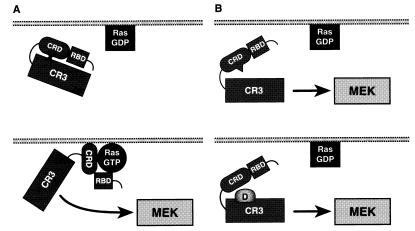
Autoinhibition is a mechanism used to regulate the activity of other serine/threonine kinases, such as protein kinase C (PKC) and the Schizosaccharomyces pombe Byr2 kinase (43, 44). As with Raf-1, the N-terminal regulatory region of these kinases inhibits the activity of the C-terminal kinase domain. For PKC, the autoinhibition has been well characterized and is mediated by the binding of a pseudosubstrate sequence in the regulatory region to the substrate binding site of the catalytic domain (43). The exact mechanism of Byr2 autoinhibition is not as well defined, yet interesting parallels can be observed between the autoregulation of Byr2 and Raf-1. Byr2 is a MEK kinase in S. pombe and therefore occupies the same position as does Raf-1 in the MAPK cascade (45). Like Raf-1, deletion of the N terminus activates Byr2, an interaction between the Byr2 regulatory and catalytic domains has been observed, and mutations disrupting this interaction enhance Byr2 activity (44). In addition, both Raf-1 and Byr2 contain Ras-binding domains, and mutational analysis indicates that the region required for the autoinhibitory effect is located adjacent to the RBD in both kinases (44). Raf-1 and Byr2, as well as PKC, translocate to the plasma membrane in response to signaling events, and for PKC and Byr2, relief of autoinhibition and kinase activation occurs at the membrane. In the case of PKC, the pseudosubstrate sequence is removed from the catalytic pocket when the cysteine-rich C1 and C2 domains contact diacylglycerol and phosphatidylserine, respectively (43). Byr2 localizes to the membrane by binding to activated Ras. The activation of Byr2 and the disruption of the intramolecular interactions then occur at the membrane by a mechanism that has not been completely elucidated (44). As with PKC and Byr2, membrane localization also activates Raf-1. The translocation of Raf-1 is mediated by the binding of the Raf-1 RBD to GTP-bound Ras. This interaction then allows the CRD either to contact Ras directly or to interact with membrane phospholipids. Because the CRD is a critical region for the autorepressor activity, the binding of the CRD to membrane components may stably relieve the autorepression of the regulatory region and allow the catalytic domain to contact its activators and substrates (Fig. 6). Additional phosphorylation of Raf-1 at tyrosines 340 and 341 may further help to keep Raf-1 in an activated and derepressed conformation. Consistent with this model and the findings obtained from this study, we propose that relief of autorepression is an important step in Raf-1 activation.
Figure 6.
Model of Raf-1 regulation. (A) In quiescent cells, Ras is in the inactive GDP-bound form and Raf-1 is in an inactive state in the cytosol. The regulatory region inhibits the activity of the catalytic domain, perhaps through intramolecular interactions involving the CRD (Upper). Under signaling conditions, Ras becomes GTP-loaded and activated. Raf-1 translocates to the plasma membrane, where both the RBD and CRD contact membrane components, thereby relieving the repression by the regulatory region (Lower). (B) In the absence of Ras activation, activating mutations in the CRD (Upper) or the catalytic domain (Lower) relieve the repression of the regulatory region and allow the catalytic domain to contact its downstream target, MEK1.
Acknowledgments
We thank members of the Morrison laboratory, Jonathan Cooper, and David Winkler for helpful comments and critical reading of the manuscript. This work was supported by the National Cancer Institute, Department of Health and Human Services, under contract with Advance BioSciences Laboratories.
ABBREVIATIONS
- MEK1
mitogen and extracellular regulated kinase 1
- MAPK
mitogen-activated protein kinase
- GVBD
germinal vesicle breakdown
- CRD
cysteine-rich domain
- RBD
Ras-binding domain
- WT
wild type
- PKC
protein kinase C
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Marshall C J. Curr Opin Genet Dev. 1994;4:82–89. doi: 10.1016/0959-437x(94)90095-7. [DOI] [PubMed] [Google Scholar]
- 2.Moodie S A, Wolfman A. Trends Genet. 1994;10:44–48. doi: 10.1016/0168-9525(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 3.Morrison D K, Cutler R E., Jr Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 4.Bonner T I, Opperman H, Seeburg P, Kerby S B, Gunnell M A, Young A C, Rapp U R. Nucleic Acids Res. 1986;14:1009–1015. doi: 10.1093/nar/14.2.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck T W, Huleihel M, Gunnell M A, Bonner T I, Rapp U R. Nucleic Acids Res. 1987;15:595–609. doi: 10.1093/nar/15.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukui M, Yamamoto T, Kawai S, Maruo T, Toyoshima K. Proc Natl Acad Sci USA. 1985;82:5954–5958. doi: 10.1073/pnas.82.17.5954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimizu K, Nakatsu Y, Sekisuchi M, Hokamura K, Tanaka K, Terada M, Sugimura T. Proc Natl Acad Sci USA. 1985;82:5641–5645. doi: 10.1073/pnas.82.17.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stanton V P, Jr, Cooper G M. Mol Cell Biol. 1987;7:1171–1179. doi: 10.1128/mcb.7.3.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ikawa S, Fukui M, Ueyama Y, Tamaoki N, Yamamoto T, Toyoshima K. Mol Cell Biol. 1988;8:2651–2654. doi: 10.1128/mcb.8.6.2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ishikawa F, Takaku F, Nagao M, Sugimura T. Mol Cell Biol. 1987;7:1226–1232. doi: 10.1128/mcb.7.3.1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitsunobu F, Fukui M, Oda T, Yamamoto T, Toyoshima K. Oncogene. 1989;4:437–442. [PubMed] [Google Scholar]
- 12.Rapp U R, Goldsborough M D, Mark G E, Bonner T I, Groffen J, Reynolds F J, Stephenson J R. Proc Natl Acad Sci USA. 1983;80:4218–4222. doi: 10.1073/pnas.80.14.4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Heidecker G, Huleihel M, Cleveland J L, Kolch W, Beck T W, Lloyd P, Pawson T, Rapp U R. Mol Cell Biol. 1990;10:2503–2512. doi: 10.1128/mcb.10.6.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanton V P, Nichols D W, Laudano A P, Cooper G M. Mol Cell Biol. 1989;9:639–647. doi: 10.1128/mcb.9.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daum G, Eisenmann-Tappe I, Fries H W, Troppmair J, Rapp U R. Trends Biol Sci. 1994;19:474–479. doi: 10.1016/0968-0004(94)90133-3. [DOI] [PubMed] [Google Scholar]
- 16.Morrison D K. Cancer Cells. 1990;2:377–382. [PubMed] [Google Scholar]
- 17.Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. Nature (London) 1995;375:554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- 18.Scheffler J E, Waugh D S, Bekesi E, Kiefer S E, LoSardo J E, Neri A, Prinzo K M, Tsao K, Wegrzynski B, Emerson S D, Fry D C. J Biol Chem. 1994;269:22340–22346. [PubMed] [Google Scholar]
- 19.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 20.Mott H, Carpenter J, Zhong S, Ghosh S, Bell R, Campbell S. Proc Natl Acad Sci USA. 1996;93:8312–8317. doi: 10.1073/pnas.93.16.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Michaud N R, Fabian J R, Mathes K D, Morrison D K. Mol Cell Biol. 1995;15:3390–3397. doi: 10.1128/mcb.15.6.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morrison D K, Heidecker G, Rapp U R, Copeland T D. J Biol Chem. 1993;268:17309–17316. [PubMed] [Google Scholar]
- 23.Muslin A, Tanner J, Allen P, Shaw A. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 24.Fabian J R, Daar I, Morrison D K. Mol Cell Biol. 1993;13:7133–7143. doi: 10.1128/mcb.13.11.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Diaz B, Barnard D, Filson A, MacDonald S, King A, Marshall M. Mol Cell Biol. 1997;17:4509–4516. doi: 10.1128/mcb.17.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabian J R, Morrison D K, Daar I. J Cell Biol. 1993;122:645–652. doi: 10.1083/jcb.122.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cutler R E, Jr, Morrison D K. EMBO J. 1997;16:1953–1960. doi: 10.1093/emboj/16.8.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fabian J R, Vojtek A B, Cooper J A, Morrison D K. Proc Natl Acad Sci USA. 1994;91:5982–5986. doi: 10.1073/pnas.91.13.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Block C, Janknecht R, Herrmann C, Nassar N, Wittinghofer A. Nat Struct Biol. 1996;3:244–251. doi: 10.1038/nsb0396-244. [DOI] [PubMed] [Google Scholar]
- 30.Lu X, Melnick M B, Hsu J-C, Perrimon N. EMBO J. 1994;13:2592–2599. doi: 10.1002/j.1460-2075.1994.tb06549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hurley J H, Newton A C, Parker P J, Blumberg P M, Nishizuka Y. Protein Sci. 1997;6:477–480. doi: 10.1002/pro.5560060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brtva T, Drugan J, Ghosh S, Terrell R, Campbell-Burk S, Bell R, Der C. J Biol Chem. 1995;270:9809–9812. doi: 10.1074/jbc.270.17.9809. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh S, Xie W Q, Quest A F G, Mabrouk G M, Strum J C, Bell R M. J Biol Chem. 1994;269:10000–10007. [PubMed] [Google Scholar]
- 34.Ghosh S, Strum J C, Sciorra V A, Daniel L, Bell R M. J Biol Chem. 1996;271:8472–8480. doi: 10.1074/jbc.271.14.8472. [DOI] [PubMed] [Google Scholar]
- 35.Hu C, Kariya K, Tamada M, Akasaka K, Shirouzu M, Yokoyama S, Kataoka T. J Biol Chem. 1995;270:30274–30277. doi: 10.1074/jbc.270.51.30274. [DOI] [PubMed] [Google Scholar]
- 36.Drugan J, Khosravi-Far R, White M, Der C, Sung Y, Hwang Y, Campbell S. J Biol Chem. 1996;271:233–237. doi: 10.1074/jbc.271.1.233. [DOI] [PubMed] [Google Scholar]
- 37.Rommel C, Radziwill G, Lovric J, Noeldeke J, Heinicke T, Jones D, Aitken A, Moelling K. Oncogene. 1996;12:609–619. [PubMed] [Google Scholar]
- 38.Clark G J, Drugan J K, Rossman K L, Carpenter J W, Rogers-Graham K, Fu H, Der C J, Campbell S L. J Biol Chem. 1997;272:20990–20993. doi: 10.1074/jbc.272.34.20990. [DOI] [PubMed] [Google Scholar]
- 39.Marais R, Light Y, Patterson H F, Marshall C J. EMBO J. 1995;14:3136–3145. doi: 10.1002/j.1460-2075.1995.tb07316.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fridman M, Tikoo A, Varga M, Murphy A, Nur-E-Kamal M S, Maruta H. J Biol Chem. 1994;269:30105–30108. [PubMed] [Google Scholar]
- 41.Bruder J T, Heidecker G, Rapp U R. Genes Dev. 1992;6:545–556. doi: 10.1101/gad.6.4.545. [DOI] [PubMed] [Google Scholar]
- 42.Schaap D, van der Wal J, Howe L R, Marshall C J, van Blitterswijk W J. J Biol Chem. 1993;268:20232–20236. [PubMed] [Google Scholar]
- 43.Newton A C. Curr Opin Cell Biol. 1997;9:161–167. doi: 10.1016/s0955-0674(97)80058-0. [DOI] [PubMed] [Google Scholar]
- 44.Tu H, Barr M, Dong D L, Wigler M. Mol Cell Biol. 1997;17:5876–5887. doi: 10.1128/mcb.17.10.5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herskowitz I. Cell. 1995;80:187–197. doi: 10.1016/0092-8674(95)90402-6. [DOI] [PubMed] [Google Scholar]