Abstract
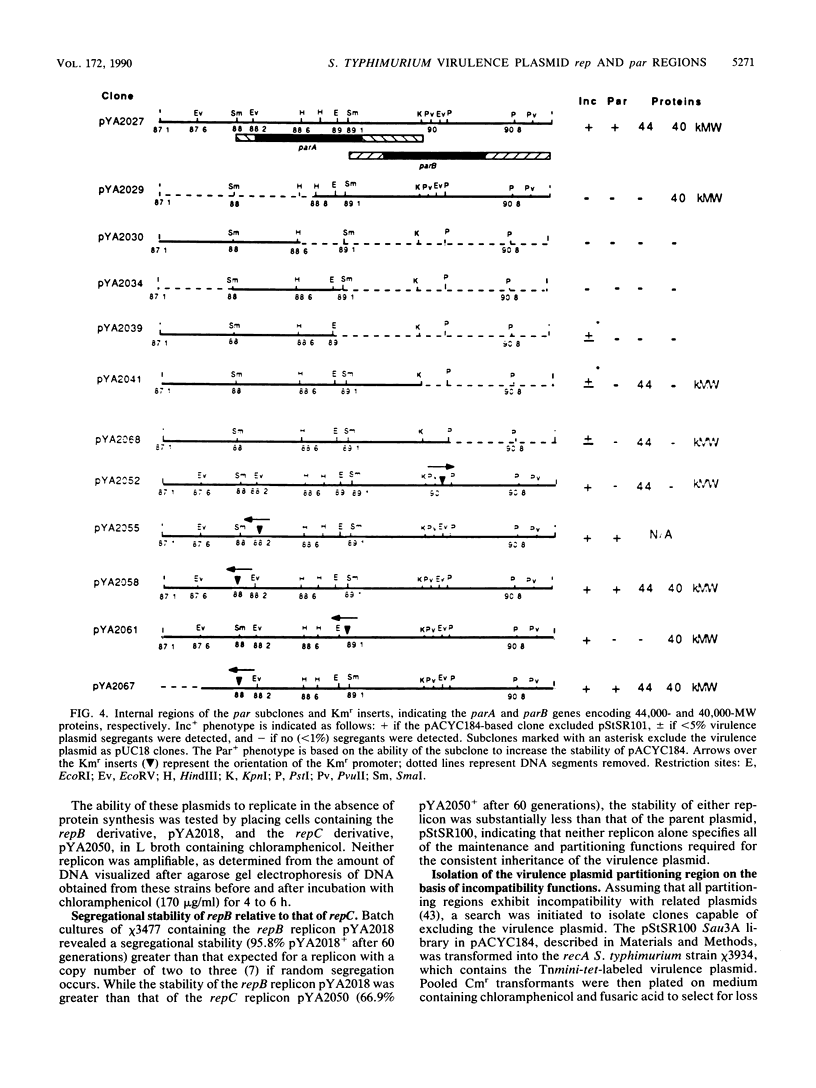
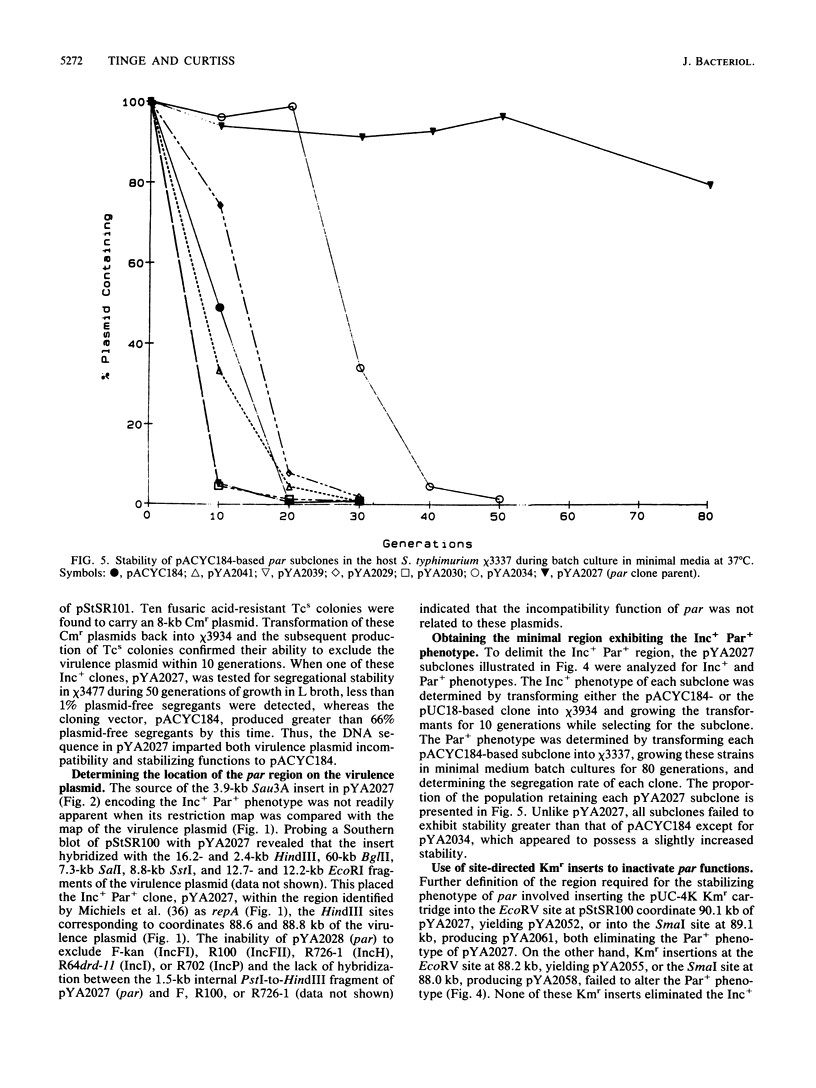
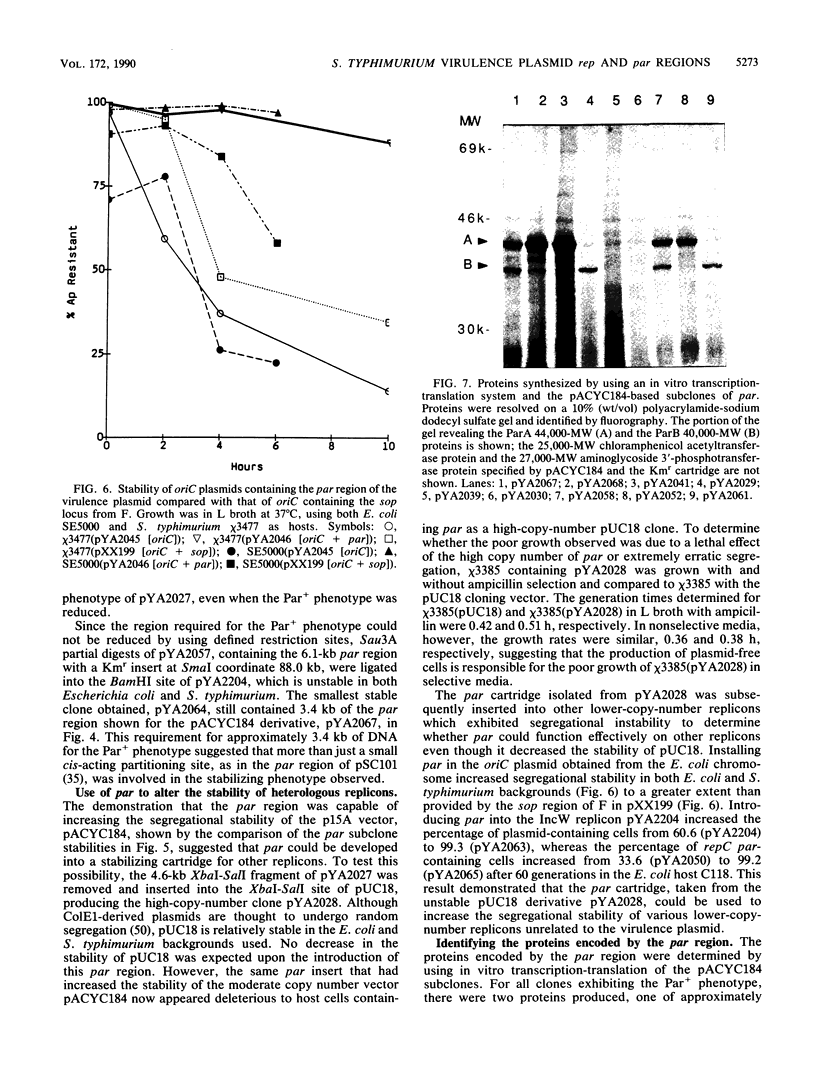
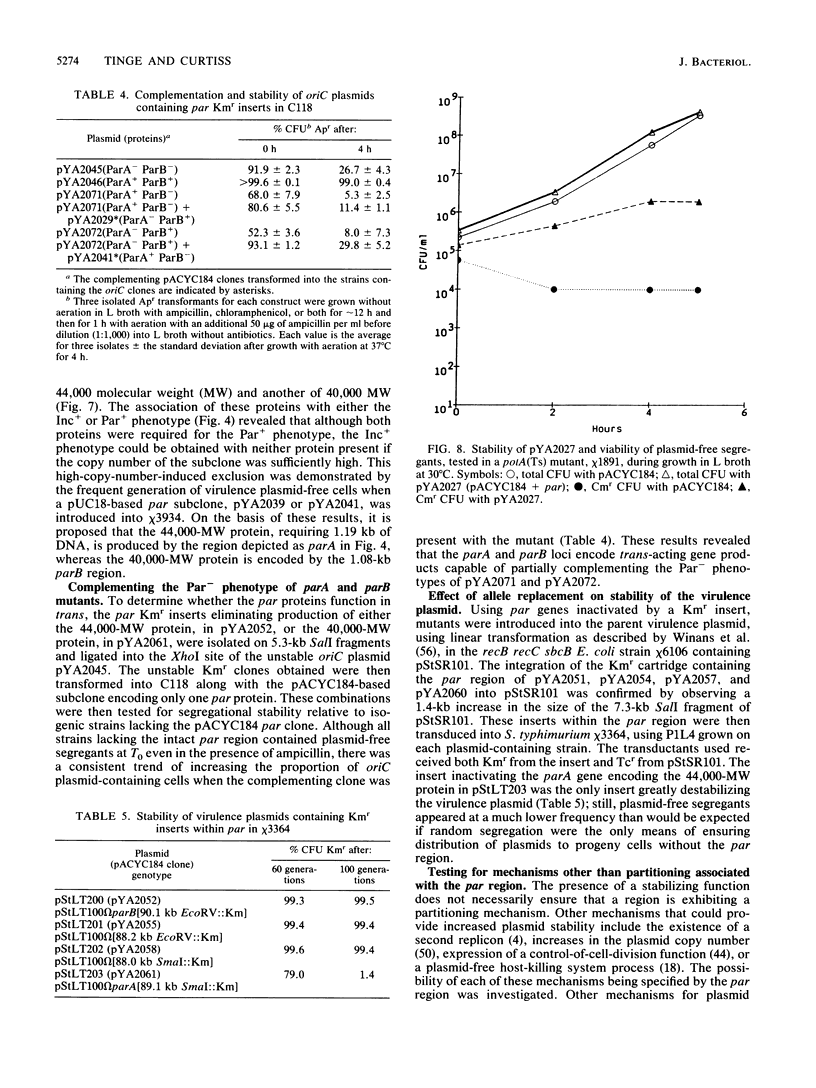
Although the virulence plasmid of Salmonella typhimurium has a copy number of one to two per chromosome, plasmid-free segregants are produced at a rate less than 10(-7) per cell per generation. Three regions appear to be involved in the maintenance of this virulence plasmid. The first two, repB and repC, are functional replicons hybridizing with IncFII and IncFI plasmids, respectively, neither exhibiting the segregational stability of the parent virulence plasmid. The third region, par, cloned as a 3.9-kilobase Sau3A fragment, is not a functional replicon but exhibits incompatibility with the virulence plasmid. Subsequent tests revealed the ability of this 3.9-kilobase par insert to increase the stability of pACYC184 in S. typhimurium from less than 34% to 99% plasmid-containing cells after 50 generations. In addition, the par region increased the stability of oriC, R388, and repC replicons in both S. typhimurium and Escherichia coli hosts. The par region encodes 44,000- and 40,000-molecular-weight proteins essential for the Par+ phenotype but not for the Inc+ phenotype. Although actual sequestering of plasmids within the cell was not demonstrated, all results indicate that the par region described is an actual partitioning locus, similar in organization to those described for plasmids F, P1, and NR1.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Austin S. J. Plasmid partition. Plasmid. 1988 Jul;20(1):1–9. doi: 10.1016/0147-619x(88)90001-7. [DOI] [PubMed] [Google Scholar]
- Austin S., Abeles A. Partition of unit-copy miniplasmids to daughter cells. II. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J Mol Biol. 1983 Sep 15;169(2):373–387. doi: 10.1016/s0022-2836(83)80056-4. [DOI] [PubMed] [Google Scholar]
- Baxter-Gabbard K. L. A simple method for the large-scale preparation of sucrose gradients. FEBS Lett. 1972 Jan 15;20(1):117–119. doi: 10.1016/0014-5793(72)80031-0. [DOI] [PubMed] [Google Scholar]
- Bergquist P. L., Saadi S., Maas W. K. Distribution of basic replicons having homology with RepFIA, RepFIB, and RepFIC among IncF group plasmids. Plasmid. 1986 Jan;15(1):19–34. doi: 10.1016/0147-619x(86)90011-9. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boe L., Gerdes K., Molin S. Effects of genes exerting growth inhibition and plasmid stability on plasmid maintenance. J Bacteriol. 1987 Oct;169(10):4646–4650. doi: 10.1128/jb.169.10.4646-4650.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D. J., Munro D. S., Platt D. J. Recognition of the cryptic plasmid, pSLT, by restriction fingerprinting and a study of its incidence in Scottish Salmonella isolates. J Hyg (Lond) 1986 Oct;97(2):193–197. doi: 10.1017/s0022172400065268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CURTIS S. R., 3rd CHROMOSOMAL ABERRATIONS ASSOCIATED WITH MUTATIONS TO BACTERIOPHAGE RESISTANCE IN ESCHERICHIA COLI. J Bacteriol. 1965 Jan;89:28–40. doi: 10.1128/jb.89.1.28-40.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerin H., Hackett J. Molecular cloning and analysis of the incompatibility and partition functions of the virulence plasmid of Salmonella typhimurium. Microb Pathog. 1989 Aug;7(2):85–99. doi: 10.1016/0882-4010(89)90028-4. [DOI] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978 Jun;134(3):1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couturier M., Bex F., Bergquist P. L., Maas W. K. Identification and classification of bacterial plasmids. Microbiol Rev. 1988 Sep;52(3):375–395. doi: 10.1128/mr.52.3.375-395.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtiss R., 3rd, Charamella L. J., Stallions D. R., Mays J. A. Parental functions during conjugation in Escherichia coli K-12. Bacteriol Rev. 1968 Dec;32(4 Pt 1):320–348. [PMC free article] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Friedman S. A., Austin S. J. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid. 1988 Mar;19(2):103–112. doi: 10.1016/0147-619x(88)90049-2. [DOI] [PubMed] [Google Scholar]
- Gerdes K., Rasmussen P. B., Molin S. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc Natl Acad Sci U S A. 1986 May;83(10):3116–3120. doi: 10.1073/pnas.83.10.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig P. A., Curtiss R., 3rd Cloning and transposon insertion mutagenesis of virulence genes of the 100-kilobase plasmid of Salmonella typhimurium. Infect Immun. 1988 Dec;56(12):3262–3271. doi: 10.1128/iai.56.12.3262-3271.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig P. A., Curtiss R., 3rd Plasmid-associated virulence of Salmonella typhimurium. Infect Immun. 1987 Dec;55(12):2891–2901. doi: 10.1128/iai.55.12.2891-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulig P. A. Virulence plasmids of Salmonella typhimurium and other salmonellae. Microb Pathog. 1990 Jan;8(1):3–11. doi: 10.1016/0882-4010(90)90003-9. [DOI] [PubMed] [Google Scholar]
- Hackett J., Wyk P., Reeves P., Mathan V. Mediation of serum resistance in Salmonella typhimurium by an 11-kilodalton polypeptide encoded by the cryptic plasmid. J Infect Dis. 1987 Mar;155(3):540–549. doi: 10.1093/infdis/155.3.540. [DOI] [PubMed] [Google Scholar]
- Hashimoto-Gotoh T., Franklin F. C., Nordheim A., Timmis K. N. Specific-purpose plasmid cloning vectors. I. Low copy number, temperature-sensitive, mobilization-defective pSC101-derived containment vectors. Gene. 1981 Dec;16(1-3):227–235. doi: 10.1016/0378-1119(81)90079-2. [DOI] [PubMed] [Google Scholar]
- Helmuth R., Stephan R., Bunge C., Hoog B., Steinbeck A., Bulling E. Epidemiology of virulence-associated plasmids and outer membrane protein patterns within seven common Salmonella serotypes. Infect Immun. 1985 Apr;48(1):175–182. doi: 10.1128/iai.48.1.175-182.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan J. E., Kline B. C., Levy S. B. Regions on the F plasmid which affect plasmid maintenance and the ability to segregate into Escherichia coli minicells. Plasmid. 1982 Jul;8(1):36–44. doi: 10.1016/0147-619x(82)90039-7. [DOI] [PubMed] [Google Scholar]
- Jones G. W., Rabert D. K., Svinarich D. M., Whitfield H. J. Association of adhesive, invasive, and virulent phenotypes of Salmonella typhimurium with autonomous 60-megadalton plasmids. Infect Immun. 1982 Nov;38(2):476–486. doi: 10.1128/iai.38.2.476-486.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleckner N., Roth J., Botstein D. Genetic engineering in vivo using translocatable drug-resistance elements. New methods in bacterial genetics. J Mol Biol. 1977 Oct 15;116(1):125–159. doi: 10.1016/0022-2836(77)90123-1. [DOI] [PubMed] [Google Scholar]
- Kline B. C. A review of mini-F plasmid maintenance. Plasmid. 1985 Jul;14(1):1–16. doi: 10.1016/0147-619x(85)90027-7. [DOI] [PubMed] [Google Scholar]
- Kline B. C., Miller J. R. Detection of nonintegrated plasmid deoxyribonucleic acid in the folded chromosome of Escherichia coli: physiochemical approach to studying the unit of segregation. J Bacteriol. 1975 Jan;121(1):165–172. doi: 10.1128/jb.121.1.165-172.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner S. R., Nagaishi H., Templin A., Clark A. J. Genetic recombination in Escherichia coli: the role of exonuclease I. Proc Natl Acad Sci U S A. 1971 Apr;68(4):824–827. doi: 10.1073/pnas.68.4.824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Martinez E., Bartolomé B., de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZ alpha reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988 Aug 15;68(1):159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- Meacock P. A., Cohen S. N. Partitioning of bacterial plasmids during cell division: a cis-acting locus that accomplishes stable plasmid inheritance. Cell. 1980 Jun;20(2):529–542. doi: 10.1016/0092-8674(80)90639-x. [DOI] [PubMed] [Google Scholar]
- Michiels T., Popoff M. Y., Durviaux S., Coynault C., Cornelis G. A new method for the physical and genetic mapping of large plasmids: application to the localisation of the virulence determinants on the 90 kb plasmid of Salmonella typhimurium. Microb Pathog. 1987 Aug;3(2):109–116. doi: 10.1016/0882-4010(87)90069-6. [DOI] [PubMed] [Google Scholar]
- Miki T., Easton A. M., Rownd R. H. Cloning of replication, incompatibility, and stability functions of R plasmid NR1. J Bacteriol. 1980 Jan;141(1):87–99. doi: 10.1128/jb.141.1.87-99.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordström K., Molin S., Aagaard-Hansen H. Partitioning of plasmid R1 in Escherichia coli. II. Incompatibility properties of the partitioning system. Plasmid. 1980 Nov;4(3):332–339. doi: 10.1016/0147-619x(80)90071-2. [DOI] [PubMed] [Google Scholar]
- Norel F., Coynault C., Miras I., Hermant D., Popoff M. Y. Cloning and expression of plasmid DNA sequences involved in Salmonella serotype typhimurium virulence. Mol Microbiol. 1989 Jun;3(6):733–743. doi: 10.1111/j.1365-2958.1989.tb00222.x. [DOI] [PubMed] [Google Scholar]
- Novick R. P., Hoppensteadt F. C. On plasmid incompatibility. Plasmid. 1978 Sep;1(4):421–434. doi: 10.1016/0147-619x(78)90001-x. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Plasmid incompatibility. Microbiol Rev. 1987 Dec;51(4):381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4784–4788. doi: 10.1073/pnas.80.15.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T., Hiraga S. Partition mechanism of F plasmid: two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 1983 Feb;32(2):351–360. doi: 10.1016/0092-8674(83)90454-3. [DOI] [PubMed] [Google Scholar]
- Pardon P., Popoff M. Y., Coynault C., Marly J., Miras I. Virulence-associated plasmids of Salmonella serotype Typhimurium in experimental murine infection. Ann Inst Pasteur Microbiol. 1986 Jul-Aug;137B(1):47–60. doi: 10.1016/s0769-2609(86)80093-x. [DOI] [PubMed] [Google Scholar]
- Poppe C., Curtiss R., 3rd, Gulig P. A., Gyles C. L. Hybridization studies with a DNA probe derived from the virulence region of the 60 Mdal plasmid of Salmonella typhimurium. Can J Vet Res. 1989 Oct;53(4):378–384. [PMC free article] [PubMed] [Google Scholar]
- Smith H. R., Humphreys G. O., Grindley N. D., Grindley J. N., Anderson E. S. Molecular studies of an fi+ plasmid from strains of Salmonella typhimurium. Mol Gen Genet. 1973 Nov 2;126(2):143–151. doi: 10.1007/BF00330989. [DOI] [PubMed] [Google Scholar]
- Summers D. K., Sherratt D. J. Multimerization of high copy number plasmids causes instability: CoIE1 encodes a determinant essential for plasmid monomerization and stability. Cell. 1984 Apr;36(4):1097–1103. doi: 10.1016/0092-8674(84)90060-6. [DOI] [PubMed] [Google Scholar]
- Tabuchi A., Min Y. N., Kim C. K., Fan Y. L., Womble D. D., Rownd R. H. Genetic organization and nucleotide sequence of the stability locus of IncFII plasmid NR1. J Mol Biol. 1988 Aug 5;202(3):511–525. doi: 10.1016/0022-2836(88)90282-3. [DOI] [PubMed] [Google Scholar]
- Terakado N., Hamaoka T., Danbara H. Plasmid-mediated serum resistance and alterations in the composition of lipopolysaccharides in Salmonella dublin. J Gen Microbiol. 1988 Jul;134(7):2089–2093. doi: 10.1099/00221287-134-7-2089. [DOI] [PubMed] [Google Scholar]
- Thomas C. M., Cross M. A., Hussain A. A., Smith C. A. Analysis of copy number control elements in the region of the vegetative replication origin of the broad host range plasmid RK2. EMBO J. 1984 Jan;3(1):57–63. doi: 10.1002/j.1460-2075.1984.tb01761.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinge S. A., Curtiss R., 3rd Conservation of Salmonella typhimurium virulence plasmid maintenance regions among Salmonella serovars as a basis for plasmid curing. Infect Immun. 1990 Sep;58(9):3084–3092. doi: 10.1128/iai.58.9.3084-3092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Winans S. C., Elledge S. J., Krueger J. H., Walker G. C. Site-directed insertion and deletion mutagenesis with cloned fragments in Escherichia coli. J Bacteriol. 1985 Mar;161(3):1219–1221. doi: 10.1128/jb.161.3.1219-1221.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Womble D. D., Rownd R. H. Genetic and physical map of plasmid NR1: comparison with other IncFII antibiotic resistance plasmids. Microbiol Rev. 1988 Dec;52(4):433–451. doi: 10.1128/mr.52.4.433-451.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]