Abstract
A split intein capable of protein trans-splicing is identified in a DnaE protein of the cyanobacterium Synechocystis sp. strain PCC6803. The N- and C-terminal halves of DnaE (catalytic subunit α of DNA polymerase III) are encoded by two separate genes, dnaE-n and dnaE-c, respectively. These two genes are located 745,226 bp apart in the genome and on opposite DNA strands. The dnaE-n product consists of a N-extein sequence followed by a 123-aa intein sequence, whereas the dnaE-c product consists of a 36-aa intein sequence followed by a C-extein sequence. The N- and C-extein sequences together reconstitute a complete DnaE sequence that is interrupted by the intein sequences inside the β- and τ-binding domains. The two intein sequences together reconstitute a split mini-intein that not only has intein-like sequence features but also exhibited protein trans-splicing activity when tested in Escherichia coli cells.
Inteins have been defined as protein sequences embedded in-frame within a precursor protein sequence and excised during a maturation process termed protein splicing (1, 2). Protein splicing is a post-translational event involving precise excision of the intein sequence and concomitant ligation of the flanking sequences (N- and C-exteins) by a normal peptide bond (3–5). Most reported inteins are thought to be bifunctional elements, possessing a protein splicing activity and an endonuclease activity (6–9). Crystal structure of the Sce VMA1 intein revealed a two-domain structure, with domain I consisting of the N- and C-terminal regions of the intein sequence and domain II formed by the middle part of the intein sequence (10). Domain I (or a part of it) was suggested to be the splicing domain, whereas domain II corresponded to the endonuclease domain. Such a bipartite structure may be applicable to many other inteins, as has been suggested by studies including mutagenesis (11, 12) and sequence statistical modeling (7–9). Functional studies of mini-inteins, either found in nature or engineered in vitro, also confirmed such a two-domain model (13–15), further suggesting that the N- and C-terminal regions of an intein make up a functional splicing domain. Molecular mechanisms of protein splicing involve an N→S (or N→O) acyl shift at the N-terminal splice site (16–18), formation of a branched intermediate (19, 20), and cyclization of an invariant Asn residue at the C terminus of intein to form succinimide (21), leading to excision of the intein. The ligated exteins undergo an S→N (or O→N) acyl shift to form a native peptide bond (21). Amino acid residues that are implicated in the splicing mechanism include a nucleophilic amino acid (Cys, Ser, or Thr) both at the beginning of the intein sequence and at the beginning of the C-extein sequence, an internal His, and a His–Asn dipeptide at the end of the intein sequence. In crystal structures of two inteins, these amino acids are indeed positioned at or near the active site of protein splicing (10, 22).
Approximately 50 intein-coding sequences have been found in >20 different genes distributed among the nuclear and organellar genomes of eukaryotes, archaebacteria (archaea), and eubacteria, suggesting a wide distribution of inteins (see the Intein Registry at http://www.neb.com/neb/inteins.html). Inteins, like many introns (23), are thought to be mobile genetic elements that can be transmitted through horizontal transfer (intein homing), and the intein endonuclease activity is thought to initiate this process (24–26). Known inteins share little overall sequence identity, except between homologous inteins found at the same insertion site in homologous proteins of different organisms (6). A number of short sequence motifs do show a low but significant degree of conservation among inteins (6, 27), suggesting similarities in intein structure, function, and evolutionary origin. Previously reported inteins all have continuous sequences, most are 400–500 aa in size with a protein splicing domain and an endonuclease domain, whereas a few mini-inteins are ≈150 aa in size with a splicing domain only. Three intein sequences were found previously in the cyanobacterium Synechocystis sp. strain PCC6803 (Ssp), including the Ssp DnaB intein in a DNA helicase (28), the Ssp DnaX intein in the τ subunit of DNA polymerase III (29), and the Ssp GyrB intein in a DNA gyrase B subunit (7, 9). Here, we report a new intein (Ssp DnaE intein) found in this cyanobacterium and present in a DnaE protein. DnaE is the catalytic subunit of bacterial DNA polymerase III. In E. coli, DNA polymerase III holoenzyme is the replicative polymerase responsible for the synthesis of the majority of the genome. DnaE (also known as α), in addition to its catalytic role, also serves as an organization protein to hold the 18-protein holoenzyme complex together. Its C-terminal half interacts directly with the τ subunit to form a dimeric polymerase and with the β subunit that forms a sliding clamp on the DNA template, whereas its N-terminal half contains the polymerase active site (30). In this study, we show that the DnaE protein of Synechocystis sp. PCC6803 is encoded by a split gene interrupted by intein sequences. In an independent study, Gorbalenya also predicted this intein-containing split DnaE gene through sequence analysis (39). We further demonstrate that the products of the split DnaE gene can undergo protein trans-splicing to form an intact DnaE protein.
EXPERIMENTAL PROCEDURES
DNA Sequence Analysis and Cloning.
The blast search program (31) was used in GenBank searches. Protein sequence alignments were produced by using the clustal w program (32) followed by hand fitting. The Ssp dnaE-coding sequences were prepared from total DNA of Synechocystis sp. strain PCC6803 (Ssp) by a PCR using the thermostable DNA polymerase Pfu (Stratagene). The 2,694-bp dnaE-n gene was amplified by using a pair of oligonucleotide primers: 5′-ATGTCCTTCGTCGGTCYTCCATATC-3′ and 5′-ATCAATAAATCGCCTTCACATTGTAATC-3′. The 1,377-bp dnaE-c gene was amplified by using a pair of oligonucleotide primers: 5′-ATGGTTAAAGTTATCGGTCGTCGTTC-3′ and 5′-CTAGCCAACACTCTGGCTTTGG-3′. A recombinant expression plasmid was constructed as a tripartite fusion of the complete dnaE-c sequence, a portion of the dnaE-n sequence (named dnaE-n′, 1,017 bp), and the expression plasmid vector pET-32 (Novagen) without the thioredoxin gene. A cassette of termination codon followed by Shine-Dalgarno sequence followed by initiation codon was inserted between the two genes by a PCR-mediated method. First, a linear DNA fragment was amplified from the circular plasmid DNA in a PCR, using the Advantage cDNA polymerase mix (CLONTECH) and a pair of oligonucleotide primers: 5′-TTAATAATAATGGGTACCTTGAAAATGGATTTTTTAGGCTTG-3′, and 5′-ATTATTATTAACCTCCTTAACTCTGGCTTTGGGGTAACAGTGG-3′. The amplified linear DNA molecule was circularized to form the expression plasmid.
Protein Production and Splicing in E. coli Cells.
The expression plasmid containing dnaE-c and dnaE-n′ sequences was used to transform E. coli cells. The transformed cells were grown in liquid Lurie Broth medium at 37°C to late log phase (A600, 0.5). Isopropyl β-d-thiogalactoside (IPTG) was added to a final concentration of 0.8 mM to induce production of the recombinant proteins, and the induction was continued overnight at 15°C. Cells were lysed in SDS-containing loading buffer in a boiling water bath before SDS/PAGE. Antisera used in Western blots were raised in rabbits against specific antigens that had been overproduced in E. coli cells transformed with the corresponding genes. The anti-N antiserum was raised against the complete DnaE-n protein. The anti-C antiserum was raised against the first 400 aa of the DnaE-c protein. The specificity of each antiserum was confirmed by testing on the corresponding antigen. The amount of protein in individual protein bands was estimated by using a gel documentation system (Gel Doc 1000 coupled with molecular analyst software, Bio-Rad). A protein band of interest was excised from SDS-polyacrylamide gel after staining, and the protein was electro-eluted and transferred onto poly(vinylidene difluoride) membrane for protein micro-sequencing. In peptide analysis and sequencing, the protein of interest was treated with protease trypsin, the resulting peptides were resolved by HPLC chromatography, peptides of interest were screened by mass spectrometry, and selected peptides were subjected to micro-sequencing. Protein and peptide sequencing, protease digestion, and peptide analysis were all carried out at the Microchemistry Facility of Harvard University.
RESULTS
Sequence Analysis of the Split DnaE Genes.
The complete genome sequence has been determined previously for Synechocystis sp. PCC6803 (33), and a list of the gene content can be seen at the CyanoBase web site (http://www.kazusa.or.jp/cyano/cyano.html). In browsing through this CyanoBase, we noticed that there are two separate ORFs (ORFs slr0603 and sll1572) showing significant sequence similarities to the E. coli DnaE protein (DNA polymerase III α subunit). Further analysis revealed that ORF slr0603 and ORF sll1572 are two members of a discontinuous (split) DnaE gene, and these ORFs subsequently were named dnaE-n and dnaE-c, respectively (Fig. 1). The dnaE-n-coding sequence is 2,694 bp long and spans from base 3,561,946 to 3,564,639 of the genome. The dnaE-c-coding sequence is 1,377 bp long and spans from base 737,811 to 736,435 of the genome. These two genes are separated by 745,226 bp of sequence and numerous unrelated genes on the 3,573,470-bp circular genome. In addition to distance, coding sequences of these two genes are located on opposite DNA strands. There is no indication of intron sequence either downstream of dnaE-n or upstream of dnaE-c. In fact, the dnaE-n gene is followed immediately downstream by a lepA gene that encodes a GTP-binding protein unrelated to DnaE, with a 199-bp intergenic spacer between them. The dnaE-c gene is flanked upstream by an unidentified ORF that is unrelated to DnaE and has some similarity to lysostaphin, with a 215-bp intergenic spacer between them. There is no additional DnaE-like gene listed in the CyanoBase. We also were unable to find an additional DnaE gene (complete or in fragments) either by extensive blast searches of the complete Ssp genome sequence or by Southern blot analysis of the total Ssp DNA by using the Ssp DnaE gene and the E. coli DnaE gene as DNA probes (data not shown).
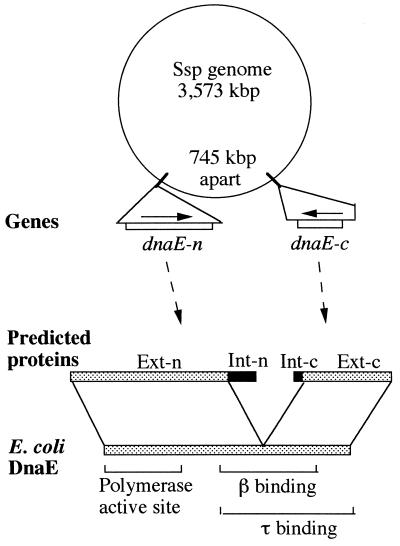
Figure 1.
Gene map and protein structure. Two members of the split DnaE gene, dnaE-n and dnaE-c, are shown on the genome of Synechocystis sp. PCC6803 (Ssp genome). In the predicted proteins, DnaE-related sequences (░⃞) are specified as exteins Ext-n and Ext-c, whereas intein-related sequences (■) are specified as Int-n and Int-c. The exteins are related to E. coli DnaE protein whose functional domains are marked.
Protein sequence deduced from the dnaE-n gene can be divided into two regions: a 774-aa extein region named Ext-n followed by a 123-aa intein region named Int-n. Similarly, protein sequences deduced from the dnaE-c gene can be divided into an intein region (Int-c, 36 aa) followed by an extein region (Ext-c, 423 aa). The Ext-n and Ext-c sequences correspond to the N- and C- terminal halves of a DnaE protein, respectively, and together they reconstitute a complete DnaE sequence. This Ssp DnaE sequence, although discontinuous or split, resembles the continuous DnaE sequences of other organisms both in length and in sequence (Fig. 2A). The Ssp DnaE sequence is 36%, 37%, and 35% identical to DnaE proteins of E. coli, Bacillus subtilis, and Mycobacterium tuberculosis, respectively, over the entire 1,196 aa sequence. These degrees of sequence identity are comparable with the 35–36% sequence identities found among DnaE proteins of the other three compared bacterial organisms.
Figure 2.
Sequence analysis. (A) Sequence comparison to DnaE proteins. The Ssp DnaE extein sequences (Ssp) are aligned with corresponding DnaE sequences of E. coli (Eco), Bacillus subtilis (Bsu), and Mycobacterium tuberculosis (Mtu). Only sequences proximal to the intein sequences (Int-n and Int-c) are shown, whereas the number of omitted residues at the N- and C-termini are shown in parentheses. Symbols: − represent gaps introduced to optimize the alignment; ∗ and . mark positions of identical and similar amino acids, respectively. (B) Sequence comparison to inteins. The Ssp DnaE intein sequences (Ssp DnaE), consisting of Int-n and Int-c as indicated, are aligned with corresponding sequences of Rhodothermus marinas DnaB intein (Rma DnaB), Synechocystis sp. PCC6803 DnaB intein (Ssp DnaB), and Porphyra purpurea chloroplast DnaB intein (Ppu DnaB). In the Rma DnaB intein and the Ssp DnaB intein, only sequences relating to Int-n and Int-c are shown, whereas the number of omitted residues are shown in parentheses. Putative intein motifs (Blocks A, B, F, and G) are underlined, with several critical residues marked by ∗.
The Int-n and Int-c sequences show no detectable similarity to DnaE proteins but instead have marked similarity to known intein sequences (Fig. 2B). Int-n and Int-c correspond to the N- and C-terminal halves of the intein, and together they reconstitute a mini-intein sequence (named Ssp DnaE intein) with a composite length of 159 aa. The sequence of this discontinuous (split) Ssp DnaE intein is most similar to corresponding sequences of the Rma DnaB intein found previously in a DnaB protein (DNA helicase) of the thermophilic eubacterium Rhodothermus marinus (34). The Ssp DnaE intein sequence is 30% identical to the Rma DnaB intein and 22% identical to the Ssp DnaB intein over the 159-aa sequence. Much lower sequence identities were found in comparing it with other known inteins. The Ssp DnaE intein, in addition to being split, lacks sequences for a centrally located endonuclease domain that is present in most known inteins including the Rma DnaB intein. Nevertheless, the split Ssp DnaE intein has many known sequence features of an intein splicing domain. A 50% sequence identity was found between the Ssp DnaE intein and the Rma DnaB intein over the conserved sequence blocks (A, B, F, and G, totaling 49 aa). Residues important for the catalysis of protein splicing were found in the Ssp DnaE intein, including a nucleophilic residue (Cys) at the beginning of the intein sequence, another Cys at the beginning of the C-extein, a Thr and a His in sequence block B, and an Asn at the end of the intein. An Ala precedes the C-terminal Asn in the Ssp DnaE intein, although this position is occupied by His in most, but not all, known inteins.
The insertion site of the split Ssp DnaE intein is inside the β- and τ-binding domains but outside the polymerase active site of the DnaE protein, according to a comparison with the better studied E. coli DnaE protein (Fig. 1). The Ssp DnaE intein disrupts a conserved region of the DnaE sequence (Fig. 2A), which helped to define the extein–intein boundaries. The first residue of Ext-c in the Ssp DnaE sequence is Cys, whereas this position in the other DnaE proteins is occupied by Gly or Ala. This observation is consistent with a requirement of the Cys in Ssp DnaE for protein splicing and the absence of an intein in the other DnaE proteins.
Protein Trans-Splicing.
The split Ssp DnaE intein was tested in E. coli cells for protein trans-splicing activity (Fig. 3). The DnaE-n- and DnaE-c-coding sequences were inserted into an expression plasmid vector to form a two-gene operon (Fig. 3A), allowing production of the two proteins inside the same E. coli cell and from a single inducible promoter. The construct contained the complete DnaE-c-coding sequence and a partial DnaE-n-coding sequence. Using a complete DnaE-n-coding sequence resulted in lower production and elevated degradation (fragmentation) of the protein (data not shown). The partial DnaE-n sequence is termed DnaE-n′ and consisted of a portion of the Ext-n sequence (216 aa, proximal to the intein) followed by the entire Int-n sequence. The DnaE-c- and DnaE-n′-coding sequences were separated by a small intergenic spacer that contained a Shine-Dalgarno sequence (ribosome-binding site) followed by an AT-rich sequence. The DnaE-c-coding sequence was placed in front of the DnaE-n-coding sequence, preventing accidental fusion of the split intein sequences, which might arise through accidental translation of the small intergenic spacer.
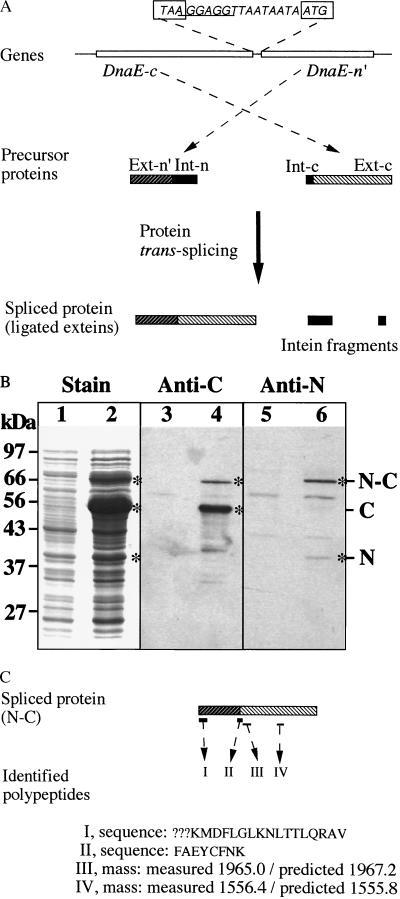
Figure 3.
Protein trans-splicing. The dnaE-n and dnaE-c genes are co-expressed in E. coli cells to observe protein trans-splicing. (A) Schematic illustration. The genes are constructed as a two-gene operon in an expression plasmid vector, with the complete DnaE-c-coding sequence followed by a partial DnaE-n-coding sequence (DnaE-n′). In the intergenic spacer, the termination codon (TAA) of DnaE-c and the initiation codon of DnaE-n′ are boxed, and the Shine-Dalgarno sequence (ribosome-binding site) is underlined. Products of the two genes are shown as precursor proteins, with their extein regions (Ext-n′ and Ext-c) and intein regions (Int-n and Int-c) as indicated. Protein trans-splicing produces a spliced protein and excised intein fragments. (B) Protein gels. Total proteins of uninduced cells (lanes 1, 3, 5) and induced cells (lanes 2, 4, 6) were resolved by SDS/PAGE and visualized by staining (lanes 1 and 2), by Western blotting with anti-C (DnaE-c) antiserum (lanes 3 and 4), or by Western blotting with anti-N (DnaE-n) antiserum (lanes 5 and 6). Positions of precursor proteins (N and C) and the spliced protein (N-C) are marked. (C) Identification of the spliced protein. Peptides I and II were identified by sequencing, and the determined sequences are shown (? marks undetermined residues). Peptides III and IV were identified by mass, with the measured value compared with predicted value.
E. coli cells containing the above recombinant plasmid were induced to produce the DnaE-c protein, the DnaE-n′ protein, and possibly a spliced protein. Three protein products (C, N, and N-C) were observed after the induction (Fig. 3B). Protein C and protein N were identified as the precursor proteins DnaE-c and DnaE-n′, respectively. Their apparent sizes matched closely the predicted sizes (51 kDa for C and 38 kDa for N), and each of them was recognized specifically by antiserum raised against that protein. The third protein, N-C, was identified as a spliced protein (ligated exteins). First, its apparent size matched closely the predicted size of a spliced protein (71 kDa). Second, protein N-C was recognized by both the anti-N and the anti-C antisera, indicating that it contains both DnaE-n and DnaE-c sequences. Finally, protein N-C was firmly identified as the spliced protein by protein sequencing and peptide analysis (Fig. 3C). N-terminal protein sequencing of protein N-C revealed a 17-aa sequence, KMDFLGLKNLTTLQRAV, which matched precisely the predicted DnaE-n′ sequence at amino acid positions 5–21. Amino acids at positions 2–4 were not determined, because of sequencing failures at these positions, and the N-terminal f-Met apparently had been removed in the E. coli cell. The protein N-C was further treated with protease trypsin, and the resulting polypeptides were selectively analyzed. Two polypeptides (peptides III and IV) inside the DnaE-c sequence were identified by matching their molecular masses to predicted molecular masses. Peptide III corresponded to the sequence SHSTAYAYVTYQTAYLK (amino acid positions 220–236), whereas peptide IV corresponded to the sequence EHLGFYVSEHPLK (amino acid positions 428–440). Most importantly, a polypeptide (peptide II) spanning the spliced junction was identified and sequenced. Its sequence, FAEYCFNK, matches precisely the predicted sequence in a spliced protein, with the sequence FAEY being the last four residues of Ext-n′ and the sequence CFNK being the first four residues of Ext-c. This shows precise excision of the intein sequences (Int-n and Int-c) and joining of the extein sequences (Ext-n′ and Ext-c) by a normal peptide bond. The two excised intein fragments were predicted but not observed, most likely because of their small sizes (14 kDa for Int-n and 4 kDa for Int-c), weak binding by the anti-N and anti-C antisera, and/or rapid degradation in the E. coli cell. Nevertheless, production of the spliced protein (protein N-C) demonstrates that protein trans-splicing had occurred. Comparing the amount of protein N-C and the amount of protein N indicates that ≈80% of the precursor protein N was incorporated into the spliced protein. The remaining protein N may have misfolded. Protein C accumulated much more than protein N, indicating that the dnaE-c gene was expressed much more than the downstream dnaE-n′ gene. This may be because of inefficient translational coupling of the two-gene operon or a more rapid degradation of protein N.
DISCUSSION
The Ssp DnaE intein is identified as a naturally occurring split mini-intein in Synechocystis sp. PCC6803, and it is shown to be capable of protein trans-splicing. The two DnaE-like genes, dnaE-n and dnaE-c, are clearly two members of an intein-containing split DnaE gene, with the split being inside the intein-coding sequence. Protein sequences deduced from the split DnaE gene, after excluding the intein sequences, reconstitute a complete DnaE protein that has neither gap nor overlapping sequences at the split point. It also has the expected degrees of sequence identity to the continuous DnaE sequences of other bacterial organisms. The two intein sequences, Int-n and Int-c, not only have intein-like sequence features but also are proven to be two parts of a split intein by demonstrating a protein trans-splicing activity in E. coli cells. This Ssp DnaE intein, consisting of two separate polypeptides with a composite size of 159 aa, represents a split mini-intein that is apparently capable of forming a functional splicing domain. Four conserved sequence blocks (A, B, F, G) have been previously localized in the splicing domain of inteins (6, 10, 15, 22, 27, 37). All of the four sequence blocks appear to exist in the Ssp DnaE intein (Fig. 2B), with blocks A and B located on Int-n, with blocks F and G located on Int-c. The Ssp DnaE intein lacks a highly conserved His residue (replaced by Ala) immediately before the C-terminal Asn. Four other inteins (Ceu ClpP, Mja PEP, Mja KlbA, and Mja RpolA′) also lack this penultimate His, in which the His is replaced by Gly, Ser, or Phe. This His has been shown to assist in Asn cyclization leading to cleavage of the peptide bond between intein and C-extein (17), and efficient splicing of the Ceu ClpP intein in E. coli cells required a restoration of this His residue (35). The observation of trans-splicing activity with the Ssp DnaE intein shows that this His residue is not required for protein splicing of this intein.
The finding of a split mini-intein has implications on intein evolution. The Ssp DnaE intein likely evolved from a continuous intein that later lost its sequence continuity. This result probably occurred through one or more genomic rearrangement events that separated the two halves of the DnaE gene (dnaE-n and dnaE-c) to different parts of the genome. A possible progenitor DnaE intein has not been found, and the 30% sequence identity between Ssp DnaE intein and the Rma DnaB intein (present in a DNA helicase) may be a coincidence, considering that the two inteins have nonhomologous exteins and dissimilar insertion sites. Emergence of a split intein requires that it possesses protein trans-splicing activity, unless the exteins can function without ligation and without removing the intein sequences. Other inteins also may possess a potential of becoming split inteins, as protein trans-splicing has been demonstrated with intein fragments engineered from several continuous inteins (36, 37, 40, 41). The Ssp DnaE intein (in fragments) has a total size of a mini-intein (splicing domain only) and lacks any of the endonuclease sequence motifs. The Ssp DnaE intein, like other inteins that lack an endonuclease domain, may once have had and lost the endonuclease domain (13), or alternatively it may never have acquired an endonuclease domain. The split site in the Ssp DnaE intein coincides with predicted endonuclease insertion site, indicating that this site of the intein is tolerant of both insertion and cleavage. If the Ssp DnaE intein once had and lost its endonuclease domain, this could have occurred before or after the loss of sequence continuity. An intein presumably loses the ability of intein homing once the endonuclease domain is lost. As for the Ssp DnaE intein, having the two intein fragments on different parts of the genome would prevent intein homing even if the endonuclease domain were present.
The Ssp DnaE intein likely does protein trans-splicing in its native cyanobacterial cell, as it did so in E. coli cells. A DnaE protein, either a spliced protein or precursors, has not been detected in the total protein of Synechocystis sp. PCC6803 by using the available anti-DnaE antisera (data not shown). This is most likely because of a combination of weak antisera and low levels of the DnaE protein. DnaE has been known to exist at very low levels in other bacterial cells. The E. coli DnaE protein was estimated at 10–12 molecules per cell (38), which is sufficient to replicate the E. coli genome approximately every 0.5 hr. In comparison, Synechocystis sp. PCC6803 has a smaller genome that needs to be duplicated only every 10 hr (approximate cell-doubling time). It is therefore not unreasonable for this organism to have extremely low levels of the DnaE protein for DNA replication. Nevertheless, a DnaE protein is essential for the cell, and there is no other DnaE-like gene (complete or partial) beside dnaE-n and dnaE-c in this genome. These two genes, unlike pseudo genes, maintain long ORFs (2,694 bp for dnaE-n and 1,377 bp for dnaE-c), whereas their noncoding frames have numerous termination codons. Production of a functional DnaE protein likely requires protein trans-splicing to remove the intein sequences and ligate the extein sequences. It is less likely, although possible, for the two precursor proteins (DnaE-n and DnaE-c) to reconstitute a functional protein without splicing, considering that the intein sequences interrupt both the β-binding domain and the τ-binding domain. Although the polymerase active site is contained within the DnaE-n precursor protein, both the β-binding domain and the τ-binding domain are interrupted by the intein sequences and split between the DnaE-n and DnaE-c precursor proteins. There is no indication that the half intein sequences (Int-n and Int-c) can be cleaved off the precursor proteins without undergoing protein trans-splicing. Such a cleavage product was not observed with the DnaE-n and DnaE-c proteins in E. coli. Half inteins engineered in vitro from other inteins also lack such a cleavage activity (36, 37). Functional β- and τ-binding domains are essential, because interactions of DnaE with the β subunit (DNA clamp) and the τ subunit are critical for the function of DNA polymerase III (30).
Protein trans-splicing has been demonstrated with engineered inteins in vivo and in vitro (36, 37, 40, 41) and has produced insights into the structural requirements for protein splicing. The discovery of the Ssp DnaE intein, a natural split intein that does protein trans-splicing, provides a new perspective on this phenomenon. In terms of structural requirements for protein splicing, the size and sequence of this naturally evolved split mini-intein are in close agreement with those of the smallest functional mini-inteins that have been engineered so far in a laboratory (15, 41). In terms of possible biological function, the trans-splicing reaction between the DnaE-n and DnaE-c precursor proteins may present a step in which the synthesis of a functional DnaE protein is regulated. Absence of the penultimate C-terminal His residue (replaced by Ala) in the Ssp DnaE intein, although not preventing protein trans-splicing, may slow down the splicing reaction, as was the case for other inteins (16, 17, 35). A slow and regulated splicing step may be a mechanism for assuring very low levels of production of the mature DnaE protein. The β and τ subunits of DNA polymerase III bind strongly with the DnaE protein and may therefore affect the trans-splicing reaction by bringing together the two precursor polypeptides of DnaE. It is interesting that the τ subunit of this organism also has an intein (Ssp DnaX intein), although the Ssp DnaX intein has a continuous sequence and is not specifically related to the Ssp DnaE intein in sequence and insertion site (29).
Acknowledgments
This work was supported by a grant from the Medical Research Council of Canada.
ABBREVIATIONS
- Ssp
Synechocystis sp. PCC6803
- DnaE
catalytic subunit α of DNA polymerase III
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Perler F B, Davis E O, Dean G E, Gimble F S, Jack W E, Neff N, Noren C J, Thorner J, Belfort M. Nucleic Acids Res. 1994;22:1125–1127. doi: 10.1093/nar/22.7.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perler F B. Cell. 1998;92:1–4. doi: 10.1016/s0092-8674(00)80892-2. [DOI] [PubMed] [Google Scholar]
- 3.Kane P M, Yamashiro C T, Wolczyk D F, Neff N, Goebl M, Stevens T H. Science. 1990;250:651–657. doi: 10.1126/science.2146742. [DOI] [PubMed] [Google Scholar]
- 4.Colston M J, Davis E O. Mol Microbiol. 1994;12:359–363. doi: 10.1111/j.1365-2958.1994.tb01025.x. [DOI] [PubMed] [Google Scholar]
- 5.Cooper A A, Stevens T H. Trends Biochem Sci. 1995;20:351–356. doi: 10.1016/s0968-0004(00)89075-1. [DOI] [PubMed] [Google Scholar]
- 6.Perler F B, Olsen G J, Adam E. Nucleic Acids Res. 1997;25:1087–1093. doi: 10.1093/nar/25.6.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalgaard J Z, Moser M J, Hughey R, Mian I S. J Comp Biol. 1997;4:193–214. doi: 10.1089/cmb.1997.4.193. [DOI] [PubMed] [Google Scholar]
- 8.Dalgaard J Z, Klar A J, Moser M J, Holley W R, Chatterjee A, Mian I S. Nucleic Acids Res. 1997;25:4626–4638. doi: 10.1093/nar/25.22.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pietrokovski S. Protein Sci. 1998;7:64–71. doi: 10.1002/pro.5560070106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duan X, Gimble F S, Quiocho F A. Cell. 1997;89:555–564. doi: 10.1016/s0092-8674(00)80237-8. [DOI] [PubMed] [Google Scholar]
- 11.Kawasaki M, Nogami S, Satow Y, Ohya Y, Anraku Y. J Biol Chem. 1997;272:15668–15674. doi: 10.1074/jbc.272.25.15668. [DOI] [PubMed] [Google Scholar]
- 12.Nogami S, Satow Y, Ohya Y, Anraku Y. Genetics. 1997;147:73–85. doi: 10.1093/genetics/147.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Telenti A, Southworth M, Alcaide F, Daugelat S, Jacobs W R, Jr, Perler F B. J Bacteriol. 1997;179:6378–6382. doi: 10.1128/jb.179.20.6378-6382.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chong S, Xu M-Q. J Biol Chem. 1997;272:15587–15590. doi: 10.1074/jbc.272.25.15587. [DOI] [PubMed] [Google Scholar]
- 15.Derbyshire V, Wood D W, Wu W, Dansereau J T, Dalgaard J Z, Belfort M. Proc Natl Acad Sci USA. 1997;94:11466–11471. doi: 10.1073/pnas.94.21.11466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao Y, Xu M Q, Paulus H. Biochemistry. 1996;35:3810–3815. doi: 10.1021/bi952592h. [DOI] [PubMed] [Google Scholar]
- 17.Xu M-Q, Perler F B. EMBO J. 1996;15:5146–5153. [PMC free article] [PubMed] [Google Scholar]
- 18.Chong, S., Shao, Y., Paulus, H., Benner, J. & Perler, F. B. (1996) J. Biol. Chem. 271. [DOI] [PubMed]
- 19.Xu M Q, Southworth M W, Mersha F B, Hornstra L J, Perler F B. Cell. 1993;75:1371–1377. doi: 10.1016/0092-8674(93)90623-x. [DOI] [PubMed] [Google Scholar]
- 20.Xu M Q, Comb D G, Paulus H, Noren C J, Shao Y, Perler F B. EMBO J. 1994;13:5517–5522. doi: 10.1002/j.1460-2075.1994.tb06888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shao Y, Xu M Q, Paulus H. Biochemistry. 1995;34:10844–10850. doi: 10.1021/bi00034a017. [DOI] [PubMed] [Google Scholar]
- 22.Klabunde T, Sharma S, Telenti A, Jacobs Jr W R, Sacchettini J C. Nat Struct Biol. 1998;5:31–36. doi: 10.1038/nsb0198-31. [DOI] [PubMed] [Google Scholar]
- 23.Lambowitz A M, Belfort M. Annu Rev Biochem. 1993;62:587–622. doi: 10.1146/annurev.bi.62.070193.003103. [DOI] [PubMed] [Google Scholar]
- 24.Gimble F S, Thorner J. Nature (London) 1992;357:301–306. doi: 10.1038/357301a0. [DOI] [PubMed] [Google Scholar]
- 25.Shub D A, Goodrich-Blair H. Cell. 1992;71:183–186. doi: 10.1016/0092-8674(92)90345-d. [DOI] [PubMed] [Google Scholar]
- 26.Doolittle R F. Proc Natl Acad Sci USA. 1993;90:5379–5381. doi: 10.1073/pnas.90.12.5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pietrokovski S. Protein Sci. 1994;3:2340–2350. doi: 10.1002/pro.5560031218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pietrokovski S. Trends Genet. 1996;12:287–288. doi: 10.1016/0168-9525(96)20005-8. [DOI] [PubMed] [Google Scholar]
- 29.Liu X-Q, Hu Z. FEBS Lett. 1997;408:311–314. doi: 10.1016/s0014-5793(97)00393-1. [DOI] [PubMed] [Google Scholar]
- 30.Kim D R, McHenry C S. J Biol Chem. 1996;271:20699–20704. doi: 10.1074/jbc.271.34.20699. [DOI] [PubMed] [Google Scholar]
- 31.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 34.Liu X-Q, Hu Z. Proc Natl Acad Sci USA. 1997;94:7851–7856. doi: 10.1073/pnas.94.15.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang S, Liu X-Q. J Biol Chem. 1997;272:11869–11873. doi: 10.1074/jbc.272.18.11869. [DOI] [PubMed] [Google Scholar]
- 36.Southworth M W, Adam E, Panne D, Byer R, Kautz R, Perler F B. EMBO J. 1998;17:918–926. doi: 10.1093/emboj/17.4.918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shingledecker K, Jiang S-Q, Paulus H. Gene. 1998;207:187–195. doi: 10.1016/s0378-1119(97)00624-0. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y H, Franden M A, Hawker J R, McHenry C S. J Biol Chem. 1984;259:12117–12122. [PubMed] [Google Scholar]
- 39.Gorbalenya A E. Nucleic Acids Res. 1998;26:1741–1748. doi: 10.1093/nar/26.7.1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mills K V, Lew B M, Jiang S-Q, Paulus H. Proc Natl Acad Sci USA. 1998;95:3543–3548. doi: 10.1073/pnas.95.7.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu, H., Xu, M.-Q. & Liu X.-Q. (1998) Biochim. Biophys. Acta, in press.