Abstract
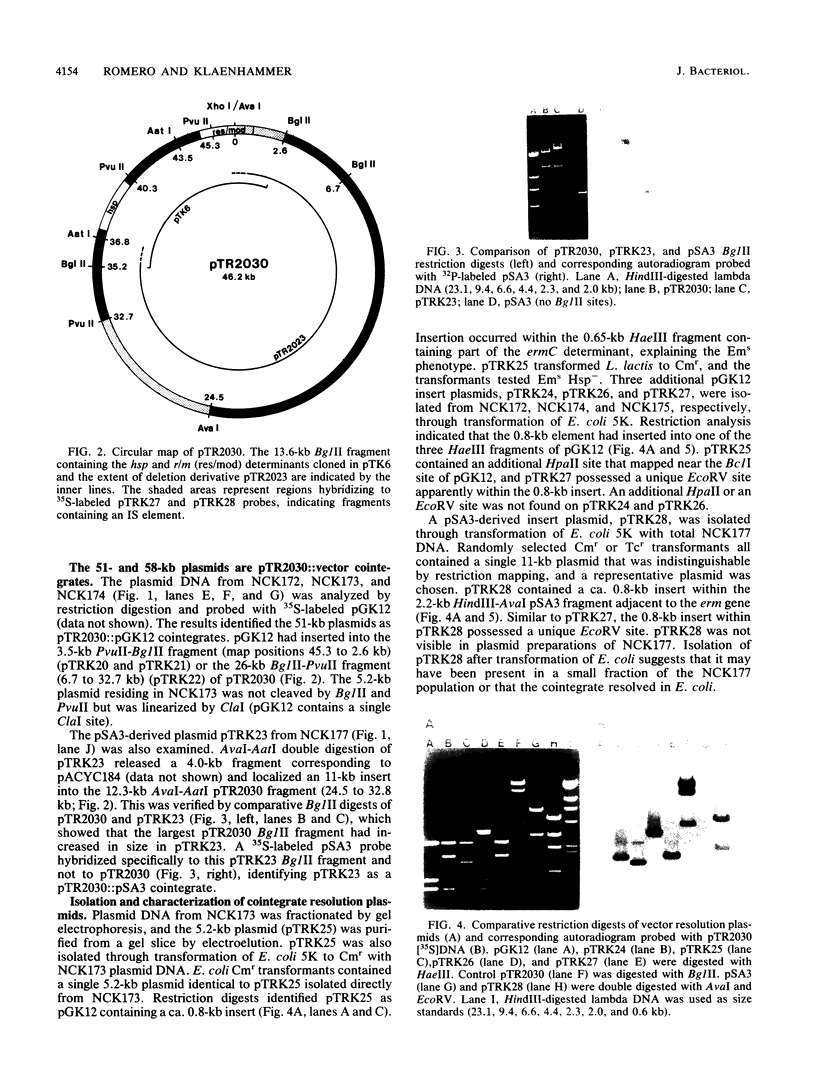
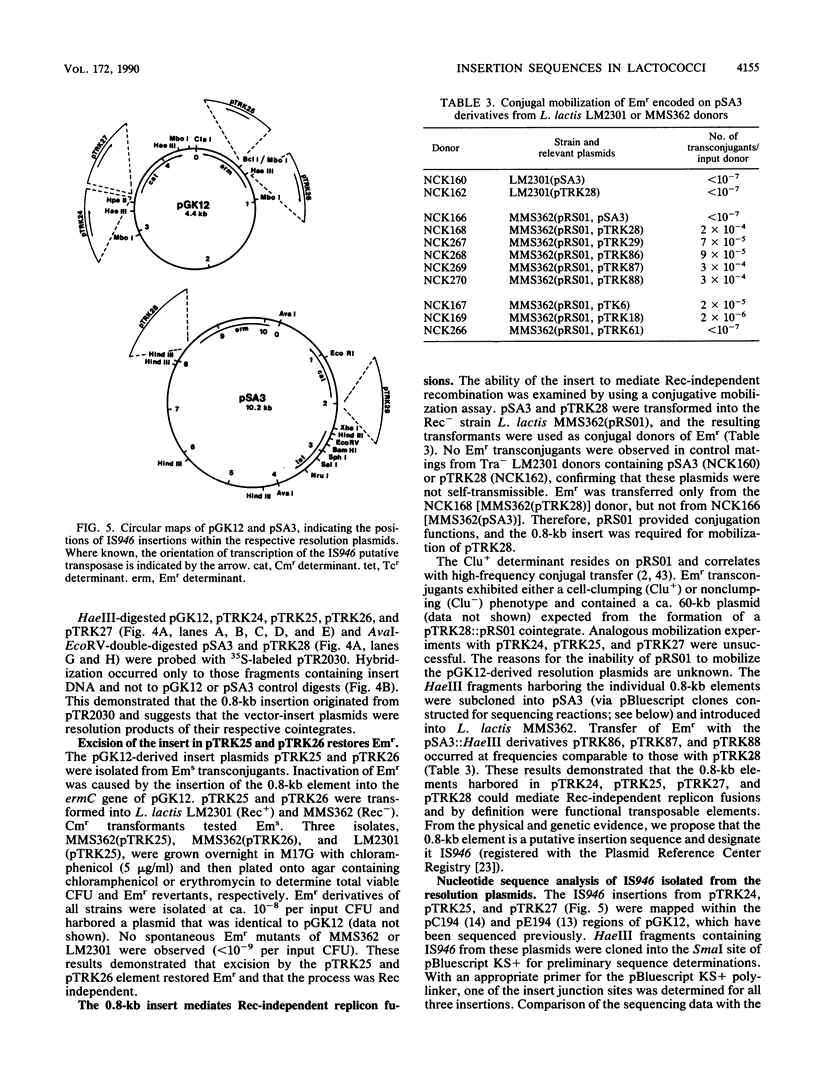
The self-transmissible plasmid pTR2030 mobilized nonconjugative heterologous cloning vectors pGK12 (Cmr Emr) and pSA3 (Emr) at frequencies of 10(-5) to 10(-6) per input donor. Transconjugants harbored a 51- or 58-kilobase (kb) plasmid not found in the parental strains that cotransferred at high frequency with Cmr Emr and pTR2030-encoded phage resistance (Hsp+) in second-round matings (10(-1) per input donor). Restriction endonuclease mapping and DNA-DNA hybridization identified the 51- to 58-kb plasmids as pTR2030::vector cointegrates. Examination of four cointegrates indicated that pGK12 and pSA3 had inserted within two locations on pTR2030. Resolution of the cointegrates generated vector derivatives containing a 0.8-kb insert of pTR2030 DNA. Restriction analyses of several resolution plasmids indicated that the 0.8-kb element had inserted into various positions within pGK12 and pSA3 and in certain cases had inactivated the Cmr or Emr marker of pGK12. A conjugative mobilization assay demonstrated that the 0.8-kb element, designated IS946, mediated transpositional recombination. Nucleotide sequence determination identified IS946 as an 808-base-pair (bp) insertion sequence sharing ca. 96% homology with lactococcal insertion sequence ISS1. IS946 differed by 27 and 31 bp from ISS1S and ISS1T, respectively, and in 2 of 226 amino acids in the deduced sequence of the putative transposase. IS946 has perfect 18-bp terminal inverted repeats, identical to ISS1, and similarly generated 8-bp direct repeats of the target site upon insertion.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson D. G., McKay L. L. Genetic and physical characterization of recombinant plasmids associated with cell aggregation and high-frequency conjugal transfer in Streptococcus lactis ML3. J Bacteriol. 1984 Jun;158(3):954–962. doi: 10.1128/jb.158.3.954-962.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson D. G., McKay L. L. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl Environ Microbiol. 1983 Sep;46(3):549–552. doi: 10.1128/aem.46.3.549-552.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calos M. P., Miller J. H. Transposable elements. Cell. 1980 Jul;20(3):579–595. doi: 10.1016/0092-8674(80)90305-0. [DOI] [PubMed] [Google Scholar]
- Clark A. J., Warren G. J. Conjugal transmission of plasmids. Annu Rev Genet. 1979;13:99–125. doi: 10.1146/annurev.ge.13.120179.000531. [DOI] [PubMed] [Google Scholar]
- Dao M. L., Ferretti J. J. Streptococcus-Escherichia coli shuttle vector pSA3 and its use in the cloning of streptococcal genes. Appl Environ Microbiol. 1985 Jan;49(1):115–119. doi: 10.1128/aem.49.1.115-119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grindley N. D., Reed R. R. Transpositional recombination in prokaryotes. Annu Rev Biochem. 1985;54:863–896. doi: 10.1146/annurev.bi.54.070185.004243. [DOI] [PubMed] [Google Scholar]
- Guyer M. S. The gamma delta sequence of F is an insertion sequence. J Mol Biol. 1978 Dec 15;126(3):347–365. doi: 10.1016/0022-2836(78)90045-1. [DOI] [PubMed] [Google Scholar]
- Guyer M. S. Uses of the transposon gamma delta in the analysis of cloned genes. Methods Enzymol. 1983;101:362–369. doi: 10.1016/0076-6879(83)01027-7. [DOI] [PubMed] [Google Scholar]
- Higgins D. L., Sanozky-Dawes R. B., Klaenhammer T. R. Restriction and modification activities from Streptococcus lactis ME2 are encoded by a self-transmissible plasmid, pTN20, that forms cointegrates during mobilization of lactose-fermenting ability. J Bacteriol. 1988 Aug;170(8):3435–3442. doi: 10.1128/jb.170.8.3435-3442.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Pierce K., Klaenhammer T. R. The conjugative plasmid pTR2030 encodes two bacteriophage defense mechanisms in lactococci, restriction modification (R+/M+) and abortive infection (Hsp+). Appl Environ Microbiol. 1989 Sep;55(9):2416–2419. doi: 10.1128/aem.55.9.2416-2419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill C., Romero D. A., McKenney D. S., Finer K. R., Klaenhammer T. R. Localization, cloning, and expression of genetic determinants for bacteriophage resistance (Hsp) from the conjugative plasmid pTR2030. Appl Environ Microbiol. 1989 Jul;55(7):1684–1689. doi: 10.1128/aem.55.7.1684-1689.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J Bacteriol. 1982 May;150(2):815–825. doi: 10.1128/jb.150.2.815-825.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Weisblum B. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibodies. J Bacteriol. 1982 May;150(2):804–814. doi: 10.1128/jb.150.2.804-814.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubacek J., Glover S. W. Complementation analysis of temperature-sensitive host specificity mutations in Escherichia coli. J Mol Biol. 1970 May 28;50(1):111–127. doi: 10.1016/0022-2836(70)90108-7. [DOI] [PubMed] [Google Scholar]
- Klaenhammer T. R., McKay L. L., Baldwin K. A. Improved lysis of group N streptococci for isolation and rapid characterization of plasmid deoxyribonucleic acid. Appl Environ Microbiol. 1978 Mar;35(3):592–600. doi: 10.1128/aem.35.3.592-600.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaenhammer T. R., Sanozky R. B. Conjugal transfer from Streptococcus lactis ME2 of plasmids encoding phage resistance, nisin resistance and lactose-fermenting ability: evidence for a high-frequency conjugative plasmid responsible for abortive infection of virulent bacteriophage. J Gen Microbiol. 1985 Jun;131(6):1531–1541. doi: 10.1099/00221287-131-6-1531. [DOI] [PubMed] [Google Scholar]
- Kleckner N. Translocatable elements in procaryotes. Cell. 1977 May;11(1):11–23. doi: 10.1016/0092-8674(77)90313-0. [DOI] [PubMed] [Google Scholar]
- Kok J., van der Vossen J. M., Venema G. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl Environ Microbiol. 1984 Oct;48(4):726–731. doi: 10.1128/aem.48.4.726-731.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lederberg E. M. Plasmid Reference Center Registry of transposon(Tn) and insertion sequence (IS) allocations through December 1986. Gene. 1987;51(2-3):115–118. doi: 10.1016/0378-1119(87)90299-x. [DOI] [PubMed] [Google Scholar]
- Liu L., Whalen W., Das A., Berg C. M. Rapid sequencing of cloned DNA using a transposon for bidirectional priming: sequence of the Escherichia coli K-12 avtA gene. Nucleic Acids Res. 1987 Nov 25;15(22):9461–9469. doi: 10.1093/nar/15.22.9461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchansky J. B., Benson A. K., Atherly A. G. Construction, transfer and properties of a novel temperature-sensitive integrable plasmid for genomic analysis of Staphylococcus aureus. Mol Microbiol. 1989 Jan;3(1):65–78. doi: 10.1111/j.1365-2958.1989.tb00105.x. [DOI] [PubMed] [Google Scholar]
- Macrina F. L., Kopecko D. J., Jones K. R., Ayers D. J., McCowen S. M. A multiple plasmid-containing Escherichia coli strain: convenient source of size reference plasmid molecules. Plasmid. 1978 Jun;1(3):417–420. doi: 10.1016/0147-619x(78)90056-2. [DOI] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Walsh P. M. Conjugal transfer of genetic information in group N streptococci. Appl Environ Microbiol. 1980 Jul;40(1):84–89. doi: 10.1128/aem.40.1.84-91.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay L. L., Baldwin K. A., Zottola E. A. Loss of lactose metabolism in lactic streptococci. Appl Microbiol. 1972 Jun;23(6):1090–1096. doi: 10.1128/am.23.6.1090-1096.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novel M., Huang D. C., Novel G. Transposition of the Streptococcus lactis ssp. lactis Z270 lactose plasmid to pVA797: demonstration of an insertion sequence and its relationship to an inverted repeat sequence isolated by self-annealing. Biochimie. 1988 Apr;70(4):543–551. doi: 10.1016/0300-9084(88)90091-0. [DOI] [PubMed] [Google Scholar]
- Polzin K. M., Shimizu-Kadota M. Identification of a new insertion element, similar to gram-negative IS26, on the lactose plasmid of Streptococcus lactis ML3. J Bacteriol. 1987 Dec;169(12):5481–5488. doi: 10.1128/jb.169.12.5481-5488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein S. J., Reznikoff W. S. The functional differences in the inverted repeats of Tn5 are caused by a single base pair nonhomology. Cell. 1981 Jan;23(1):191–199. doi: 10.1016/0092-8674(81)90284-1. [DOI] [PubMed] [Google Scholar]
- Rouch D. A., Skurray R. A. IS257 from Staphylococcus aureus: member of an insertion sequence superfamily prevalent among gram-positive and gram-negative bacteria. Gene. 1989;76(2):195–205. doi: 10.1016/0378-1119(89)90160-1. [DOI] [PubMed] [Google Scholar]
- Sanders M. E., Leonhard P. J., Sing W. D., Klaenhammer T. R. Conjugal strategy for construction of fast Acid-producing, bacteriophage-resistant lactic streptococci for use in dairy fermentations. Appl Environ Microbiol. 1986 Nov;52(5):1001–1007. doi: 10.1128/aem.52.5.1001-1007.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders M. E. Phage resistance in lactic acid bacteria. Biochimie. 1988 Mar;70(3):411–422. doi: 10.1016/0300-9084(88)90215-5. [DOI] [PubMed] [Google Scholar]
- Sing W. D., Klaenhammer T. R. Conjugal Transfer of Bacteriophage Resistance Determinants on pTR2030 into Streptococcus cremoris Strains. Appl Environ Microbiol. 1986 Jun;51(6):1264–1271. doi: 10.1128/aem.51.6.1264-1271.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steele J. L., McKay L. L. Conjugal transfer of genetic material by Lactococcus lactis subsp. lactis 11007. Plasmid. 1989 Jul;22(1):32–43. doi: 10.1016/0147-619x(89)90033-4. [DOI] [PubMed] [Google Scholar]
- Walsh P. M., McKay L. L. Recombinant plasmid associated cell aggregation and high-frequency conjugation of Streptococcus lactis ML3. J Bacteriol. 1981 Jun;146(3):937–944. doi: 10.1128/jb.146.3.937-944.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wright A., Taimisto A. M., Sivelä S. Effect of Ca2+ ions on plasmid transformation of Streptococcus lactis protoplasts. Appl Environ Microbiol. 1985 Oct;50(4):1100–1102. doi: 10.1128/aem.50.4.1100-1102.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]