Abstract
The Drosophila GATA factor Serpent interacts with the RUNX factor Lozenge to activate the crystal cell program, whereas SerpentNC binds the Friend of GATA protein U-shaped to limit crystal cell production. Here, we identified a lozenge minimal hematopoietic cis-regulatory module and showed that lozenge-lacZ reporter-gene expression was autoregulated by Serpent and Lozenge. We also showed that upregulation of u-shaped was delayed until after lozenge activation, consistent with our previous results that showed u-shaped expression in the crystal cell lineage is dependent on both Serpent and Lozenge. Together, these observations describe a feed forward regulatory motif, which controls the temporal expression of u-shaped. Finally, we showed that lozenge reporter-gene activity increased in a u-shaped mutant background and that forced expression of SerpentNC with U-shaped blocked lozenge- and u-shaped-lacZ reporter-gene activity. This is the first demonstration of GATA:FOG regulation of Runx and Fog gene expression. Moreover, these results identify components of a Serpent cross-regulatory sub-circuit that can modulate lozenge expression. Based on the sub-circuit design and the combinatorial control of crystal cell production, we present a model for the specification of a dynamic bi-potential regulatory state that contributes to the selection between a Lozenge-positive and Lozenge-negative state.
INTRODUCTION
Hematopoiesis is a dynamic process that produces the various blood cell lineages from a single hematopoietic stem cell and is regulated by key lineage-specific factors (Orkin, 2000; Zhu and Emerson, 2002; Warren and Rothenberg, 2003). When viewed in the context of all known genetic interactions, the complexity of the processes that control hematopoiesis can be appreciated, but not readily understood (Swiers et al., 2006). An understanding of the interactions in terms of gene activation or repression, coupled with information about cis-regulatory inputs, has revealed mechanistic details about the modular sub-circuits that together describe these processes and development in general (Swiers et al., 2006; Oliveri and Davidson, 2007). Moreover, simple genetic model organisms provide an opportunity to analyze these interactions in vivo, thereby providing a direct link between the genomic programs that encode them and the biological functions they control. The Drosophila model system has been used to identify conserved key factors and investigate their function during hematopoiesis (Dearolf, 1998; Fossett and Schulz, 2001; Evans et al., 2003; Meister and Lagueux, 2003; Sorrentino et al., 2005; Hartenstein, 2006; Crozatier and Meister, 2007). In order to more fully understand Drosophila hematopoiesis, we characterized the role of Serpent (Srp) cross-regulation of Lozenge (Lz) and U-shaped (Ush) in the crystal cell lineage.
The blood system of the fly lacks the complexity seen in vertebrates. Nevertheless, cross-species comparisons have shown that fundamental aspects of hematopoiesis are conserved across taxa (Fossett and Schulz, 2001; Evans et al., 2003; Fossett et al., 2003; Meister and Lagueux, 2003; Sorrentino et al., 2005; Hartenstein, 2006; Crozatier and Meister, 2007). Drosophila has two primary blood cell types, plasmatocytes and crystal cells, which have similar functions to the vertebrate myeloid lineages (Rizki, 1978; Dearolf, 1998; Evans et al., 2003). Crystal cells, named for their crystalline inclusion bodies, are required for wound repair and xenobiotic encapsulation (Rizki, 1978). Plasmatocytes are operational macrophages and synthesize antimicrobials (Rizki, 1978; Tepass et al., 1994; Dearolf, 1998). Like their vertebrate counterparts, these cells develop from a common hematopoietic progenitor (Rizki, 1978; Tepass et al., 1994; Dearolf, 1998; Lebestky et al., 2000; Lanot et al., 2001). Both vertebrate and Drosophila hematopoiesis consists of two spatially and temporally distinct periods or waves. In Drosophila, the first hematopoietic wave begins in the early embryonic head mesoderm. The second wave begins in embryogenesis and continues throughout larval development within a specialized hematopoietic organ, called the lymph gland (Dearolf et al., 1998; Lebestky et al., 2000; Lanot et al., 2001; Fossett and Schulz, 2001; Evans et al., 2003; Hartenstein, 2006; Crozatier and Meister, 2007).
Srp, similar to vertebrate GATA-2, is positioned at the apex of hematopoiesis and, as such, is required for the production of hemocyte precursors (Rehorn et al., 1996; Sam et al., 1996). In this role, Srp acts upstream of Glial cells missing (Gcm) and the RUNX factor Lz, which are required later for plasmatocyte and crystal cell production, respectively (Bernardoni et al., 1997; Lebestky et al., 2000; Kammerer and Giangrande, 2001; Alfonso and Jones, 2002). Of the conserved hematopoietic regulators, the GATA, Friend of GATA (FOG), and RUNX protein families are of particular interest because pair-wise interactions between GATA and FOG or GATA and RUNX regulate both vertebrate and Drosophila hematopoiesis (Tsang et al., 1997; 1998; Querfurth et al, 2000; Elagib et al., 2003; Fossett et al., 2003; Waltzer et al., 2003; Cantor and Orkin, 2005, Ferjoux et al., 2007). Moreover, Srp acts as a contextual switch between RUNX activation and FOG repression of the crystal cell lineage (Fossett et al., 2003). GATA transcriptional regulators generally have two zinc-finger domains. The C-terminal zinc-finger binds the DNA recognition sequence, WGATAR (Cantor and Orkin, 2005). The N-terminal zinc-finger interacts with FOG proteins; and the GATA:FOG complex modifies transcription by either activating or antagonizing activity, depending upon the gene regulatory context (Crispino et al., 1999; Lu et al., 1999; Svensson et al., 1999; Tevosian et al., 1999; Chang et al., 2002; Letting et al., 2004; Hong et al., 2005; Cantor and Orkin, 2005; Lowry and Mackay, 2006). The srp gene is alternatively spliced to produce either a single C-terminal zinc-finger isoform (SrpC) or the canonical dual zinc-finger protein (SrpNC). The FOG protein Ush interacts with SrpNC, but not SrpC, which lacks the N-terminal zinc-finger (Walter et al., 2002; Fossett et al., 2003). RUNX proteins bind DNA through the conserved Runt domain (Tracy and Speck, 2000; Speck and Gilliland, 2002; Rennert et al., 2003; Anglin and Passaniti, 2004). In general, RUNX activity is influenced by a variety of interacting factors, including GATA factors (Coffman, 2003; Elagib et al., 2003; Fossett et al., 2003; Waltzer et al., 2003, Ferjoux et al., 2007). Of the three mammalian Runx genes, Runx1 is required for hematopoiesis (Otto et al., 2003), and is one of the most frequent targets of chromosomal translocations associated with human leukemia (Okuda et al., 1996; Speck and Gilliland, 2002; deBruijn and Speck, 2004). Currently, there is a lack of information about the role of Runx1 in hematopoietic gene regulatory networks (Swiers et al., 2006; Otto et al., 2003).
Srp, Lz and Ush act combinatorially to regulate crystal cell production. Both SrpC and SrpNC can interact with Lz to activate the crystal cell program (Fossett et al., 2003; Waltzer et al., 2003), whereas only SrpNC interacts with Ush to repress crystal cell production (Fossett et al., 2003). This suggests that Srp acts as a contextual switch, mediating cross-talk between crystal cell activation and repression pathways. In order to increase our understanding of the mechanistic basis for this contextual switch, and how it regulates crystal cell production, we investigated the cis- and trans-regulation of lz and ush. Collectively, our data provide evidence for a Srp cross-regulatory sub-circuit that regulates lz and ush expression. Based on these results, we present a model for the specification of a dynamic bi-potential regulatory state that contributes to the selection between a Lz-positive and Lz-negative state.
MATERIALS AND METHODS
Fly strains
Fly stocks were maintained at 23°C on standard food, and w1118 was used as the wild-type stock. The following fly lines were used in this study and are described elsewhere: ush1/SM6, Roi, eve-lacZ; upstream activation sequence (UAS)-ush; UAS-srpNC; UAS-srpNCV421G; and twi-Gal4 (Fossett et al., 2001; 2003). The following strains were generous gifts from colleagues: lz-Gal4;UAS-GFP (J. Pollock, Duquesne University); UAS-lz (J. Canon and U. Banerjee, University of California, Los Angeles); UAS-srpC (D. K. Hoshizaki, University of Nevada, Las Vegas, NV); UAS-srpNC;UAS-lz (K. M. Gajewski, University of Texas, M. D. Anderson Cancer Center, Houston, TX). UAS-srpNC,UAS-ush stock was constructed using genetic recombination between UAS-transgenes located on chromosome II. The generation of strains carrying lz-lacZ transgenes is described below.
Generation of transgenic animals carrying lz-lacZ fusion constructs
Overlapping DNA fragments of the lz 1.5 kb 5′ UTR genomic DNA region were analyzed for their ability to direct lacZ reporter-gene expression in crystal cells. This was accomplished by generating PCR fragments that were either cloned directly into the P-element CaSpeR-Hsp43-AUG-βgal (Chab) germline transformation vector (Thummel et al., 1988), or first cloned into pCR-II TOPO cloning vector (Invitrogen) and subsequently into Chab vector. Site-directed mutations (SDM) were introduced into DNA fragments as described previously (Muratoglu et al., 2006). The oligonucleotide primers used to generate point mutations in DNA fragments are available upon request.
The DNA sequence of each recombinant vector was verified prior to injection. w1118 embryos were injected with the recombinant vectors by Model Systems Genomics, Duke University, or Rainbow Transgenic Flies, Inc., Newbury Park, CA. Germline transformants were established according to previously described methods and were screened for tissue-specific lacZ expression using immunohistochemical staining analysis as previously described (Gajewski et al., 1997). At least six independent lines were generated and tested for each construct.
Determination of the lz-Gal4 chromosomal insertion site
Plasmid rescue was used to identify the lz genomic sequences that flank the pGawB insertion (Pirrotta, 1986). Briefly, genomic DNA was isolated from lz-Gal4 fly lines. The DNA was cut with MspI or DraI, each having a single restriction site within the pGawB plasmid insert. Ligation of this restriction digest produced a circular template for PCR. Specific primers for the pGawB plasmid, but facing the flanking sequences, were used to produce a pGawB/flanking-sequence chimeric fragment. The fragment was cloned into the pCR-II TOPO cloning vector (Invitrogen) and sent to The Biopolymer Core Facility at the University of Maryland, Baltimore for sequence analysis.
Immunohistochemical and fluorescent antibody staining of embryos and larvae
Collection, fixation, and immunohistochemical staining of embryos were performed as previously described (Schulz and Fossett, 2005). The following primary antibodies were used: mouse anti-β-galactosidase, 1:1000 (Promega); rabbit anti-U-shaped, 1:750 (Fossett et al., 2001); rabbit anti-Serpent, 1:1000 (Hu, 1995). Either biotinylated (Vector Laboratories) or fluorescent (Molecular Probes, Invitrogen) conjugated secondary antibodies were used for primary antibody recognition, according to the manufacturer’s recommendation. All embryos were visualized with Zeiss Axioplan optics.
Lymph glands from third instar wandering larvae were removed by making an incision below the head region and removing the lymph gland, which was attached to the dorsal vessel and brain. Fixation and immunostaining were preformed as previously described (Schulz and Fossett, 2005). Stained preparations were mounted in PBS under cover slips and visualized using Zeiss Axioplan optics.
Gene expression analysis in mutant and Gal4/UAS embryos
Embryos were cultured and collected at 23°C. Gain of function studies were conducted using the Gal4/UAS binary system (Brand and Perrimon, 1993). lz-lacZ activity in gain of function backgrounds required a two generation cross. Each lz-lacZ line was crossed to twi-Gal4 virgin females to produce twi-Gal4/+; lz-lacZ/+ transheterozygous progeny. The F1 virgin females were collected and then crossed to one of the following: UAS-srpNC or, UAS-lz or, UAS-srpC or, UAS-srpNC;UAS-lz or, UAS-srpNC,UAS-ush homozygous males or UAS-srpC/+; UAS-lz/+ heterozygous males. To assess lz-lacZ activity in ush1 homozygous embryos also required a two generation cross. Males carrying lz-lacZ constructs on chromosome III were crossed to ush1/SM6, Roi, eve-lacZ virgin females. F1 ush1/+; lz-lacZ/+ transheterozygous progeny were then intercrossed. Because Ush is required for germ band retraction, homozygous mutants are easily identified by the altered morphology that results from failure of the germ band to retract. In all cases, the F2 progeny were collected during embryogenesis and assayed for β-galactosidase activity using immunohistochemistry as previously described (Gajewski et al., 1997).
To assess Ush expression in Srp and Lz gain of function backgrounds, homozygous twi-Gal4 virgins were crossed to homozygous UAS-srpNC;UAS-lz males or, heterozygous UAS-srpNCV421G/+;UAS-lz/+ males. ush-lacZ activity in gain of function backgrounds required a two generation cross. Each ush-lacZ line was crossed to twi-Gal4 virgin females to produce twi-Gal4/+; ush-lacZ/+ transheterozygous progeny. The F1 virgin females were collected and then crossed to one of the following: UAS-srpNC;UAS-lz or, UAS-srpNC,UAS-ush homozygous males or, UAS-srpNCV421G/+;UAS-lz/+ or, UAS-srpNC,UAS-ush/+; UAS-lz/+ heterozygous males. The progeny from these crosses were collected and immunohistochemically stained with either the Ush or β-galactosidase antibody as previously described (Gajewski et al., 1997).
RESULTS
Identification of a lz minimal crystal cell cis-regulatory module
Both Srp isoforms can interact with Lz to activate the crystal cell program (Fossett et al., 2003; Waltzer et al., 2003). Conversely, SrpNC acting with Ush converts SrpNC from an activator to a repressor of crystal cell production (Fossett et al., 2003). These transcription factors may control crystal cell production by directly regulating lz gene expression. In this case, we expect that both activation by Srp and repression by SrpNC:Ush would be mediated through GATA sites within the lz crystal cell cis-regulatory module (CRM). To test this hypothesis, we identified a 1.2 kb lz crystal cell CRM and then determined which cis-elements are required for lz expression.
We employed a three-tiered strategy to narrow our search for the lz crystal cell CRM. First, we used the lz-Gal4 fly line to determine the approximate location of the CRM. In this fly line, the promoter-less Gal4 cDNA (pGawB) is inserted into the lz locus (Crew et al., 1997), which directs reporter-gene expression in a pattern that recapitulates endogenous lz expression (Lebestky et al., 2000). We determined that the pGawB insertion site was 160 bp upstream of the transcription start site (Fig. 1A). Based on this information, we then analyzed the region upstream of the transcription start site for highly conserved sequences between the melanogaster sibling species, sechellia, yakuba and erecta. Such comparisons can help identify evolutionarily conserved cis-elements. We identified three such regions at positions −1169 to −827, −643 to −320, and −206 to the coding region (Fig. 1A). Finally, because of our interest in Srp regulation of lz gene expression, we searched this 1.2 kb region for GATA motifs. This search identified 13 GATA motifs located between positions −1207 and −768. Thus, we isolated and tested a DNA fragment between positions −1263 and −18 for its ability to drive crystal cell-specific reporter-gene (lacZ) expression in vivo (Fig. 1A). lz-lacZ expression was first detected in embryonic crystal cell precursors during stage 9 and continued throughout development in this lineage (data not shown). Together, these results indicate that a crystal cell CRM is located within the 1.2 kb region upstream of the transcription start site. However, by stage 10, we also observed ectopic lz-lacZ expression that continued throughout development in the ectoderm (data not shown). Ectopic expression is defined as fragment-directed reporter-gene activity that differed significantly from the pattern produced by lz-Gal4 driving UAS-lacZ (Lebestky et al., 2000).
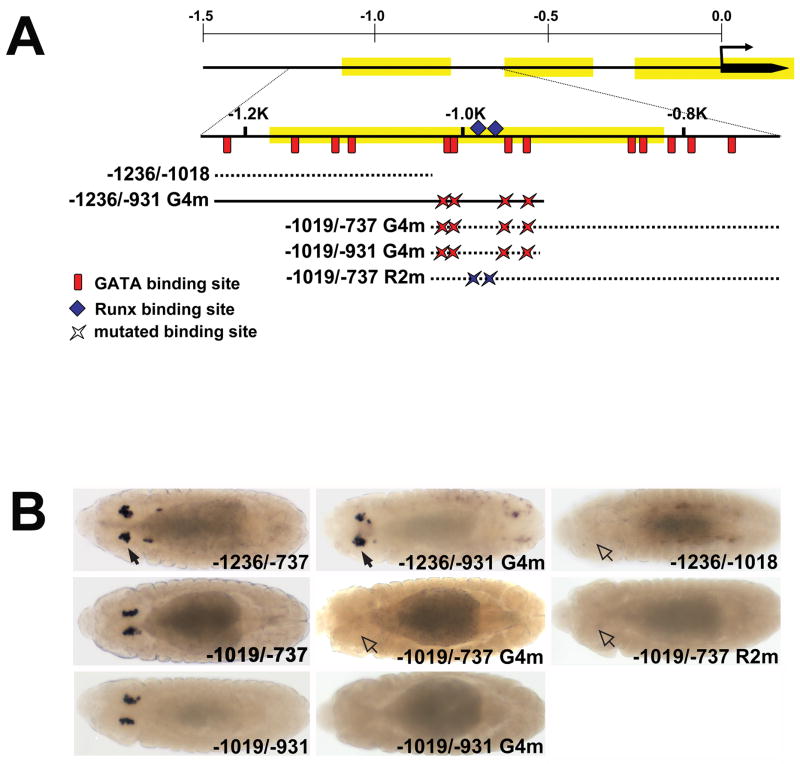
Fig. 1. Identification of the lz crystal cell cis-regulatory module.

(A) Schematic of the lz locus and screen for the crystal cell CRM. A horizontal arrow marks the transcriptional start site, designated position 0. Solid lines indicate introns, whereas boxes indicate exons. The upstream c11.1 gene is depicted in grey. Beneath the 21 kb map is an expanded map of the 1.5 kb region upstream of the lz transcription unit. Evolutionarily conserved regions are highlighted in yellow. The relative position of the P-element pGawB insert is indicated. The cryptic promoter used by this transgene likely lies just 5′ to the transcription start site. The DNA fragments used to screen for crystal cell CRMs are indicated by black lines and are positioned and numbered relative to the transcription start site. Solid lines represent fragments with crystal cell activity, whereas dashed lines represent fragments lacking crystal cell activity. The red diamonds designate representative data presented in part B. (B) Fragment-driven reporter-gene (lacZ) expression during embryogenesis and in third larval instar lymph gland. The stage of embryogenesis is indicated at the left of each row. The lz-lacZ strain designations are indicated at the top of each column. Orientation of the embryo is indicated to the right of each row. Solid arrows mark activity and open arrows mark the lack of activity in stage 8 hemocyte precursors and stage 9 crystal cell precursors. The boundaries between the cortical and medullary zones of the larval lymph glands are outlined. Abbreviations: lz, lozenge; llg, third larval instar lymph gland; cz, cortical zone; mz, medullary zone; L, lateral view; D, dorsal view.
To identify the lz minimal crystal cell CRM, we subdivided the 1.2 kb region into 5 overlapping fragments. Fragment designations are based on their positions relative to the transcription start site (Fig. 1A). Embryos harboring the −1236/−737 fragment exhibited lz-lacZ expression in the crystal cell lineage starting from stage 9 and continuing to stage 16 (Fig. 1B). We also observed lz-lacZ expression in plasmatocytes for two of the six lines tested, consistent with results seen with lz-Gal4;UAS-lacZ embryos (Lebestky et al., 2000). In addition, one line showed ectopic lz-lacZ expression in the amnioserosa and garland cells (Fig. 1B, data not shown). A similar expression pattern was detected in embryos harboring the −1019/−737 fragment (Fig. 2B). As seen with the parental fragments, the −1019/−931 fragment directed crystal cell-specific lz-lacZ expression throughout embryonic development. However, embryonic expression was first detected slightly later than with the parental fragments, beginning in stage 10 rather than stage 9 (Fig. 1B). In addition, one of the eight lines tested showed ectopic expression in the amnioserosa (Fig. 3B, data not shown). We tested two additional fragments from the 1.2 kb region. These two fragments, −1236/−1018 and −737/0, were not able to direct crystal cell-specific lz-lacZ expression (Fig. 1A). However, some of the lines generated from each fragment exhibited ectopic and plasmatocyte activity (data not shown).
Fig. 2. Clustered GATA and RUNX motifs are required for lz crystal cell CRM activity.
(A) Schematic showing fragments with mutated GATA and RUNX motifs used to evaluate lz-lacZ CRM activity. The 1.5 kb region upstream of the lz transcription start site is depicted with the evolutionarily conserved regions highlighted in yellow. Beneath the map of the 1.5 kb region is an expanded map of the region from −1250 to −727. Red vertical lines show the relative positions of the 13 GATA motifs between positions −1207 and −768. The blue diamonds mark the positions of the two RUNX motifs located at −995 and −977. Mutations in the four GATA motifs, between positions −1010 and −947, are designated G4m, whereas the mutations in the two RUNX motifs are designated R2m. The mutated fragments are numbered and positioned relative to the transcription start site, with the suffix G4m or R2m to indicate disruption of the GATA or RUNX motifs, respectively. Solid line represents a fragment with crystal cell activity, whereas dashed lines represent fragments lacking crystal cell activity. (B) Comparison of wild-type and GATA or RUNX mutant CRMs. Dorsal views of stage 13 embryos. lz-lacZ strains are indicated in the lower right hand corner of each panel. Closed arrows mark activity in crystal cells; open arrows mark lack of activity. Abbreviations: G4m, GATA Core mutants; R2m, RUNX mutants.
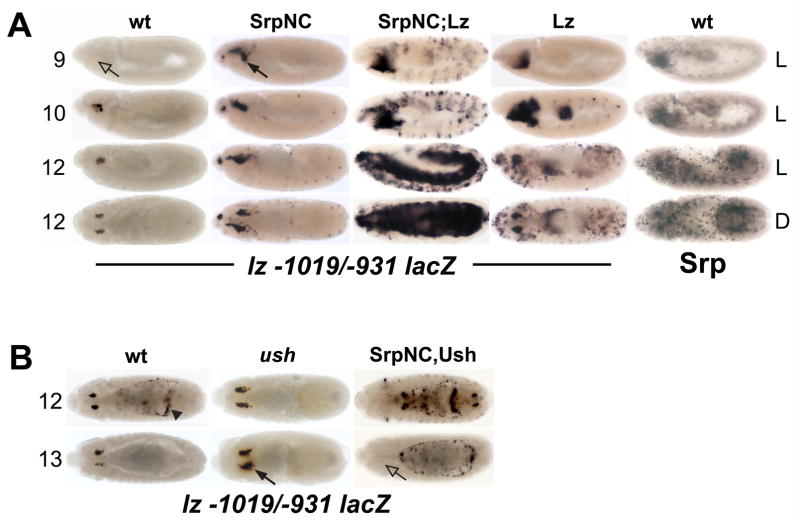
Fig. 3. SrpNC is a positive and negative regulator of lz crystal cell CRM activity.
(A) SrpNC and Lz synergistically upregulate lz −1019/−931 lacZ activity. From left to right: embryos in the first four columns are stained with α-β-galactosidase antibody; embryos in the last column are stained with α-Srp antibody. (B) SrpNC and Ush block lz −1019/−931 lacZ activity. lz −1019/−931 lacZ activity was assessed in different genetic backgrounds and developmental stages. The stage of embryogenesis is indicated at the left of each row. The genetic background is indicated at the top of each column. Orientation of the embryo is indicated at the right of each row (panel A only). SrpNC indicates twi-Gal4 driving UAS-srpNC. Lz indicates twi-Gal4 driving UAS-lz. SrpNC;Lz represents twi-Gal4 driving UAS-srpNC and UAS-lz located on chromosomes II and III, respectively. SrpNC,Ush represents twi-Gal4 driving UAS-srpNC and UAS-ush located on chromosome II. Closed arrows mark increased activity in crystal cells; open arrow marks lack of activity; arrowhead marks ectopic expression in the amnioserosa, which is used to identify embryos carrying the reporter-gene. Abbreviations: wt, wild-type; Srp, Serpent; Lz, Lozenge; Ush, U-shaped; L, lateral view; D, dorsal view.
While we were analyzing the 1.2 kb 5′ non-coding region of lz, Bataille and co-workers published the crystal cell expression pattern of a larger fragment located between −1509 and −59 (Bataille et al., 2005). They reported fragment-directed lz-lacZ expression as early as stage 7 in hemocyte precursors. We generated and analyzed 9 independent fly lines harboring this fragment. All of the lines directed expression in the crystal cell lineage throughout development. Seven lines had spatiotemporal expression similar to the −1236/−737 fragment, with expression being first detected during stage 9, whereas lz-lacZ expression was first detected during stage 8 in two of the lines (Fig. 1B). However, these two lines also exhibited ectopic expression, beginning during stage 10 and continuing throughout embryonic development (data not shown). The observed differences for the 1.5 kb-driven lz-lacZ expression pattern may be due to methodological differences between laboratories. The difficulty in detecting a signal from a low level transcript could explain the variance seen with staging in this study. In any case, the results with the −1509/−59 and −1236/−737 fragments indicate that lz expression is upregulated by stage 9. The possibility of earlier activity during stage 7 or stage 8 prompted us to determine if an additional crystal cell-specific CRM was located upstream of the 1.2 kb fragment. We tested the region between positions −1509 and −1210, but did not detect fragment-driven lz-lacZ activity in the crystal cell lineage (Fig. 1A, data not shown).
Our analysis of the lz 5′non-coding region indicated that genomic fragments lacking the −1019/−931 region were unable to drive lz-lacZ expression in the crystal cell linage. Thus, the −1019/−931 fragment most likely contains a minimal CRM that is required for lz expression during crystal cell development. However, lz −1019/−931 lacZ activity was detected during stage 10, slightly later in development than that of the larger −1509/−59 or −1236/−737 fragments (Fig. 1B). This suggests that regions outside of the −1019/−931 CRM are also required for early activation of lz expression in crystal cell precursors.
We also tested the activity of the −1509/−59, −1236/−737, and −1019/−931 fragments in the third larval instar lymph gland (Fig. 1B). The third larval instar lymph gland is a set of bilateral primary lobes and a series of smaller secondary lobes that flank the dorsal vessel (insect heart). Morphological, functional, and gene expression analyses indicate that the primary lobes are divided into three distinct regions, namely, the cortical zone, the medullary zone and the posterior signaling center. Lz is expressed in the cortical zone, but not in the medullary zone (Lebestky et al., 2000; Jung et al., 2005). Consistent with these reports, all three fragments directed lz-lacZ expression in the cortical zone, but not in the medullary zone (Fig. 1B). Together, these data show that the lz −1019/−931 lacZ expression pattern recapitulates the endogenous lz expression pattern, indicating that it is required for lz expression during both embryonic and larval lymph gland hematopoiesis.
Clustered GATA and RUNX sites are required for lz minimal crystal cell CRM activity
Our analyses of the lz cis-regulatory region showed that a minimal CRM located between positions −1019 and −931 can direct lz expression in crystal cells. In order to identify trans-acting regulators of lz expression, we surveyed the −1019 to −931 sequence for cis-regulatory elements that may be required for crystal cell activity. This sequence contains four GATA motifs with the following distribution: two overlapping motifs at positions −1010 and −1008, one at −957, and one at −947. We also identified two RUNX motifs at positions −995 and −977 (Fig. 2A). To determine if these elements are required for lz crystal cell expression, we introduced point mutations into either the GATA or RUNX motifs. We mutated all four GATA sites between positions −1010 and −947 and assigned this mutation the name G4m (Fig. 2A). The −1019/−931 G4m fragment failed to direct lz-lacZ expression in crystal cells (Fig. 2B). This indicates that Srp binds these sites to upregulate lz in the crystal cell lineage. Based on the functional requirement of these four sites, we designated this cluster the GATA Core.
Of the 13 GATA motifs within the −1236/−737 region, nine are located outside of the GATA Core. Four of the nine are located upstream of the Core, between positions −1207 and −1081, and are designated the Upstream Cluster. The remaining five are located downstream of the Core, between positions −848 and −768, and are designated the Downstream Cluster (Fig. 2A). To determine the functionality of these sites, we constructed two different fragments. Both fragments carried mutant versions of the GATA Core. Using the convention described above, the fragments were designated by their positions relative to the transcription start site, with the suffix G4m used to indicate disruption of the GATA Core. Thus, the −1019/−737 G4m fragment had a mutated GATA Core, but a wild-type version of the Downstream Cluster (Fig. 2A). This fragment was unable to direct lz-lacZ expression in crystal cells (Fig. 2B). Thus, the Downstream Cluster does not activate lz expression in crystal cells in the absence of the GATA Core. In contrast, we detected crystal cell activity in the −1236/−931 G4m fragment (Fig. 2B). This indicates that the Upstream Cluster can compensate for the loss of the GATA Core and may function in the activation of lz expression. The functional difference between the Upstream and Downstream Clusters may reflect the fact that three of the four Upstream GATA motifs are conserved consensus (WGATAR) motifs across sibling species. In contrast, the Downstream Cluster contains only one consensus motif and two conserved motifs (Fig. 2A, data not shown). Together, these observations are consistent with studies that show GATA-1 activates β-globin gene expression using a subset of GATA motifs within the locus, and that GATA occupancy occurs more often among conserved consensus motifs (Im et al., 2005; Bresnick et al., 2006). Finally, these data strongly indicate that Srp directly upregulates lz activity in crystal cells by binding to specific GATA motifs.
Although the −1236/−931 G4m fragment directed lz-lacZ expression in crystal cells, the −1236/−1018 sub-fragment failed to do so (Figs. 1A, 2). The difference in activity between these two fragments is due to the cis-elements located between positions −1019 and −931. As stated above, this region contains the GATA Core and two RUNX motifs. Because the Upstream GATA Cluster was able to compensate for the four GATA motifs within the Core, the RUNX motifs are most likely essential elements that are required to direct crystal cell activity. Furthermore, the functional Upstream Cluster is closer to the RUNX motifs than the nonfunctional Downstream Cluster, suggesting that cooperation between Lz and Srp may be necessary for lz expression. Based on these observations, we disrupted the two RUNX motifs within the −1019/−737 fragment and assigned this mutation the name R2m (Fig. 2A). This mutation completely abolished the crystal cell activity of the −1019/−737 fragment in nine out of ten lines tested, suggesting that Lz functions as an auto-activator in a positive feedback loop (Fig. 2B). Together, the mutational analysis strongly suggests that both GATA and RUNX motifs within the minimal −1019/−931 CRM are required for lz crystal cell CRM activity. Moreover, these data are consistent with studies showing that crystal cell-specific CRMs in a number of genes, including lz, contain at least one essential RUNX motif (Muratoglu et al., 2006; Gajewski et al, 2007; Ferjoux et al., 2007). However, the functional Upstream GATA Cluster may act with either the two RUNX motifs and/or the GATA Core to direct crystal cell-specific activity, and this question is the subject of continuing studies.
Dual role of SrpNC as a positive and negative regulator of lz crystal cell expression
SrpNC may function as a contextual switch by acting with Lz to activate the crystal cell program or with Ush to block activation (Fossett et al., 2003). Lz is required for crystal cell lineage commitment (Lebestky et al., 2000), and GATA and RUNX motifs within the lz minimal −1019/−931 CRM are most likely required for its expression (Fig. 2B). Together, these observations strongly suggest that SrpNC cross-regulates lz expression through the GATA and RUNX motifs within the minimal −1019/−931 CRM, thereby controlling crystal cell lineage commitment. To address this question, we assayed the lz minimal −1019/−931 CRM for activity in embryos with altered levels of SrpNC, Lz, or Ush, either individually or in combination.
Forced mesodermal expression of SrpNC increased lz −1019/−931 lacZ expression. Expression was detected in the stage 9 head mesoderm, earlier than in a wild-type background (Fig. 3A). The expanded expression pattern is reminiscent of the supernumerary crystal cell pattern produced by forced expression of both Srp isoforms (Fossett et al., 2003; Waltzer et al., 2003). Similar results were observed with SrpC (data not shown). Forced expression of Lz also increased the lz-lacZ expression domain in a pattern similar to the expression pattern of endogenous Srp (Fig. 3A). This indicates that Srp and Lz interact to activate lz −1019/−931 lacZ through the GATA and RUNX motifs. This was confirmed by showing that co-expression of SrpNC and Lz synergistically upregulated lz −1019/−931 lacZ. We observed strong lz −1019/−931 lacZ expression throughout the head mesoderm and weak expression in trunk mesoderm beginning in stage 9. By stage 12, strong expression was observed throughout the mesoderm, in stark contrast to results obtained with forced expression of either factor alone (Fig. 3A). Similar results were obtained with SrpC (data not shown). These results indicate that both Srp isoforms can interact with Lz to upregulate lz expression in crystal cells through the GATA and RUNX motifs located within the minimal CRM.
The functional role of the GATA motifs within the lz crystal cell CRM suggested that the SrpNC:Ush complex binds these motifs to block lz expression. To test this hypothesis, we examined lz −1019/−931 CRM activity in ush loss of function and SrpNC:Ush gain of function genetic backgrounds. Compared to the wild-type control, lz −1019/−931 CRM activity increased in a ush mutant background (Fig. 3B). Similar results were observed with the larger −1236/−737 fragment that contained all 13 GATA motifs (data not shown), indicating that Ush is required to limit the lz expression domain. Consistent with these results, forced expression of SrpNC and Ush blocked lz −1019/−931 CRM activity (Fig. 3B). We observed similar results with the larger −1509/−59 and −1236/−737 fragments (data not shown). Thus, the SrpNC:Ush complex blocked lz expression through the GATA sites within the minimal CRM. Together, these data indicate that SrpNC acts as cross-regulatory switch by interacting with Lz to maintain lz expression or with Ush to block lz expression. Collectively, our new data, coupled with our previously published results, indicate that the SrpNC:Ush complex blocks crystal cell production by blocking lz expression (Fig. 3B; Fossett et al., 2001; 2003). Finally, forced expression of SrpNC and Ush did not block gcm-lacZ-expressing hemocyte precursors or plasmatocytes (data not shown). These results indicate that the SrpNC:Ush complex does not limit the production of these hemocyte classes. To increase our understanding of the cross-regulatory process and how it modulates crystal cell production, we expanded our investigation of ush expression during crystal cell lineage commitment.
Ush expression is upregulated after crystal cell lineage specification
Ush is expressed in a variety of tissues throughout embryogenesis, including the following hematopoietic tissues: hemocyte precursors, plasmatocytes, and crystal cells (Fossett, et al., 2000; 2001). Previously, we identified a ush −7462/−25 CRM, which recapitulates endogenous Ush embryonic expression. Our analyses of sub-fragments from the −7462 to −25 region showed that two hematopoietic CRMs are located within 1.2 kb of the transcription start site. The distal −1243/−956 CRM drives expression in hemocyte precursors and plasmatocytes but not crystal cells, whereas the proximal −174/−25 CRM is required for expression in all three cell types. Genetic and mutational analyses of these hematopoietic CRMs showed that Srp activates expression in hemocyte precursors and plasmatocytes through the GATA motifs within both CRMs, whereas both Srp and Lz activate expression in crystal cells through clustered GATA and RUNX motifs unique to the −174/−25 CRM (Muratoglu et al., 2006).
During embryonic development, Srp is first detected in stage 5 hemocyte precursors (Rehorn et al., 1996; Sam et al., 1996). Lz is upregulated as early as stage 7 (Bataille et al., 2005) and as late as stage 9 (Fig. 1B). And, Ush is detected in hemocyte precursors during stage 8 and in crystal cell precursors again later during stage 10. During stage 13 and continuing through the end of embryogenesis, Ush is increasingly downregulated in the crystal cell lineage (Fossett et al., 2001). However, it is not known whether ush expression is maintained as hemocyte precursors develop into crystal cell precursors, or if expression is upregulated after lineage specification. To more accurately assess the timing of Ush expression, we determined the level of co-expression of Ush with lz −1236/−737 lacZ in early crystal cell precursors. We observed lz −1236/−737 lacZ expression in crystal cell precursors as early as stage 9. In contrast, Ush expression was not detected in these cells. However, beginning in stage 10 and continuing through stage 11, we observed extensive co-expression of lz −1236/−737 lacZ and Ush in crystal cell precursors (Fig. 4, data not shown), consistent with our previous observations (Fossett et al., 2001). Similar results were observed with the larger −1509/−59 fragment (data not shown). Thus, Ush expression is not maintained as hemocyte precursors develop into crystal cell precursors, but is upregulated after lineage specification.
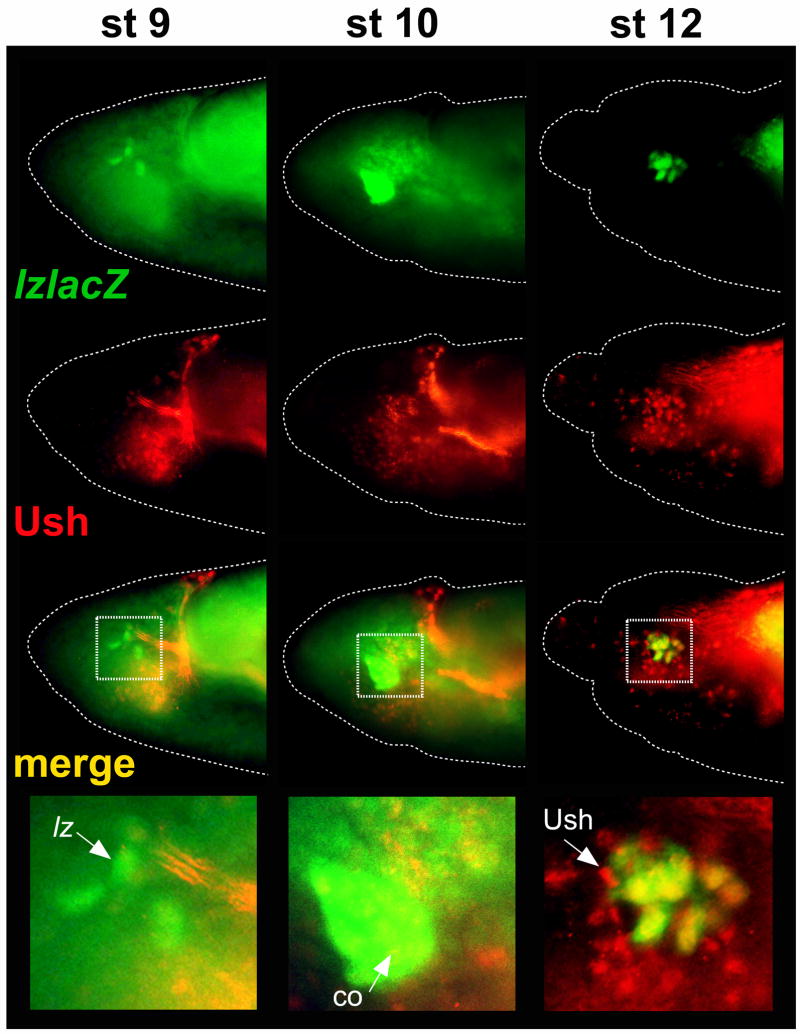
Fig. 4. lz-lacZ and Ush expression during crystal cell development.
The lz −1236/−737 CRM was used to assess lz expression and is depicted in green. Ush protein expression is depicted in red. Co-localized expression is depicted in yellow. The stage of embryogenesis is indicated at the top of each column. The enlarged area is marked by a white dotted box. Arrows mark the following: stage 9 cells expressing only lz-lacZ; stage 10 cells with co-localized expression of lz-lacZ and Ush; and stage 12 cells expressing only Ush. Abbreviations: lz-lacZ, lz −1236/−747 lacZ; Ush, U-shaped; co, co-localization.
During stage 12, we continued to observe co-expression of Ush and lz −1236/−737 lacZ. However, adjacent to lz −1236/−737 lacZ-positive cells and within the crystal cell cluster, we also observed cells that expressed only Ush (Fig. 4). The morphology and position of these cells suggest that they are derived from lz-lacZ-positive precursors. Moreover, these cells were primarily Ush-positive, lz-lacZ-positive during stages 10 and 11 (Fig. 4; Fossett et al., 2001). Together, these observations are consistent with downregulation of lz −1236/−737 lacZ by the endogenous SrpNC:Ush complex to specify a Ush-positive, lz-lacZ-negative state. This interpretation is also in agreement with our results that showed forced expression of SrpNC and Ush blocked lz CRM activity and that lz-lacZ activity increased in a ush mutant background (Fig. 3B). Finally, during stage 13 and continuing until the end of embryogenesis, we observed an increasing number of lz-lacZ-positive cells with greatly diminished Ush expression (data not shown), consistent with previous studies (Fossett et al., 2001).
Ush acting with SrpNC downregulates ush hematopoietic expression
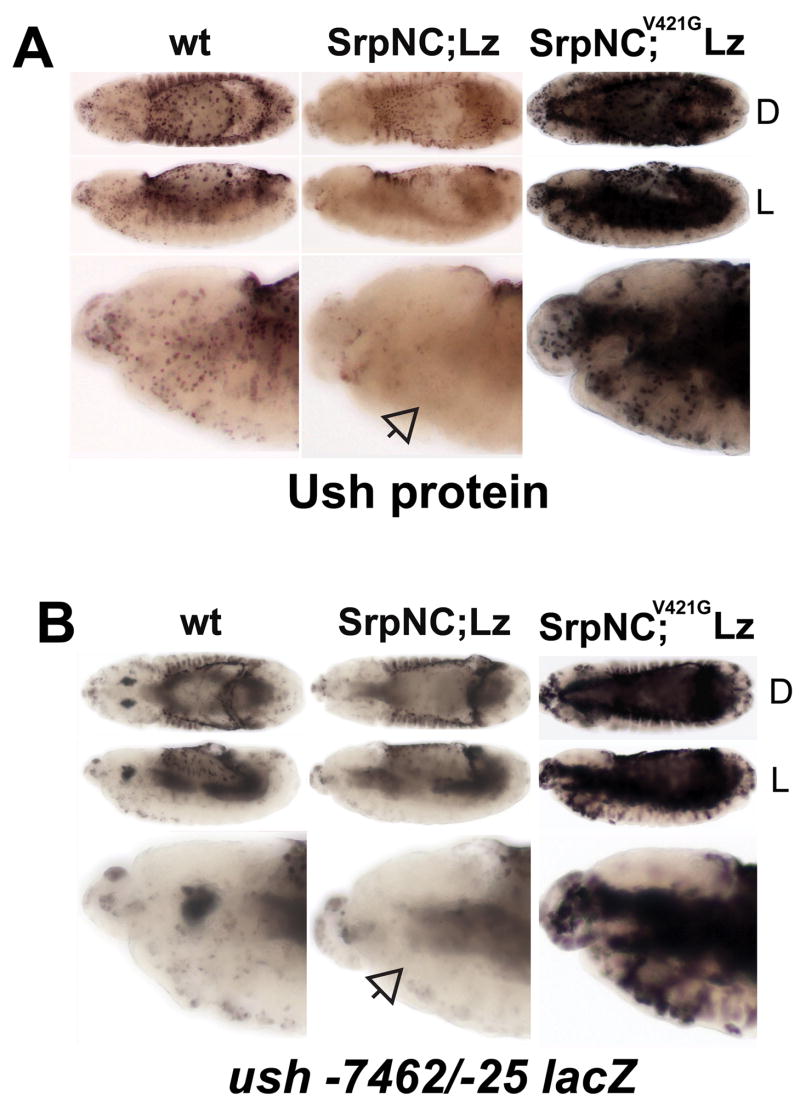
We previously showed that Ush is downregulated in embryos with forced expression of SrpNC and Lz. In contrast, forced expression of SrpC and Lz did not inhibit expression. This suggests that the N-terminal zinc-finger of Srp is essential for repression (Fossett et al., 2003). The GATA N-terminal zinc-finger stabilizes DNA binding and serves as a protein interaction domain (Pedone et al., 1997; Crispino et al., 1999; Newton et al., 2001). Ush interacts with SrpNC through a conserved valine residue at position 421 (Crispino et al., 1999; Fossett et al., 2003). To investigate the mechanism of ush gene repression, we co-expressed Lz with the N-terminal zinc-finger mutant SrpNCV421G and assayed for endogenous Ush expression. Similar to results obtained with SrpC and Lz, SrpNCV421G and Lz did not repress Ush expression (Fig. 5A). We repeated these studies using the ush −7462/−25 lacZ CRM and, as with endogenous Ush, observed downregulation of ush-lacZ in embryos with forced expression of SrpNC and Lz, but not in those with forced expression of SrpNCV421G and Lz (Fig. 5B). Together, these data suggest that Ush acting with SrpNC is an auto-repressor of its hematopoietic expression. Furthermore, forced expression of SrpNCV421G and Lz upregulated, but did not repress, endogenous Ush or ush −7462/−25 lacZ expression (Fig. 5). This is consistent with our previous work that showed at least one Srp isoform acts with Lz to upregulate ush expression in crystal cells (Muratoglu et al., 2006). Together, these data suggest that SrpNC and Lz initially activate ush expression. Subsequently, Ush acts with SrpNC to downregulate its hematopoietic expression. Consistent with a cross-regulatory process, forced expression of SrpNC and Lz produced individuals with either downregulated or upregulated ush −7462/−25 lacZ activity when compared to the wild-type control (Fig. 5B, data not shown).
Fig. 5. N-terminal zinc-finger conserved valine residue is required for SrpNC and Lz repression of ush expression.
(A) Ush protein expression and (B) ush −7462/−25 lacZ activity was assessed in stage 13 embryos with different genetic backgrounds. The genetic background is indicated at the top of each column. Orientation of the embryo is indicated at the right of each row. SrpNC;Lz represents twi-Gal4 driving UAS-srpNC and UAS-lz located on chromosomes II and III, respectively. SrpNC;V421G Lz represents twi-Gal4 driving UAS-srpNCV4216 and UAS-lz located on chromosomes II and III, respectively. Open arrows mark lack of expression in hemocytes. Abbreviations: wt, wild-type; Srp, Serpent; Lz, Lozenge; Ush, U-shaped; L, lateral view; D, dorsal view.
Ush competes with Lz to control SrpNC regulation of ush gene expression
Ush auto-repression may occur by one of two possible mechanisms. First, Ush could interact with SrpNC and Lz to form a complex that blocks ush expression. Alternatively, Ush may compete with Lz by converting SrpNC from an activator to a repressor of ush gene expression in a similar manner to control of SrpNC regulation of lz gene expression. To determine if Lz is required for inhibition, we ectopically expressed SrpNC and Lz, SrpNC and Ush, or all three factors together and assayed ush −7462/−25 lacZ activity. As with forced expression of SrpNC and Lz, forced expression of all three factors downregulated ush −7462/−25 lacZ in embryonic hemocytes (Fig. 6A). We also observed embryos with increased ush −7462/−25 lacZ activity (data not shown). In contrast, forced expression of SrpNC and Ush completely blocked ush −7462/−25 lacZ expression in hemocytes throughout the entire embryo population. Moreover, SrpNC and Ush completely blocked ush −7462/−25 lacZ activity in plasmatocytes, whereas residual activity remained in a sub-population of plasmatocytes with either forced expression of SrpNC and Lz or forced expression of all three factors (Fig. 6A). Collectively, these results show that SrpNC and Ush repressed ush −7462/−25 lacZ expression more effectively in the absence of ectopic Lz. In addition, forced expression of SrpNC and Ush did not block production of gcm-lacZ-expressing plasmatocytes but did block ush expression (data not shown). Finally, two observations show that the SrpNC:Ush complex can block ush expression without first downregulating the trans-activator Lz. First, the SrpNC:Ush complex blocked ush −7462/−25 lacZ activity in plasmatocytes, which do not express Lz. Second, the complex blocked ush −7462/−25 lacZ activity even in the presence of ectopic Lz (Fig. 6A). Together, these data support cross-regulatory control of ush expression, with activation by SrpNC and Lz and repression by SrpNC and Ush.
Fig. 6. Ush competes with Lz to control SrpNC regulation of ush expression.
(A) ush −7462/−25 lacZ and (B) ush −174/−25 lacZ was assessed in different genetic backgrounds and developmental stages. The stage of embryogenesis is indicated at the left of each row. All embryos are lateral views. The genetic background is indicated at the top of each column. SrpNC;Lz represents twi-Gal4 driving UAS-srpNC and UAS-lz located on chromosome II and III, respectively. SrpNC,Ush represents twi-Gal4 driving UAS-srpNC and UAS-ush located on chromosome II. SrpNC,Ush;Lz indicates twi-Gal4 driving UAS-srpNC,UAS-ush and UAS-lz located on chromosomes II and III, respectively. Plasmatocytes and crystal cells (outlined in panel A) are marked with arrows: closed arrows indicate wild-type CRM activity; open arrows indicate reduced activity; open dotted arrows indicate lack of activity. The level of CRM activity in the head mesoderm is marked with arrowheads: closed arrowheads indicate increased activity; the open arrowhead indicates lack of activity. Abbreviations: wt, wild-type; Srp, Serpent; Lz, Lozenge; Ush, U-shaped; cc, crystal cells; pl, plasmatocytes.
As stated above we previously identified a ush minimal −174/−25 CRM, which directs lacZ expression in hemocyte precursors, plasmatocytes, and crystal cells. This minimal hematopoietic CRM has a single RUNX motif and three GATA motifs, which are required for activity in the crystal cell lineage (Muratoglu et al., 2006). We next asked whether activation of ush expression by SrpNC and Lz and repression by SrpNC:Ush were mediated through this CRM. Forced expression of SrpNC and Lz upregulated the ush −174/−25 CRM, producing pan-mesodermal expression by stage 12. This activity was somewhat reduced when all three factors were expressed together. In contrast, forced expression of SrpNC and Ush, in the absence of ectopic Lz, inhibited ush −174/−25 lacZ expression when compared to the wild-type control (Fig. 6B). These results indicate that both activation and repression can be mediated through the ush −174/−25 CRM. Together, these results and those obtained with the larger −7462/−25 fragment indicate that SrpNC and Lz activate, whereas SrpNC and Ush repress, ush hematopoietic expression.
We observed differences between the ush minimal −174/−25 CRM and the larger −7462/−25 fragment that suggest the upstream −7462 to −174 region is important for repression of ush by SrpNC and Ush. Specifically, in contrast to the −7462/−25 CRM, forced expression of SrpNC, Lz and Ush did not downregulate the −174/−25 CRM to levels below that of wild-type (Fig. 6, compare A and B). This difference suggests that the SrpNC:Ush complex may function to repress ush hematopoietic expression through binding to GATA motifs within the −7462 to −174 region, thereby blocking activation of ush expression by competing with Lz. In this regard, there is a stark contrast between the −7462/−174 and the −174/−25 regions with respect to GATA/RUNX motif clustering. The −174/−25 region has one RUNX and three GATA motifs clustered within 83 bp. This is similar to the lz minimal CRM, which has four GATA and two RUNX motifs within 88 bp. In contrast, the −7462/−174 region has 19 GATA motifs and two RUNX motifs. However, the RUNX motifs are separated from the nearest GATA motif by at least 80 bp. The proximity of the GATA and RUNX motifs appears to be a key feature in the co-activation by Srp and Lz (Ferjoux et al., 2007). Thus, the architecture of the −7462/−174 region may favor SrpNC:Ush complex binding over Srp/Lz binding, whereas the −174/−25 region appears to accommodate both SrpNC:Ush and Srp/Lz binding. Increased binding of the SrpNC:Ush complex to the GATA motifs within the −7462 to −174 region may increase the ability of the SrpNC:Ush complex to repress ush expression in the presence of ectopic Lz. Overall, these results show that the SrpNC:Ush complex negatively regulates ush expression and that Ush competes with Lz to control SrpNC regulation of ush gene expression.
DISCUSSION
Here, we present a gene regulatory basis for combinatorial control of crystal cell production by Srp, Lz, and Ush. Our results show that Srp cross-regulates lz and ush through the GATA/RUNX motifs within the hematopoietic CRMs of both genes. This gene regulatory sub-circuit functions to modulate lz expression. The sub-circuit specifies a dynamic bi-potential regulatory state in crystal cell precursors by co-activating lz and its repressor ush. Subsequently, the sub-circuit contributes to the selection of a Lz-positive or Lz-negative state, which regulates crystal cell production.
Lz is upregulated in hemocyte precursors and is required for crystal cell lineage specification and differentiation (Lebestky et al., 2000; Bataille et al., 2005). To more fully understand the control of crystal cell production, we began by investigating the transcriptional regulation of lz expression. We identified a lz minimal CRM and showed that it directs expression in the crystal cell lineage. Our results showed that Srp activates lz-lacZ reporter-gene expression by binding to the GATA motifs within this CRM. Specifically, mutational analyses of the lz CRM showed that the GATA motifs were required for activation. In addition, forced expression of Srp produced both early activation of the lz CRM and an expanded expression domain when compared to the wild-type control. Moreover, this expression pattern was reminiscent of the supernumerary crystal cell pattern produced by forced expression of Srp (Fossett et al., 2003; Waltzer et al., 2003). Together, these observations indicate that lz is directly activated by Srp as a prerequisite for crystal cell specification.
In addition to the GATA motifs, lz CRM activity requires canonical RUNX motifs. This indicates that positive autoregulation of lz is involved during crystal cell commitment and development. Autoregulation is likely a conserved property of the Runx gene family. RUNX2 has been shown to both positively and negatively autoregulate its expression in osteoblasts (Ducy et al., 1999; Drissi et al., 2000). In addition, all three mammalian Runx genes carry canonical RUNX motifs within their respective cis-regulatory regions, suggesting that Runx1 and Runx3 are autoregulated as well (Otto et al., 2003; Smith et al., 2004). However, Lz autoregulation requires cooperative interaction with Srp, evidenced by the fact that mutation of either the GATA or RUNX motifs completely blocked lz CRM activity throughout embryonic development. Moreover, forced co-expression Srp and Lz synergistically activated the lz CRM. Together, these results and the recent report of Ferjoux and colleagues (Ferjoux, et al., 2007) indicate that both Srp and Lz interact to maintain lz expression during crystal cell commitment and development.
Srp and Lz are also required to activate ush expression in crystal cell precursors (Muratoglu et al., 2006). This gene regulatory sequence, where Srp activates lz and then both Srp and Lz are required to co-activate ush, constitutes a feed forward regulatory motif. This type of motif determines the order of gene expression and controls the temporal expression of the downstream target (Mangan et al., 2003; Swiers et al., 2006). Accordingly, we observed Ush expression in the crystal cell lineage after the onset of lz-lacZ expression. Thus, this design insures that the crystal cell precursors are specified by Lz before upregulation of the repressor Ush. This is essential because premature activation of the SrpNC:Ush complex completely blocked lz CRM expression and crystal cell production.
After the initial specification of the crystal cell precursors, Srp and Lz maintain lz and upregulate ush through clustered GATA/RUNX motifs within the CRMs of both genes. As a result, lz-lacZ and Ush were co-expressed in the crystal cell lineage from embryonic stage 10 through 14. The co-expression of Lz and its antagonist Ush forms the basis for the bi-potential nature of the crystal cell precursor. As such, this regulatory state provides the precursor with the option of progressing towards either the Lz-positive or Lz-negative state. Thus, Srp and Lz co-regulation of crystal cell development involves more than just activation of the crystal cell program, but rather includes the specification of a dynamic bi-potential regulatory state. Furthermore, Srp and Lz activate two crystal cell-specific genes (prophenoloxidases; proPOs) in stage 11 precursors through GATA/RUNX sites within the CRMs of each gene (Gajewski et al., 2007; Ferjoux et al., 2007). The co-expression of Ush with these crystal cell-specific genes may be consistent with lineage priming, showing that regulators of opposing pathways are expressed in bi-potent precursors prior to lineage commitment (Hu et al., 1997; Orkin, 2000; Warren and Rothenberg, 2003; Ye et al., 2003; Huang et al., 2007). Collectively, these results indicate that Srp and Lz direct ush expression as part of a feed forward motif, which insures the bi-potential regulatory state is established after crystal cell specification and before lineage commitment. Finally, in the dynamic bi-potential regulatory state, where Ush and Lz compete with SrpNC to cross-regulate each other, the expected phenotypic heterogeneity is reflected in the co-localization of lz-lacZ and Ush during embryonic stages 12 through 14. Initially, we observed lz-lacZ-positive, Ush-positive precursors. By embryonic stage 12, the population included cells that expressed only Ush (lz-lacZ-negative, Ush-positive). During stage 13, we observed a third cell type (lz-lacZ-positive with diminished Ush expression).
As part of the Srp cross-regulatory sub-circuit, both Srp isoforms can act with Lz to maintain lz expression and activate ush to produce the bi-potential regulatory state. However, only SrpNC can mediate cross-antagonism between Lz and Ush and, thereby, the selection of the Lz-positive or Lz-negative state. This illustrates the importance of alternative splicing of srp (Fossett et al., 2001; Waltzer et al., 2002) as a determinant of lz gene regulation. The role of the SrpNC:Ush complex in selecting the Lz-negative state is supported by our results that showed forced expression of SrpNC and Ush blocked lz CRM activity, whereas CRM activity increased in ush mutant embryos. Thus, Ush converts SrpNC from an activator to a repressor that blocks lz expression through the GATA motifs within the crystal cell CRM. This contributes to the selection of the Lz-negative state, thereby preventing the bi-potential precursor from becoming a terminally differentiated crystal cell. Overall, this limits crystal cell number. Thus, these results provide a gene regulatory basis for our previous studies that showed the SrpNC:Ush complex limits crystal cell production (Fossett et al., 2003). Moreover, Lz-expressing crystal cell precursors can give rise to either Lz-positive crystal cells or Lz-negative plasmatocytes (Lebestky et al., 2000; Bataille et al., 2005), and the modulation of lz expression is most likely a critical determinant of cell fate choice. Therefore, the SrpNC:Ush complex may contribute to selection of the plasmatocyte lineage by downregulating lz expression, thereby terminating the crystal cell developmental program to stabilize plasmatocyte cell fate choice.
SrpNC-mediated cross-antagonism of Lz and Ush is a dynamic process. SrpNC and Ush not only blocked lz expression, but the SrpNC:Ush complex can also block ush expression in crystal cells and plasmatocytes. This limits the formation of the SrpNC:Ush complex and, thereby, its capacity to regulate gene expression in these cells. In plasmatocytes the SrpNC:Ush complex also blocks expression of croquemort, an important component of the programmed cell death pathway (Franc et al., 1999; Waltzer et al., 2002). Thus, negative regulation of ush may relieve croquemort repression preparing plasmatocytes to function during programmed cell death. In crystal cells, downregulation of ush limits the capacity of the SrpNC:Ush complex to block lz expression. This may provide an additional regulatory tier that prevents the complete repression of lz expression in bi-potent crystal cell precursors. The net effect would be to increase the availability of SrpNC to act with Lz to maintain the Lz-positive state. As part of the Srp cross-regulatory sub-circuit, subtle changes in the relative levels of SrpC and SrpNC may be involved in locking on the Lz-positive, Ush-negative regulatory state. These differences would be mediated through the distinct characteristics of the individual CRMs, within the overall architecture of the cross-regulatory sub-circuit, resulting in the shift to the Lz-positive, Ush-negative state. This subject is also being investigated in our laboratory as part of the larger gene regulatory network that controls crystal cell commitment and development.
Together, these studies provide the basis for a model of Drosophila hematopoiesis that describes the regulation of lz expression and, as a result, blood cell production. Although the totality of the regulatory process remains to be determined, this model builds on previous studies of crystal cell specification and development and describes the events that occur after the initial activation of lz by Srp. Subsequently, the Lz-positive state is maintained by cooperative interaction between Srp and Lz. These transcriptional regulators also activate ush expression in Lz-positive crystal cell precursors to produce a dynamic, bi-potential regulatory state. This provides for the selection of either a Lz-positive or Lz-negative state, which is determined in part by SrpNC-mediated cross-antagonism of Lz and Ush (Fig. 7). This model is also consistent with the observation that a number of factors involved in cell fate control function as components of cross-regulatory and auto-regulatory sub-circuits (Orkin, 2000; Swiers et al., 2006; Huang et al., 2007). Furthermore, the Srp cross-regulatory sub-circuit controls blood cell development as one part of a larger gene network. This sub-circuit is undoubtedly interlinked to additional sub-circuits that control related functions, such as the regulation of alternative splicing of the srp transcript to produce either the SrpNC or SrpC isoform (Fossett et al., 2001; Waltzer et al., 2002). Finally, the dynamic regulation of these hematopoietic factors is consistent with the observed population heterogeneity as crystal cell precursors progress from a bi-potential regulatory state to either a Lz-positive or Lz-negative state.
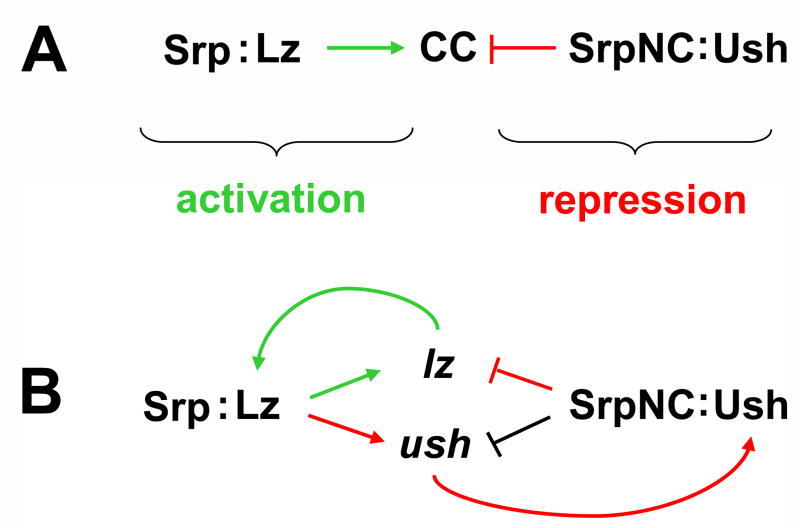
Fig. 7. Model of Srp cross-regulatory control of crystal cell lineage commitment.
(A) Combinatorial control of crystal cell lineage commitment by Srp, Lz, and Ush. Srp acts as a contextual switch, interacting with Lz to activate crystal cell lineage commitment and with Ush to block crystal cell production. (B) Srp cross-regulatory control of lz and ush expression. Srp and Lz interact to maintain the Lz-positive regulatory state. These transcriptional regulators also activate ush expression in Lz-positive crystal cell precursors to produce a dynamic, bi-potential regulatory state. SrpNC mediates cross-antagonism of Lz and Ush, acting with Lz to maintain the Lz-positive state or with Ush to select the Lz-negative regulatory state. Additionally, the SrpNC:Ush complex can downregulate ush expression, thereby limiting the capacity of the complex to block lz expression. Srp cross-regulatory control of lz and ush is mediated through the GATA and RUNX binding sites within the minimal hematopoietic CRMs of each locus. Green arrows indicate the activation pathway. Red arrows and blocked red lines indicate the repression pathway. The blocked black line indicates relief of repression. Abbreviations: Srp, Serpent; Lz, Lozenge, CC, crystal cells; Ush, U-shaped.
Aspects of GATA, FOG and RUNX functions are conserved between the fly and vertebrates. Our studies provide new information describing how these factors may interact to regulate Runx and Fog gene expression in Drosophila. Specifically, this study presents the first demonstration of GATA:FOG regulation of Runx gene expression. This may be particularly important to the understanding of vertebrate hematopoiesis because GATA, FOG and RUNX factors are co-expressed and required for megakaryopoiesis (Tsang et al., 1997; 1998; Song et al., 1999; Gaines et al., 2000; Nichols et al., 2000; Freson, et al., 2001; Mehaffey et al., 2001; Cantor et al., 2002; Chang et al., 2002; Michaud et al., 2002; Walker et al., 2002; Yu et al., 2002; Elagib et al., 2003; Tanaka et al., 2004). In addition, little is known about how Runx gene expression is regulated during this process (Otto et al., 2003; Swiers et al., 2006). We also present the first demonstration of FOG autoregulation. This finding may provide insights into regulation of vertebrate Fog genes, thereby increasing our understanding of the mechanisms by which these transcriptional regulators modify the function of GATA factors.
During vertebrate hematopoiesis, multipotent progenitors express lineage specific factors prior to lineage commitment (Hu et al., 1997; Orkin, 2000; Warren and Rothenberg, 2003; Ye et al., 2003). This constitutes a dynamic regulatory state that provides for multiple developmental options (Orkin, 2000). As a result, lineage commitment can involve cross-antagonism to select one developmental program and repress alternative lineage programs (Orkin, 2000; Cantor and Orkin, 2002). Our studies have identified a gene regulatory sub-circuit that produces a dynamic, bi-potential regulatory state that contributes to selection of one of two possible regulatory pathways, consistent with these models of vertebrate hematopoiesis. Finally, this provides a mechanistic basis for the combinatorial control of blood cell development by Srp, Lz, and Ush and a framework for future investigations of Drosophila hematopoiesis.
Acknowledgments
We are grateful to the following colleagues for providing fly strains: J. Pollock (Duquesne University) for lz-Gal4;UAS-GFP; J. Canon and U. Banerjee (University of California, Los Angeles) for UAS-lz; D. K. Hoshizaki (University of Nevada, Las Vegas, NV) for UAS-srpC; K. M. Gajewski (University of Texas, M. D. Anderson Cancer Center, Houston, TX) for UAS-srpNC;UAS-lz. We also thank the Bloomington stock center for fly stocks. We are grateful to Betsy Garratt and Kamal Deol for excellent technical assistance. This research was supported by grants to N. F. from the American Heart Association and the National Institutes of Health (DK072229). B. H. was supported by NIH postdoctoral training grant T32-HL07698.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alfonso TB, Jones BW. gcm2 promotes glial cell differentiation and is required with glial cells missing for macrophage development in Drosophila. Dev Biol. 2002;248:369–383. doi: 10.1006/dbio.2002.0740. [DOI] [PubMed] [Google Scholar]
- Anglin I, Passaniti A. Runx protein signaling in human cancers. Cancer Treat Res. 2004;119:189–215. doi: 10.1007/1-4020-7847-1_10. [DOI] [PubMed] [Google Scholar]
- Bataille L, Auge B, Ferjoux G, Haenlin M, Waltzer L. Resolving embryonic blood cell fate choice in Drosophila: interplay of GCM and RUNX factors. Development. 2005;132:4635–4644. doi: 10.1242/dev.02034. [DOI] [PubMed] [Google Scholar]
- Bernardoni R, Vivancos V, Giangrande A. glide/gcm is expressed and required in the scavenger cell lineage. Dev Biol. 1997;191:118–130. doi: 10.1006/dbio.1997.8702. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Bresnick EH, Johnson KD, Kim SI, Im H. Establishment and regulation of chromatin domains: mechanistic insights from studies of hemoglobin synthesis. Prog Nucleic Acid Res Mol Biol. 2006;81:435–471. doi: 10.1016/S0079-6603(06)81011-1. [DOI] [PubMed] [Google Scholar]
- Cantor AB, Katz SG, Orkin SH. Distinct domains of the GATA-1 cofactor FOG-1 differentially influence erythroid versus megakaryocytic maturation. Mol Cell Biol. 2002;22:4268–4279. doi: 10.1128/MCB.22.12.4268-4279.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantor AB, Orkin SH. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin Cell Dev Biol. 2005;16:117–128. doi: 10.1016/j.semcdb.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Chang A, Cantor A, Fujiwara Y, Droho S, Lodish M, Crispino JD, Orkin SH. FOG-1 requires an interaction with either GATA-1 or GATA-2 for its essential role in early megakaryopoiesis. Proc Natl Acad Sci U S A. 2002;99:9237–9242. doi: 10.1073/pnas.142302099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman JA. Runx transcription factors and the developmental balance between cell proliferation and differentiation. Cell Biol Int. 2003;27:315–324. doi: 10.1016/s1065-6995(03)00018-0. [DOI] [PubMed] [Google Scholar]
- Crew JR, Batterham P, Pollock JA. Developing compound eye in lozenge mutants of Drosophila: lozenge expression in the R7 equivalence group. Dev Gene Evol. 1997;206:481–493. doi: 10.1007/s004270050079. [DOI] [PubMed] [Google Scholar]
- Crispino JD, Lodish MB, MacKay JP, Orkin SH. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol Cell. 1999;3:219–228. doi: 10.1016/s1097-2765(00)80312-3. [DOI] [PubMed] [Google Scholar]
- Crozatier M, Meister M. Drosophila haematopoiesis. Cell Micro. 2007;9:1117–1126. doi: 10.1111/j.1462-5822.2007.00930.x. [DOI] [PubMed] [Google Scholar]
- Dearolf CR. Fruit fly “leukemia”. BBA. 1998;1377:M13–23. doi: 10.1016/s0304-419x(97)00031-0. [DOI] [PubMed] [Google Scholar]
- de Bruijn MF, Speck NA. Core-binding factors in hematopoiesis and immune function. Oncogene. 2004;23:4238–4248. doi: 10.1038/sj.onc.1207763. [DOI] [PubMed] [Google Scholar]
- Drissi H, Luc Q, Shakoori R, Chuva De Sousa Lopes S, Choi JY, Terry A, Hu M, Jones S, Neil JC, Lian JB, Stein JL, Van Wijnen AJ, Stein GS. Transcriptional autoregulation of the bone related CBFA1/RUNX2 gene. J Cell Physiol. 2000;184:341–350. doi: 10.1002/1097-4652(200009)184:3<341::AID-JCP8>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Ducy P, Starbuck M, Priemel M, Shen J, Pinero G, Geoffroy V, Amling M, Karsenty G. A Cbfa1-dependent genetic pathway controls bone formation beyond embryonic development. Genes Dev. 1999;13:1025–1036. doi: 10.1101/gad.13.8.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elagib KE, Racke FK, Mogass M, Khetawat R, Delehanty LL, Goldfarb AN. RUNX1 and GATA-1 coexpression and cooperation in megakaryocytic differentiation. Blood. 2003;101:4333–4341. doi: 10.1182/blood-2002-09-2708. [DOI] [PubMed] [Google Scholar]
- Evans CJ, Hartenstein V, Banerjee U. Thicker than blood: conserved mechanisms in Drosophila and vertebrate hematopoiesis. Dev Cell. 2003;5:673–690. doi: 10.1016/s1534-5807(03)00335-6. [DOI] [PubMed] [Google Scholar]
- Ferjoux G, Benoit A, Boyer K, Haenlin M, Waltzer L. A GATA/RUNX cis-regulatory module couples Drosophila blood cell commitment and differentiation into crystal cells. Dev Biol. 2007 doi: 10.1016/j.ydbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Fossett N, Tevosian SG, Gajewski K, Zhang Q, Orkin SH, Schulz RA. The Friend of GATA proteins U-shaped, FOG-1, and FOG-2 function as negative regulators of blood, heart, and eye development in Drosophila. Proc Natl Acad Sci U S A. 2001;98:7342–7347. doi: 10.1073/pnas.131215798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fossett N, Schulz RA. Functional conservation of hematopoietic factors in Drosophila and vertebrates. Differentiation. 2001;69:83–90. doi: 10.1046/j.1432-0436.2001.690202.x. [DOI] [PubMed] [Google Scholar]
- Fossett N, Hyman K, Gajewski K, Orkin SH, Schulz RA. Combinatorial interactions of Serpent, Lozenge, and U-shaped regulate crystal cell lineage commitment during Drosophila hematopoiesis. Proc Natl Acad Sci U S A. 2003;100:11451–11456. doi: 10.1073/pnas.1635050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franc NC, Heitzler P, Ezekowitz RA, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. 1999;284:1991–1994. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- Freson K, Devriendt K, Matthijs G, Van Hoof A, De Vos R, Thys C, Minner K, Hoylaerts MF, Vermylen J, Van Geet C. Platelet characteristics in patients with X-linked macrothrombocytopenia because of a novel GATA1 mutation. Blood. 2001;98:185–192. doi: 10.1182/blood.v98.1.85. [DOI] [PubMed] [Google Scholar]
- Gaines P, Geiger JN, Knudsen G, Seshasayee D, Wojchowski DM. GATA-1- and FOG-dependent activation of megakaryocytic alpha IIB gene expression. J Biol Chem. 2000;275:34114–34121. doi: 10.1074/jbc.M006017200. [DOI] [PubMed] [Google Scholar]
- Gajewski K, Kim Y, Lee YM, Olson EN, Schulz RA. D-mef2 is a target for Tinman activation during Drosophila heart development. EMBO J. 1997;16:515–522. doi: 10.1093/emboj/16.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski KM, Sorrentino RP, Lee JH, Zhang Q, Russell M, Schulz RA. Identification of a crystal cell-specific enhancer of the black cells prophenoloxidase gene in drosophila. Genesis. 2007;45:200–207. doi: 10.1002/dvg.20285. [DOI] [PubMed] [Google Scholar]
- Hartenstein V. Blood cells and blood cell development in the animal kingdom. Annu Rev Cell Dev Biol. 2006;22:677–712. doi: 10.1146/annurev.cellbio.22.010605.093317. [DOI] [PubMed] [Google Scholar]
- Hong W, Nakazawa M, Chen YY, Kori R, Vakoc CR Rakowski C, Blobel GA. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005;24:2367–2378. doi: 10.1038/sj.emboj.7600703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J. PhD Thesis. The University of Louisville; Louisville, KY: 1995. The role of DNA/protein interactions in transcription from the larval promoter of the D. affinidisjuncta alcohol dehydrogenase gene. [Google Scholar]
- Hu M, Krause D, Greaves M, Sharkis S, Dexter M, Heyworth C, Enver T. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997;11:774–785. doi: 10.1101/gad.11.6.774. [DOI] [PubMed] [Google Scholar]
- Huang S, Guo YP, May G, Enver T. Bifurcation dynamics in lineage-commitment in bipotent progenitor cells. Dev Biol. 2007 doi: 10.1016/j.ydbio.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Im H, Grass JA, Johnson KD, Kim SI, Boyer ME, Imbalzano AN, Bieker JJ, Bresnick EH. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc Natl Acad Sci U S A. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SH, Evans CJ, Uemura C, Banerjee U. The Drosophila lymph gland as a developmental model of hematopoiesis. Development. 2005;132:2521–2533. doi: 10.1242/dev.01837. [DOI] [PubMed] [Google Scholar]
- Kammerer M, Giangrande A. Glide2, a second glial promoting factor in Drosophila melanogaster. EMBO J. 2001;20:4664–4673. doi: 10.1093/emboj/20.17.4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila. Dev Biol. 2001;230:243–257. doi: 10.1006/dbio.2000.0123. [DOI] [PubMed] [Google Scholar]
- Lebestky T, Chang T, Hartenstein V, Banerjee U. Specification of Drosophila hematopoietic lineage by conserved transcription factor. Science. 2000;288:146–149. doi: 10.1126/science.288.5463.146. [DOI] [PubMed] [Google Scholar]
- Letting DL, Chen YY, Rakowski C, Reedy S, Blobel GA. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc Natl Acad Sci U S A. 2004;101:476–481. doi: 10.1073/pnas.0306315101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowry JA, Mackay JP. GATA-1: one protein, many partners. Int J Biochem Cell Biol. 2006;38:6–11. doi: 10.1016/j.biocel.2005.06.017. [DOI] [PubMed] [Google Scholar]
- Lu JR, McKinsey TA, Xu H, Wang DZ, Richardson JA, Olson EN. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol Cell Biol. 1999;19:4495–502. doi: 10.1128/mcb.19.6.4495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehaffey MG, Newton AL, Gandhi MJ, Crossley M, Drachman JG. X-linked thrombocytopenia caused by a novel mutation of GATA-1. Blood. 2001;989:2681–2688. doi: 10.1182/blood.v98.9.2681. [DOI] [PubMed] [Google Scholar]
- Meister M, Lagueux M. Drosophila blood cells. Cell Microbiol. 2003;5:573–580. doi: 10.1046/j.1462-5822.2003.00302.x. [DOI] [PubMed] [Google Scholar]
- Michaud J, Wu F, Osato M, Cottles GM, Yanagida M, Asou N, Shigesada K, Ito Y, Benson KF, Raskind WH, Rossier C, Antonarakis SE, Israels S, McNicol A, Weiss H, Horwitz M, Scott HS. In vitro analyses of known and novel RUNX1/AML1 mutations in dominant familial platelet disorder with predisposition to acute myelogenous leukemia: implications for mechanisms of pathogenesis. Blood. 2002;99:1364–1372. doi: 10.1182/blood.v99.4.1364. [DOI] [PubMed] [Google Scholar]
- Muratoglu S, Garratt B, Hyman K, Gajewski K, Schulz RA, Fossett N. Regulation of Drosophila Friend of GATA gene, u-shaped, during hematopoiesis: A direct role for Serpent and Lozenge. Dev Biol. 2006;296:561–579. doi: 10.1016/j.ydbio.2006.04.455. [DOI] [PubMed] [Google Scholar]
- Newton A, Mackay J, Crossley M. The N-terminal zinc finger of the erythroid transcription factor GATA-1 binds GATC motifs in DNA. J Biol Chem. 2001;276:35794–35801. doi: 10.1074/jbc.M106256200. [DOI] [PubMed] [Google Scholar]
- Nichols KE, Crispino JD, Poncz M, White JG, Orkin SH, Maris JM, Weiss MJ. Familial dyserythropoietic anaemia and thrombocytopenia due to an inherited mutation in GATA1. Nat Genet. 2000;24:266–270. doi: 10.1038/73480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda T, van Deursen J, Hiebert SW, Grosveld G, Downing JR. AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell. 1996;84:321–330. doi: 10.1016/s0092-8674(00)80986-1. [DOI] [PubMed] [Google Scholar]
- Oliveri P, Davidson EH. Built to run, not fail. Science. 2007;315:1510–1511. doi: 10.1126/science.1140979. [DOI] [PubMed] [Google Scholar]
- Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet. 2000;1:57–64. doi: 10.1038/35049577. [DOI] [PubMed] [Google Scholar]
- Otto F, Lubbert M, Stock M. Upstream and downstream targets of RUNX proteins. J Cell Biochem. 2003;89:9–18. doi: 10.1002/jcb.10491. [DOI] [PubMed] [Google Scholar]
- Pedone PV, Omichinski JG, Nony P, Trainor C, Gronenborn AM, Clore GM, Felsenfeld G. The N-terminal fingers of chicken GATA-2 and GATA-3 are independent sequence-specific DNA binding domains. EMBO J. 1997;16:2874–2882. doi: 10.1093/emboj/16.10.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrotta V. Cloning Drosophila genes. In: Roberts DB, editor. Drosophila: A practical approach. IRL Press; Oxford: 1986. pp. 83–110. [Google Scholar]
- Querfurth E, Schuster M, Kulessa H, Crispino JD, Doderlein G, Orkin SH, Graf T, Nerlov C. Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 2000;14:2515–2525. doi: 10.1101/gad.177200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehorn KP, Thelen H, Michelson AM, Reuter R. A molecular aspect of hematopoiesis and endoderm development common to vertebrates and Drosophila. Development. 1996;122:4023–4031. doi: 10.1242/dev.122.12.4023. [DOI] [PubMed] [Google Scholar]
- Rennert J, Coffman JA, Mushegian AR, Robertson AJ. The evolution of Runx genes I. A comparative study of sequences from phylogenetically diverse model organisms. BMC Evol Biol. 2003;3:4. doi: 10.1186/1471-2148-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizki TM. The circulatory system and associated cells and tissues. In: Ashburner M, Wright TRF, editors. The Genetics and Biology of Drosophila. Academic Press; New York: 1978. pp. 397–452. [Google Scholar]
- Sam S, Leise W, Hoshizaki DK. The serpent gene is necessary for progression through the early stages of fat-body development. Mech Dev. 1996;60:197–205. doi: 10.1016/s0925-4773(96)00615-6. [DOI] [PubMed] [Google Scholar]
- Schulz RA, Fossett N. Hemocyte development during Drosophila embryogenesis. In: Baron MH, editor. Developmental Regulation of Hematopoiesis: Methods and Practical Approaches, Methods in Molecular Medicine Series. Humana Press; New York: 2005. pp. 109–122. [DOI] [PubMed] [Google Scholar]
- Smith N, Dong Y, Lian JB, Pratap J, Kingsley PD, van Wijnen AJ, Stein JL, Schwarz EM, O’Keefe RJ, Stein GS, Drissi MH. Overlapping expression of Runx1(Cbfa2) and Runx2(Cbfa1) transcription factors supports cooperative induction of skeletal development. J Cell Physiol. 2005;203:133–143. doi: 10.1002/jcp.20210. [DOI] [PubMed] [Google Scholar]
- Song WJ, Sullivan MG, Legare RD, Hutchings S, Tan X, Kufrin D, Ratajczak J, Resende IC, Haworth C, Hock R, Loh M, Felix C, Roy DC, Busque L, Kurnit D, Willman C, Gewirtz AM, Speck NA, Bushweller JH, Li FP, Gardiner K, Poncz M, Maris JM, Gilliland DG. Haploinsufficiency of CBFA2 causes familial thrombocytopenia with propensity to develop acute myelogenous leukaemia. Nat Genet. 1999;23:166–175. doi: 10.1038/13793. [DOI] [PubMed] [Google Scholar]
- Sorrentino RP, Gajewski KM, Schulz RA. GATA factors in Drosophila heart and blood cell development. Semin Cell Dev Biol. 2005;16:107–116. doi: 10.1016/j.semcdb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- Speck NA, Gilliland DG. Core-binding factors in haematopoiesis and leukaemia. Nat Rev Cancer. 2002;2:502–513. doi: 10.1038/nrc840. [DOI] [PubMed] [Google Scholar]
- Svensson EC, Tufts RL, Polk CE, Leiden JM. Molecular cloning of FOG-2: a modulator of transcription factor GATA-4 in cardiomyocytes. Proc Natl Acad Sci U S A. 1999;96:956–961. doi: 10.1073/pnas.96.3.956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiers G, Patient R, Loose M. Genetic regulatory networks programming hematopoietic stem cells and erythroid lineage specification. Dev Biol. 2006;294:525–540. doi: 10.1016/j.ydbio.2006.02.051. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Zheng J, Kitajima K, Kita K, Yoshikawa H, Nakano T. Differentiation status dependent function of FOG-1. Genes Cells. 2004;9:1213–1226. doi: 10.1111/j.1365-2443.2004.00796.x. [DOI] [PubMed] [Google Scholar]
- Tepass U, Fessler LI, Aziz A, Hartenstein V. Embryonic origin of hemocytes and their relationship to cell death in Drosophila. Development. 1994;120:1829–1837. doi: 10.1242/dev.120.7.1829. [DOI] [PubMed] [Google Scholar]
- Tevosian SG, Deconinck AE, Cantor AB, Rieff HI, Fujiwara Y, Corfas G, Orkin SH. FOG-2: A novel GATA-family cofactor related to multitype zinc-finger proteins Friend of GATA-1 and U-shaped. Proc Natl Acad Sci U S A. 1999;96:950–955. doi: 10.1073/pnas.96.3.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel CS, Boulet AM, Lipshitz HD. Vectors for Drosophila P-element-mediated transformation and tissue culture transfection. Gene. 1988;74:445–456. doi: 10.1016/0378-1119(88)90177-1. [DOI] [PubMed] [Google Scholar]
- Tracey WD, Speck NA. Potential roles for RUNX1 and its orthologs in determining hematopoietic cell fate. Semin Cell Dev Biol. 2000;11:337–342. doi: 10.1006/scdb.2000.0186. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell. 1997;90:109–119. doi: 10.1016/s0092-8674(00)80318-9. [DOI] [PubMed] [Google Scholar]
- Tsang AP, Fujiwara Y, Hom DB, Orkin SH. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 1998;12:1176–1188. doi: 10.1101/gad.12.8.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker LC, Stevens J, Campbell H, Corbett R, Spearing R, Heaton D, Macdonald DH, Morris CM, Ganly P. A novel inherited mutation of the transcription factor RUNX1 causes thrombocytopenia and may predispose to acute myeloid leukaemia. Br J Haematol. 2002;117:878–881. doi: 10.1046/j.1365-2141.2002.03512.x. [DOI] [PubMed] [Google Scholar]
- Waltzer L, Bataille L, Peyrefitte S, Haenlin M. Two isoforms of Serpent containing either one or two GATA zinc fingers have different roles in Drosophila haematopoiesis. EMBO J. 2002;21:5477–5486. doi: 10.1093/emboj/cdf545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waltzer L, Ferjoux G, Bataille L, Haenlin M. Cooperation between the GATA and RUNX factors Serpent and Lozenge during Drosophila hematopoiesis. EMBO J. 2003;22:6516–6525. doi: 10.1093/emboj/cdg622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren LA, Rothenberg EV. Regulatory coding of lymphoid lineage choice by hematopoietic transcription factors. Curr Opin Immunol. 2003;15:166–175. doi: 10.1016/s0952-7915(03)00011-6. [DOI] [PubMed] [Google Scholar]
- Ye M, Iwasaki H, Laiosa CV, Stadtfeld M, Xie H, Heck S, Clausen B, Akashi K, Graf T. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilineage repopulation potential. Immunity. 2003;19:689–699. doi: 10.1016/s1074-7613(03)00299-1. [DOI] [PubMed] [Google Scholar]
- Yu C, Niakan KK, Matsushita M, Stamatoyannopoulos G, Orkin SH, Raskind WH. X-linked thrombocytopenia with thalassemia from a mutation in the amino finger of GATA-1 affecting DNA binding rather than FOG-1 interaction. Blood. 2002;100:2040–2045. doi: 10.1182/blood-2002-02-0387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Emerson SG. Hematopoietic cytokines, transcription factors and lineage commitment. Oncogene. 2002;21:3295–3313. doi: 10.1038/sj.onc.1205318. [DOI] [PubMed] [Google Scholar]