Abstract
Peroxisomes in living CV1 cells were visualized by targeting the green fluorescent protein (GFP) to this subcellular compartment through the addition of a COOH-terminal peroxisomal targeting signal 1 (GFP–PTS1). The organelle dynamics were examined and analyzed using time-lapse confocal laser scanning microscopy. Two types of movement could be distinguished: a relatively slow, random, vibration-like movement displayed by the majority (∼95%) of the peroxisomes, and a saltatory, fast directional movement displayed by a small subset (∼5%) of the peroxisomes. In the latter instance, peak velocities up to 0.75 μm/s and sustained directional velocities up to 0.45 μm/s over 11.5 μm were recorded. Only the directional type of motion appeared to be energy dependent, whereas the vibrational movement continued even after the cells were depleted of energy. Treatment of cells, transiently expressing GFP–PTS1, with microtubule-destabilizing agents such as nocodazole, vinblastine, and demecolcine clearly altered peroxisome morphology and subcellular distribution and blocked the directional movement. In contrast, the microtubule-stabilizing compound paclitaxel, or the microfilament-destabilizing drugs cytochalasin B or D, did not exert these effects. High resolution confocal analysis of cells expressing GFP–PTS1 and stained with anti-tubulin antibodies revealed that many peroxisomes were associated with microtubules. The GFP–PTS1–labeled peroxisomes were found to distribute themselves in a stochastic, rather than ordered, manner to daughter cells at the time of mitosis.
The mammalian peroxisome is a versatile and ubiquitous subcellular organelle involved in numerous catabolic and anabolic pathways: most importantly, peroxide metabolism, the β-oxidation of (very long chain) fatty acids, and the biosynthesis of etherphospholipids (see van den Bosch et al., 1992). The vital importance of the peroxisome is stressed by the existence of a number of severely debilitating, and often lethal, disorders in humans in which the biogenesis of the organelle is impaired (Lazarow and Moser, 1994).
Proteins resident in the peroxisomal membrane and matrix are encoded by nuclear genes and imported posttranslationally (Lazarow and Fujiki, 1985). Genetic and biochemical evidence in yeast and humans supports the notion that there are at least two pathways for the import of proteins into the peroxisomal matrix, each dependent on the use of a specific peroxisomal targeting signal (PTS)1 and a cognate receptor (Subramani, 1993). Whether these import pathways are completely distinct or converge to use the same membrane translocation machinery is unknown at present. Earlier work in mammalian cells identified a carboxy-terminal, tripeptide sequence (S/A/C-K/R/ H-L/M), called PTS1, as the major targeting signal used for the sorting of proteins into the peroxisomal matrix (Gould et al., 1989; Keller et al., 1991). A second sequence R/K-L/I/V-X5-H/Q-L/A, called PTS2, is used by a smaller subset of proteins and is found at the NH2-terminal end of mammalian (Osumi et al., 1991; Swinkels et al., 1991) and yeast (Erdmann, 1994; Glover et al., 1994) 3-oxoacyl-CoA thiolase, watermelon malate dehydrogenase (Gietl, 1990), and amine oxidase of Hansenula polymorpha (Faber et al., 1994). Peroxisomal membrane proteins are targeted to the organelle via the use of different targeting signals termed membrane PTSs (mPTSs), which have been defined in two proteins (McNew and Goodman 1996; Wiemer et al., 1996).
In mammalian cells, peroxisomes usually appear as single membrane-bound spherical vesicles evenly dispersed throughout the cytoplasm. However, a more complex peroxisomal architecture has been encountered when the peroxisomal compartment is going through a phase of rapid growth or is actively engaged in the synthesis of special kinds of lipids (Yamamoto and Fahimi, 1987; Gorgas, 1987). Not much is known yet about the nature of the factor(s) that determine and sustain peroxisome morphogenesis and their characteristic subcellular distribution. It is reasonable to assume that cytoskeletal elements, in particular microtubules, are a major determinant in these processes. The cellular organization of various cytoplasmic organelles such as the Golgi and ER seems to be generated and maintained by microtubules (for reviews see Heuser et al., 1989; Kelly, 1990; Cole and Lippincott-Schwartz, 1995). Recently Schrader et al. (1996) provided in vitro and in vivo evidence for a direct interaction of microtubules with peroxisomes.
In this study we describe the use of the green fluorescent protein (GFP)–PTS1 fusion protein (Monosov et al., 1996) to study peroxisome movement, organization, and segregation in living cells.
Materials and Methods
Cell Lines and Culture Conditions
African green monkey kidney (CV1) cells were obtained from American Type Culture Collection (Rockville, MD). The cells were maintained in DME supplemented with 10% (vol/vol) FCS, 2 mM l-glutamine, 250 U/ml penicillin, and 250 μg/ml streptomycin under a humidified atmosphere containing 5% CO2.
GFP Expression Plasmids
The S65T mutant version of the GFP gene from Aequorea victoria (see Cubitt et al., 1995) was amplified by PCR from plasmid pBIIKS-GFP/ S65T using primers GCTgaattcATGAGTAAAGGAGAA (5′ end, coding strand) and AGAgaattcTTATTATTTGTATAGTTC (3′ end, noncoding strand). Similarly, GFP–PTS1 was obtained using primers GCTgaattcATGAGTAAAGGAGAA (5′ end, coding strand) and AGAgaattcTTATAATTTGGATAGTTCATCCAT (3′ end, noncoding strand). All primers were flanked by EcoRI sites (lower case). Start and stop codons, ATG and TTA (complementary strand), respectively, are underlined. Nucleotides altered to create the PTS1 (serine-lysine-leucine) codons are shown in bold. PCR fragments were cloned in the EcoRI site of the mammalian expression vector pcDNA3 (Invitrogen, San Diego, CA) under control of the cytomegalovirus (CMV) promoter, yielding the plasmids pGFP and pGFP-PTS1.
Transient Transfections
CV1 cells at 50–60% confluence were transfected with pGFP and pGFPPTS1 using lipofectamine (GIBCO BRL, Gaithersburg, MD). GFP was expressed at detectable levels between 24 and 72 h after transfection. Routinely cells were used for further experimentation 48 h after transfection.
Fluorescence Microscopy
CV1 cells were cultured in 9-cm petri dishes on glass coverslips and processed for immunofluorescence essentially as described by Keller et al. (1987). In brief, cells were fixed for 20 min in 3% (wt/vol) formaldehyde (from paraformaldehyde) and permeabilized by a 5-min incubation in 1% (vol/vol) Triton X-100 in PBS. Subsequently, the cells were washed four times with PBS and incubated with the appropriate antiserum dilutions made up in PBS containing 5 μg/ml BSA as a blocking agent. For multiple labeling, cells were incubated with species-specific anti-IgG antibodies conjugated to rhodamine or CY5. Coverslips were mounted on microscope slides in glycerol containing 4% n-propyl gallate as the antifade agent. The fluorescent staining pattern was viewed using a Photomicroscope III fluorescence microscope (Zeiss AG, Oberkochen, Germany) equipped with a Planapochromat 63/1.4 NA objective. The S65T mutant of the green fluorescent protein (490 nm excitation wavelength, 510 nm emission wavelength) was visualized either in living cells or in fixed and permeabilized cells using a standard FITC filter set. For high resolution analysis, fixed and stained cells were visualized using an Axiovert 35M microscope (Zeiss) equipped with an MRC-1024 laser scanning confocal microscope (Bio Rad Laboratories, Hercules, CA) using a krypton/argon laser.
Antibodies, DNA, and Microfilament Staining
Anti-PMP70 antisera were prepared in a rabbit using a peroxisomal membrane protein (PMP) 70 peptide conjugated to human serum albumin as described by Kamijo et al. (1990). A mouse mAb against tubulin was obtained from Sigma Chemical Co. (St. Louis, MO). Species-specific antiIgG antibodies conjugated to rhodamine or CY5 were obtained from Jackson ImmunoResearch Laboratories (West Grove, PA). DNA was stained using propidium iodide, and microfilaments were labeled using phalloidin conjugated to rhodamine from Molecular Probes, Inc. (Eugene, OR).
Treatment with Drugs Affecting the Cytoskeleton
CV1 cells were cultured on glass coverslips and transfected with the GFP– PTS1 expression construct. After 48 h various cytoskeleton-affecting drugs were added to the culture medium: the microtubule-destabilizing agents nocodazole (20 μM final concentration), vinblastine (10 μM final concentration), and demecolcine (5 μM final concentration); the microtubule-stabilizing compound paclitaxel (taxol) (20 μM final concentration); and the actin-depolymerizing drugs cytochalasin B and D at a final concentration of 2 μM and 0.5 μM, respectively. Stock solutions were prepared in DMSO and stored at −20°C. The concentration of the solvent never exceeded 0.1% (vol/vol) in the actual incubations.
Live Cell Imaging
For studies involving the detection of GFP in living cells by confocal microscopy, CV1 cells were cultured in 3.5-cm glass-bottom microwell dishes with a #0 thickness coverslip (Mattek Corp., Ashland, MA). 10 mM Hepes, pH 7.5, was added to the culture medium to stabilize the pH. An MRC-1024 confocal scanning laser microscope was used, equipped with a thermostatted stage that was kept at 35°C during the course of the experiment. Cells with a high expression of GFP were selected and monitored at various time intervals for a total of 120–180 min. (A sampling rate of 1 frame per 6.4 s was found to be optimal and was used exclusively for this study.) Excitation illumination was from an argon ion laser (488 nm) operating on low power mode and using a 0.3% transmittance neutral density filter, a ×40/1.3 objective, and ×2.5–3.5 zoom setting. Time-lapse images (single scan 512 × 512 pixels) were stored using an optical disc drive (Pinnacle Micro, Irvine, CA). Imaging conditions were determined not to be detrimental to the cells by recording a time series of images for 90 min, putting the cells back in the incubator for 2 h, and reexamining the same cells for viability.
Computer Analysis
Time-lapse digital images were transferred to an LVR 5000A video disc recorder (Sony, Montvale, NJ) for viewing as movies. For motion analysis of peroxisomes, selected sequential digital images were transferred to an Indigo2 computer (Silicon Graphics, Mountain View, CA), and the coordinates of the brightest pixels associated with individual peroxisomes were tracked over time using the program Fido (Soto et al., 1993). 85 individual peroxisomes that remained mainly in the plane of focus over the time course were selected so as to minimize the impact of z-dimension motion. Data were imported into the program KaleidaGraph (Synergy Software, Redding, PA) run on a Macintosh Quadra 950 computer (Apple Computer Inc., Cupertino, CA) and graphed as peak velocity vs count, mean average velocity, vs count, and as coordinate vector plots.
Cell Cycle Synchronization
CV1 cells were cultured to 50–75% confluence, washed twice with DME, and synchronized in the G0 phase by an incubation for 48 h in DME supplemented with 2% (vol/vol) dialyzed FCS. Transfection of the cells with the pGFP–PTS1 expression construct was performed, using the standard procedure, during the last 6 h of this starvation period. Subsequently, the cells were washed with DME and incubated in DME supplemented with 10% (vol/vol) FCS and containing 0.5 mM hydroxyurea to arrest the cells in the early S phase. After 16 h, the hydroxyurea was removed, and the cells were incubated in fully supplemented medium. Approximately 40– 60% of the cells went through mitosis within 9–11 h after release of the hydroxyurea block.
Results
GFP–PTS1 Fusion Protein Is Targeted to the Peroxisomal Compartment
A cDNA encoding the S65T mutant version (Cubitt et al., 1995) of GFP and a GFP–PTS1 fusion construct were cloned in the mammalian expression vector pcDNA3 (Fig. 1) under control of the human CMV promoter/enhancer region. After transfection of CV1 cells with pGFP-PTS1, a punctate fluorescent signal was detected (Fig. 2 A). The GFP–PTS1 fusion protein was in peroxisomes, as indicated by its colocalization with a genuine peroxisomal marker protein PMP70 (Kamijo et al., 1990) (see Fig. 2, C and D). A similar observation was made previously when GFP–PTS1 was expressed in the yeast Pichia pastoris (Monosov et al., 1996). In control cells expressing GFP, a diffuse cytosolic fluorescence was observed (Fig. 2 B). Not all the cells expressed GFP or GFP–PTS1 equally well; some cells only displayed a weak green fluorescence, while others stained very brightly, probably reflecting differences in expression levels.
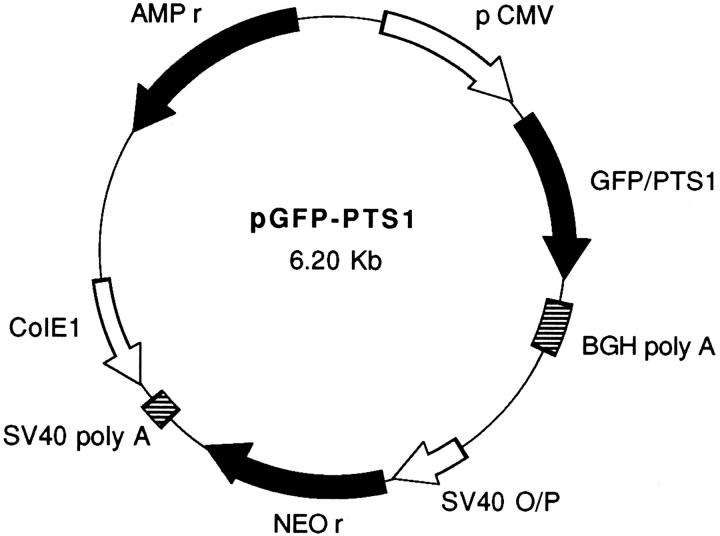
Figure 1.
GFP–PTS1 expression construct. A mutant version (S65T) of the GFP cDNA was cloned into the vector pcDNA3 as described in Materials and Methods. The resulting plasmids either expressed GFP (pGFP) alone or GFP onto which a peroxisomal type I targeting signal (Ser-Lys-Leu-COOH) was fused at its COOH-terminal end (pGFP-PTS1). Depicted is pGFP-PTS1 containing the GFP–PTS1 construct under control of the human cytomegalovirus intermediate early promoter/enhancer region (pCMV). BGH poly A, bovine growth hormone polyadenylation signal; SV40 O/P, SV40 origin and early promoter; NEO r, neomycin-resistance gene; SV40 poly A, SV40 T-antigen polyadenylation signal; Col E1, col E1 origin of replication; AMP r, β-lactamase gene.
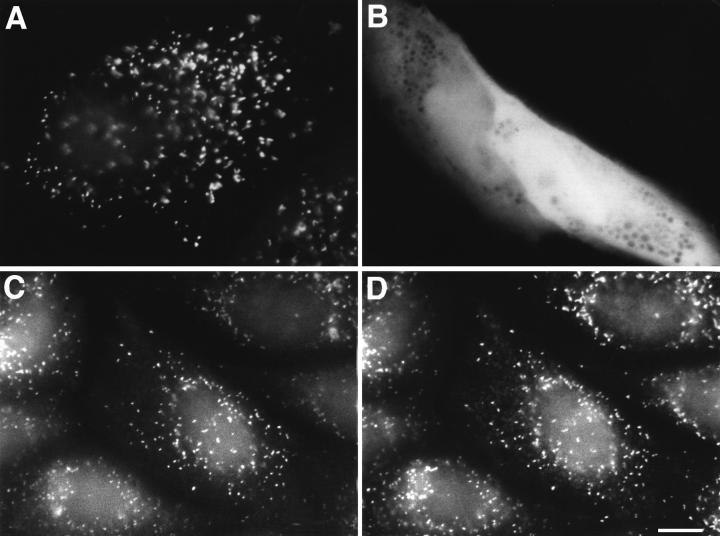
Figure 2.
Subcellular localization of GFP and GFP–PTS1 in CV1 cells. CV1 cells were cultured on coverslips, transfected with pGFP and pGFP-PTS1. After 48 h the cells were examined directly for GFP–PTS1 (A) or GFP (B) expression. Alternatively, pGFP-PTS1– transfected CV1 cells were fixed, permeabilized, and processed for immunofluorescence. In a double-labeling experiment, the cells were incubated with rabbit anti–PMP70, and subsequently with goat anti–rabbit IgG conjugated to rhodamine. The cells were monitored for the subcellular localization of GFP–PTS1 (C) in the FITC channel and PMP70 (D) in the rhodamine channel. Bar, 10 μm.
The targeting of GFP–PTS1 to peroxisomes was remarkably accurate. In Fig. 2 C, of 203 GFP–PTS1–containing peroxisomes, 201 (99.2%) also expressed PMP70 (Fig. 2 D). Using data from this and other experiments in which a total of 570 GFP–PTS1–containing peroxisomes were analyzed, we found that, of the organelles expressing GFP–PTS1 strongly, 100% contained PMP70. In an unbiased analysis of these organelles, irrespective of whether they expressed GFP–PTS1 strongly or weakly, only 1% failed to coexpress PMP70. All our analyses of peroxisome movement were done with organelles strongly expressing GFP–PTS1. These data demonstrate that the fidelity of import of GFP–PTS1 to peroxisomes is remarkably efficient, and hence the movements described below are those of peroxisomes, and not of other organelles.
Dynamics of the Peroxisomal Compartment in Living CV1 Cells
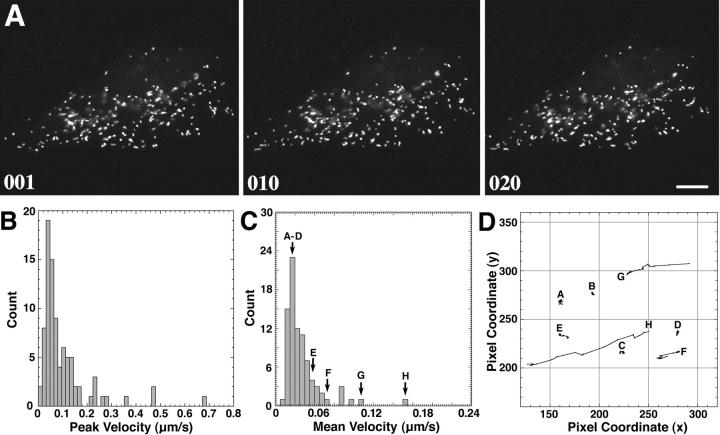
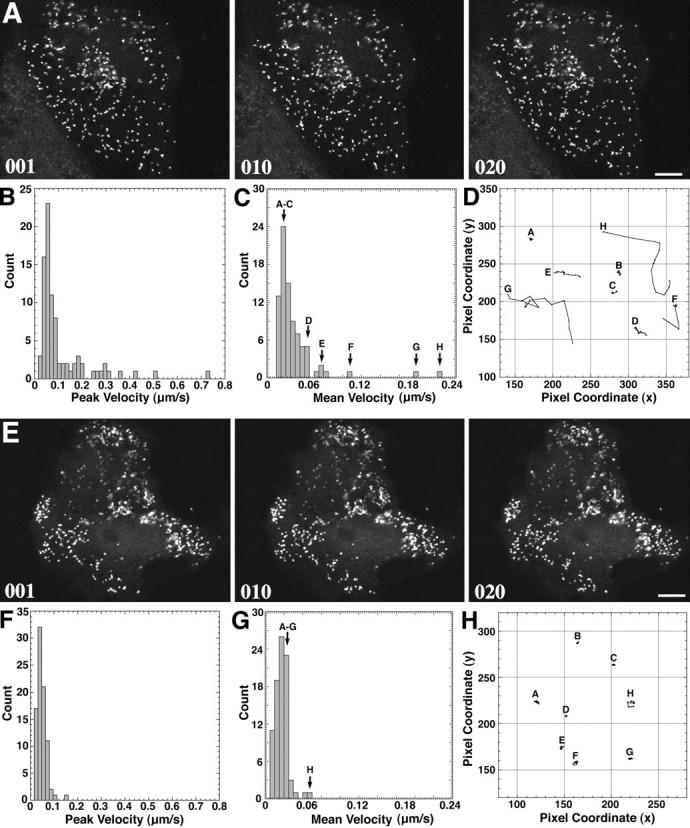
CV1 cells expressing GFP–PTS1 were examined using a confocal laser scanning microscope. To provide optimal physiological conditions during imaging, the temperature was kept constant and the laser illumination was kept to a minimum to reduce the risk of phototoxicity. It was not uncommon to observe vitality signs such as membrane ruffling and cell migration during imaging. Time-lapse experiments showed that peroxisomes are motile organelles. The expression of GFP–PTS1 did not affect the growth properties of the cells, suggesting that the movement of GFP–PTS1–containing peroxisomes was not aberrant in any way. Two types of peroxisomal movement could be discerned: a relatively slow, random, vibrational movement exhibited continuously by most (∼95%) peroxisomes, and a fast, unidirectional movement displayed by a subset (∼5%) of peroxisomes over long distances (sometimes traversing the entire cell length). Since the former type of movement exhibited by most of the peroxisomes resulted in little net movement of the peroxisomes, images of cells taken over ∼2 min (∼20 frames) showed very little displacement of most of the peroxisomes (Fig. 3 A, frames 001–020). Motion analysis of individual peroxisomes revealed that the slow movement was characterized by peak velocities of <0.2 μm/s (Fig. 3 B), mean average velocities of <0.05 μm/s (Fig. 3 C), and an average overall displacement of <0.5 μm after 2 min (Fig. 3 D). In contrast, the fast, directional movement of peroxisomes showed peak velocities that exceeded 0.2 μm/s (Fig. 3 B), mean average velocities of >0.05 μm/s (Fig. 3 C), and an average overall displacement of >0.5 μm after 2 min (Fig. 3 D). When viewed in time lapse, this type of movement was often saltatory (discontinuous and erratic), but highly directional in most instances. The rapid but discontinuous nature of the movement can be seen in the vector plots of individual representative peroxisomes (Fig. 3 D). The sporadic nature of the rapid movements led to a significant reduction in measured velocities when averaged over time (Fig. 3 B vs 3 C). The peroxisomes belonging to the two populations appeared to be interchangeable in that seemingly slow moving peroxisomes could spontaneously exhibit rapid directional movements, and vice versa (Fig. 3 D, peroxisome G). Thus, the movement behavior described is a general property of all peroxisomes with two different motility states, and not that of a fixed subset that only moved in a slow or fast manner.
Figure 3.
Time-lapse motion analysis of the dynamics of peroxisomes in living CV1 cells expressing GFP–PTS1. A representative single cell was selected, and single-frame confocal images were recorded at 6.4-s intervals for a total of 500 frames. 20 consecutive frames were chosen for motion analysis. (A) Three confocal images from the series are shown at time = 0 (frame 001), time = 64 s (frame 010), and time = 128 s (frame 020). Most peroxisomes displayed a slow, random movement within a limited space, leaving their overall distribution relatively unchanged after ∼2 min. Bar, 10 μm. (B) Plot of the peak velocities of 85 individual peroxisomes (∼50% of the cell population) observed over 20 consecutive frames revealed two distinct types of motion. The majority (95%) exhibited a localized, random type of Brownian motion with peak velocities of <0.2 μm/s. A smaller number (∼5%) exhibited a faster motion characterized by peak velocities of >0.2 μm/s and as great as 0.68 μm/s. (C) Plot of the mean average velocity of 85 individual peroxisomes over 20 consecutive frames also showed two distinct types of motion. The majority exhibited a localized, random type of Brownian motion with an average velocity of <0.05 μm/s, while the faster moving peroxisomes displayed an average velocity of >0.05 μm/s. (D) Vector diagram plot tracking the motion of eight representative individual peroxisomes (A–H) corresponding to those marked in C. A–E exhibited very little net displacement and were characteristic of the majority of peroxisomes. Peroxisome F exhibited sporadic motion, G was relatively stationary for several frames before exhibiting rapid directional motion, and H had a more continuous, rapid directional motion over the time course (1 pixel = 0.13 μm).
The slow-type random movement was independent of the energy status of the cell because it persisted in cells depleted of energy or killed by the addition of 0.05 M sodium azide (data not shown), indicating that it was probably due to Brownian motion. In contrast the fast, long-distance transport was completely abolished under these conditions.
Microtubule-based Spatial Organization of Peroxisomes
Depolymerization of the microtubules by nocodazole disrupted the intracellular organization and fast transport of the peroxisomal compartment in CV1 cells expressing GFP– PTS1, whereas treatment with the microtubule-stabilizing drug paclitaxel (taxol) had little effect on either intracellular organization or motion dynamics. After a 1.5–3-h incubation in culture media containing 20 μM paclitaxel, a sampling of a time series reveals a normal distribution of peroxisomes (Fig. 4 A, frames 001–020). Two groups of individual peroxisomes could be distinguished in cells treated with paclitaxel, and their behavior closely resembled that found in normal cells. In addition, the actin-depolymerizing cytochalasins did not affect the peroxisomal compartment, at least not in the first 3 h of the incubation in the presence of cytochalasin B or cytochalasin D (data not shown).
Figure 4.
Time-lapse analysis of the effects of paclitaxel and nocodazole on the dynamics of peroxisomes in living CV1 cells expressing GFP–PTS1. Representative single cells were selected and confocal images were taken at 6.4-s intervals for a total of 500 frames. 20 frames for each condition were chosen for motion analysis. (A) Three confocal images of a cell treated with paclitaxel at time = 0 (frame 001), time = 64 s (frame 010), and time = 128 s (frame 020) are shown. The overall distribution of peroxisomes was similar to that in normal cells. (B) Plot of the peak velocity of 85 individual peroxisomes (∼50% of the cell population) over 20 consecutive frames appeared similar to that found in normal cells (Fig. 3 B). (C) Plot of the mean average velocity of 85 individual peroxisomes over 20 consecutive frames appeared similar to that found in normal cells (Fig. 3 C). (D) Vector diagram plot tracking the motion of eight representative peroxisomes (A–H) corresponding to those marked in C. A–D were relatively motionless, while E and F exhibited sporadic motion and G and H exhibited more continuous rapid motion (1 pixel = 0.13 μm). The average velocities and mean changes in position were similar to those found in normal cells (Fig. 3 D). (E) Three frames from a series as in A of a cell treated with nocodazole showed an abnormal clustering of peroxisomes. (F) Plot of the peak velocity of 85 individual peroxisomes (∼50% of the cell population) over 20 consecutive frames appeared abnormal. All peroxisomes exhibited only a localized, random type of Brownian motion with none exceeding a peak velocity 0.2 μm/s. (G) Plot of the mean average velocity showed that only a few peroxisomes exceeded 0.05 μm/s. (H) Vector diagram plot tracking the motion of eight representative peroxisomes (A–H) corresponding to those marked in G. None of the peroxisomes exhibited rapid motion or displacements >0.5 μm over the time course (1 pixel = 0.13 μm).
In sharp contrast, incubation for 1.5–3-h culture media containing 20 μM nocodazole resulted in the formation of abnormal large peroxisomal clusters in the perinuclear region as well as in other regions of the cell (Fig. 4 E), indicating that microtubules are required for the proper distribution of peroxisomes throughout the cytoplasm. Comparable morphological changes were observed when the cells were incubated with demecolcine and vinblastine (data not shown). Several other organelles such as the ER (Terasaki and Fujiwara, 1986), mitochondria (Freed and Lebowitz, 1970), and lysosomes (Hollenbeck and Swanson, 1990) also cluster in the perinuclear region in the presence of microtubule-depolymerizing agents, but the basis for this is unknown. Analysis of the motion of individual peroxisomes revealed the loss of all peak velocities that exceeded 0.2 μm/s (Fig. 4 F). Very few were seen with an average velocity of >0.05 μm/s, and their net displacement after 2 min was <0.2 μm. These data suggest that the fast, but not the slow, movement of peroxisomes is microtubule based.
High Resolution Analysis of Peroxisomal Dynamics
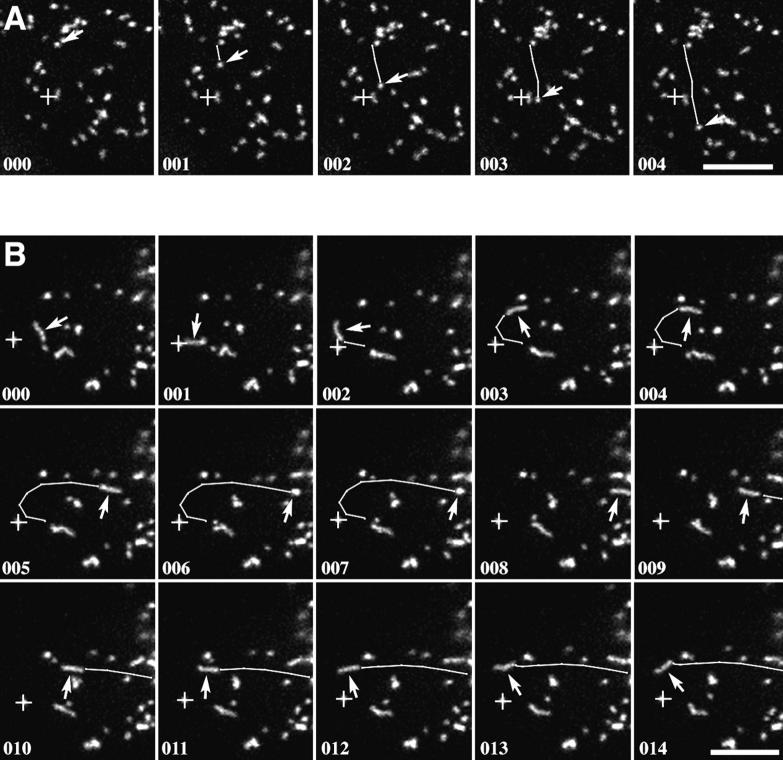
The behavior of individual peroxisomes was studied by time-lapse high resolution confocal microscopy. In Fig. 5 A, an individual peroxisome (arrow) was tracked over five consecutive frames taken at 6.4-s intervals. This peroxisome had an average velocity of 0.45 μm/s measured over 11.5 μm (frames 000–004) and a peak velocity of 0.75 μm/s (frames 003–004). This type of highly directional motion was characteristic of rapidly moving peroxisomes. Fig. 5 B shows an example of a peroxisome that started rapid motion in frame 000 and made a rapid looping motion until frame 006, at which time it rotated in the vertical axis and paused before moving back along the same path (frames 008–014). This type of bidirectional movement was rare and suggested that the peroxisome was moving along a preexisting track, most likely a microtubule. Similar bidirectional movement along microtubules has been documented for other organelles (Baumann and Murphy, 1995).
Figure 5.
High resolution confocal analysis of the movement of individual peroxisomes. (A) Five frames (000–004) taken at 6.4-s intervals showed the rapid unidirectional movement of a single peroxisome (arrows). The peak velocity of this peroxisome was 0.75 μm/s with an average velocity measured over 11.5 μm of 0.45 μm/s. (B) 15 frames (000–014) taken at 6.4-s intervals showed the bidirectional movement of a single elongated peroxisome. In frames 001–006, a peroxisome could be seen making a rapid looping motion and coming to a complete stop (arrows). In frames 006 and 007, the long axis of the peroxisome was oriented in the vertical dimension. In frames 007–014, the same peroxisome reversed course and returned along the same path with a similar velocity (arrows). Bars, 10 μm.
Association of Peroxisomes with Microtubules
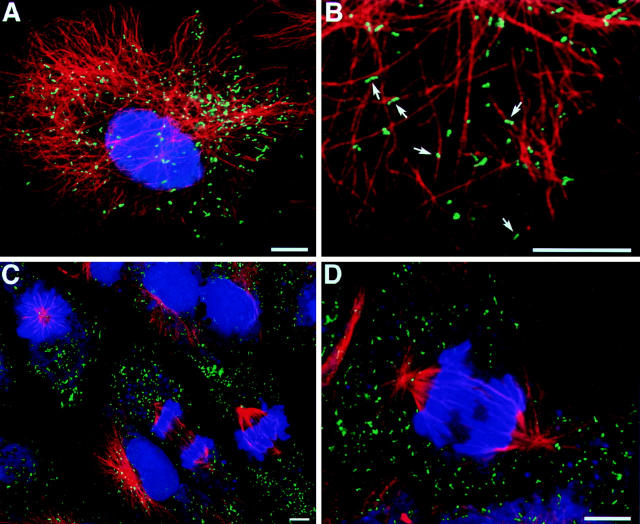
CV1 cells expressing GFP–PTS1 were fixed, permeabilized, and processed for immunofluorescence. Microtubules were specifically identified using anti-tubulin antibodies. High magnification imaging using a confocal laser scanning microscope revealed that peroxisomes, identified by the GFP, were found in many instances to be in close proximity to the microtubules (Fig. 6, A and B).
Figure 6.
Relationship of peroxisomes to microtubules and peroxisomal distribution during mitosis. Triple fluorescence confocal imaging of CV1 cells expressing GFP in peroxisomes (green), propidium iodide staining of the nucleus (pseudo-colored blue), and immunolabeling of microtubules using mouse anti–β-tubulin followed by goat anti–mouse IgG-CY5 (pseudo-colored red). (A) Low power image of the overall distribution of peroxisomes related to microtubules. (B) Higher power image of the same cell showing the close association of many of the individual peroxisomes (arrows) with individual microtubules. (C) Low power image of cells at metaphase and anaphase stages of mitosis. (D) Higher power image of an individual cell during anaphase stage of mitosis showing the random distribution of peroxisomes. Bars, 10 μm.
Behavior of the Peroxisomal Compartment during Mitosis
An intriguing question is how the peroxisomal compartment behaves during mitosis, knowing that gross cytoskeletal rearrangements take place during this event. CV1 cells expressing GFP–PTS1 were synchronized in the early S phase of the cell cycle using hydroxyurea. After the hydroxyurea block had been removed, the cells went through the S and G2 phase and entered mitosis after ∼9–11 h. At 9.5 and 10.5 h after the hydroxyurea release, the cells were fixed and processed for immunofluorescence. Multiple labeling and triple fluorescence imaging was performed after staining for DNA, microtubules, or microfilaments (data not shown) in relation to the GFP-labeled peroxisomes. The DNA staining by propidium iodide was used to identify the major steps in mitosis. Multiple cells in several mitotic stages were examined (Fig. 6, C and D). During mitosis, when cytoplasmic microtubules were absent, the peroxisomes appeared to be randomly distributed in the cytoplasm, and the majority were not associated with microtubules of the mitotic spindle. They seemed to distribute themselves more or less randomly by segregation of the bulk cytoplasm between the daughter and mother cells.
Discussion
In Vivo Labeling of Peroxisomes Using the GFP–PTS1 Fusion
The ability to target GFP–PTS1 to peroxisomes provides a vital stain for this compartment. We have used this to study the dynamics, spatial organization, and the association of peroxisomes with microtubules in CV1 cells. The fidelity of the targeting is >99% accurate, and the expression of GFP–PTS1 does not appear to perturb the growth of the cells or the behavior of the peroxisomes. Furthermore, during the visualization of the peroxisomes by confocal laser scanning microscopy, normal cellular processes such as membrane ruffling, migration, and mitosis proceeded normally. These results suggest that the motility behavior documented here for the GFP–PTS1–containing structures is characteristic of normal peroxisomes.
Microtubule-dependent and -independent Peroxisome Movements
Peroxisomes of CV1 cells exhibit two types of movement. The majority (∼95%) of the peroxisomes visualized exhibited slow, random movements that result in little net displacement. This movement was independent of energy, microtubules, and microfilaments, and it was deduced to be due to Brownian motion of the organelles in the cytosol. The other type of movement exhibited by ∼5% of the peroxisomes was fast and highly directional (with peak velocities as great as 0.75 μm/s) and could occur over many microns. The rapid movement was energy dependent and inhibited by drugs that depolymerized microtubules but not by drugs that stabilized them. The movement was independent of disruption of the actin cytoskeleton. The dependence of this movement on microtubules suggests that the microtubules provide the tracks for the movement of the peroxisomes. This is consistent with the notion that most organelle transport in mammalian cells is microtubule dependent, although actin-based movement of organelles such as mitochondria has also been documented (Morris and Hollenbeck, 1995; Simon et al., 1995). Evidence from different cell types demonstrated that movement of the Golgi apparatus (Ho et al., 1989), ER (Terasaki et al., 1986), lysosomes (Hollenbeck and Swanson, 1990), mitochondria (Morris and Hollenbeck, 1995), and intracellular transport vesicles (Allan, 1994) is microtubule dependent. It should be noted that the velocities measured were from peroxisomes that remained mainly in the plane of focus and ignored minor movements in the vertical dimension. The measured velocities correspond well to those reported for similar types of saltatory movements observed for mitochondria and organelles. Analyses by timelapse imaging in previous studies demonstrated that the velocities of the saltatory movements range from 0.5–5.0 μm/s, with an average speed of 1–2 μm/s (Freed and Lebowitz, 1970; Wang and Goldman, 1978; Schliwa, 1984). Furthermore, the distribution and directionality of saltations of other organelles has been observed to conform, as described above for peroxisomes, with the organization of the microtubule system (Freed and Lebowitz, 1970). Our results confirm those of an independent, recent study on microtubule-based peroxisome movement in CHO cells by Rapp et al. (1996).
One possible explanation for the movement of only a small percentage of the peroxisomes is that, in the presence of microtubules, as seen in Fig. 6 B, all the peroxisomes are tethered or docked with microtubules, and that only a subset of these then uses the microtubules as tracks for the fast movement at any given moment. The microtubules may serve to disperse the peroxisomes uniformly in the cell, and perhaps to restrict their movement. The activities and/or organelle binding of motor proteins are regulated by protein phosphorylation, and this may be the basis of the heterogenous motility behavior (Thaler and Haimo, 1996). Although very few peroxisomes move in the CV1 cells, it is premature to generalize this to all other cell types because we have not yet examined other tissue types to determine whether the peroxisome movement is more pronounced.
Two classes of microtubule motors, dynein and kinesin, are known. The former is involved in general intracellular movement toward the “minus,” slow growing end of microtubules, which are normally located at the microtubuleorganizing center. Kinesins involved in both minus and plus end–directed movement have been described (Vale et al., 1985; McDonald et al., 1990). At present, the nature of the motor protein involved in peroxisome movement is unknown. Cytoplasmic linker proteins (CLIPs) linking organelle membranes to microtubules have also been described (Rickard and Kreis, 1996), but none so far are involved in linking peroxisomes to microtubules.
It is also not clear at present why only a subset (∼5%) of peroxisomes moves rapidly at any given time, and what physiological relevance this directional movement might have. One possibility is that the movement may be in response to metabolic requirements in different microenvironments of the cell. In this context, it will be interesting to see whether growth of cells in peroxisome-requiring conditions, or microinjection of peroxisomal proteins into subregions of the cell, will stimulate the movement of the peroxisomes. An alternative possibility is that the organelle movement is related to peroxisome turnover, which is known to involve lysosomes.
Involvement of Microtubules in the Spatial Organization of Peroxisomes
In addition to the role that microtubules play in the movement of peroxisomes, the association of peroxisomes with microtubules (Fig. 6 B) and the clustering of the peroxisomes in the presence of microtubule-depolymerizing drugs (Fig. 4 E) support a role for microtubules in the spatial organization of this organelle (see also Schrader et al., 1996). Binding of purified peroxisomes to microtubules has been demonstrated in vitro, and the binding is sensitive to protease treatment of the peroxisomes, suggesting the involvement of some unidentified peroxisomal membrane protein/s in the association with the microtubules (Schrader et al., 1996).
Inheritance of the Peroxisomal Compartment during Mitosis
Previous studies of the modes of organelle inheritance in eukaryotic cells have revealed the use of two general strategies. The particular strategy exploited by a given organelle is dependent both on the cell type and the organism of origin (Warren and Wickner, 1996).
The first of these strategies is stochastic and relies on the presence of multiple, randomly distributed copies of the organelle in the cytoplasm of the dividing cell. The process of cytokinesis then distributes a certain number of the organelles to the two daughter cells (Birky, 1983). Mitochondria of many, but not all, organisms are segregated to daughter cells by this mechanism (Birky, 1983; Rizzuto et al., 1995).
The second strategy is exemplified by ordered inheritance in which the organelle is associated with some cytoskeletal or structural element (such as the mitotic spindle) and is partitioned along with the division and segregation of the underlying structure. Division of the vacuole in yeast cells follows this mode of inheritance (Weissman et al., 1987).
Our data suggest that the inheritance of peroxisomes in CV1 cells is stochastic rather than ordered. At the time of mitosis, when cytoplasmic microtubules were clearly absent (Fig. 6 C), many microperoxisomes were randomly distributed in the cytoplasm, and the majority were not associated with the microtubules in the mitotic spindle. As the cell divided, the peroxisomes were also distributed randomly, along with the bulk cytoplasm, to the daughter cells.
In summary, the use of peroxisome-targeted GFP fusions has provided a vital stain for the peroxisomal compartment and allowed, for the first time, an analysis of the dynamics of the organelle in living cells and during cell division. It should also be possible to use these derivatives to study other aspects of peroxisomes, such as the proliferation and degradation of the organelle.
Acknowledgments
This work was supported by grants from the National Institutes of Health to S. Subramani (DK41737) and to M.H. Ellisman (RR04050 and NS14718) and from the National Science Foundation to S. Subramani (MCB 9316018); postdoctoral fellowships from the Human Frontier Science Program Organization and the American Heart Association to E.A.C. Wiemer; and a postdoctoral fellowship (ALTF 402-1993) from the European Molecular Biology Organization to T. Wenzel.
Abbreviations used in this paper
- CMV
cytomegalovirus
- CV1
African green monkey kidney cells
- GFP
green fluorescent protein
- PMP
peroxisomal membrane protein
- PTS
peroxisomal targeting signal
Footnotes
The authors express their gratitude to Dr. Roger Tsien for making the GFP mutant (S65T) construct available for our studies, to Dr. Jon Singer for the use of his fluorescence microscope, and to Steve Lamont for technical assistance.
Address all correspondence to Dr. Suresh Subramani, Department of Biology, University of California at San Diego, 9500 Gilman Drive, La Jolla, CA 92039-0322. Tel.: (619) 534-2327. Fax: (619) 534-0053. e-mail: ssubramani@ucsd.edu
References
- Allan V. Organelle movement. Dynactin: portrait of a dynein regulator. Curr Biol. 1994;4:1000–1002. doi: 10.1016/s0960-9822(00)00225-6. [DOI] [PubMed] [Google Scholar]
- Baumann O, Murphy DB. Microtubule-associated movement of mitochondria and small particles in Acanthamoeba castellanii. . Cell Motil Cytoskeleton. 1995;32:305–317. doi: 10.1002/cm.970320407. [DOI] [PubMed] [Google Scholar]
- Birky CW. The partitioning of cytoplasmic organelles at cell division. Int Rev Cytol. 1983;15:49–89. doi: 10.1016/b978-0-12-364376-6.50009-0. [DOI] [PubMed] [Google Scholar]
- Cole NB, Lippincott-Schwartz J. Organization of organelles and membrane traffic by microtubules. Curr Opin Cell Biol. 1995;7:55–64. doi: 10.1016/0955-0674(95)80045-x. [DOI] [PubMed] [Google Scholar]
- Cubitt AB, Heim R, Adams SR, Boyd AE, Gross LA, Tsien RY. Understanding, improving and using green fluorescent proteins. Trends Biochem Sci. 1995;20:448–455. doi: 10.1016/s0968-0004(00)89099-4. [DOI] [PubMed] [Google Scholar]
- Erdmann R. The peroxisomal targeting signal of 3-oxoacyl-CoA thiolase from Saccharomyces cerevisiae. . Yeast. 1994;10:935–944. doi: 10.1002/yea.320100708. [DOI] [PubMed] [Google Scholar]
- Faber KN, Haima P, Gietl C, Harder W, AB G, Veenhuis M. The methylotrophic yeast Hansenula polymorphacontains an inducible import pathway for peroxisomal matrix proteins with an N-terminal targeting signal (PTS2 proteins) Proc Natl Acad Sci USA. 1994;91:12985–12989. doi: 10.1073/pnas.91.26.12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed JJ, Lebowitz MM. The association of a class of saltatory movements with microtubules in cultured cells. J Cell Biol. 1970;45:334–354. doi: 10.1083/jcb.45.2.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietl C. Glyoxysomal malate dehydrogenase from watermelon is synthesized with an amino-terminal transit peptide. Proc Natl Acad Sci USA. 1990;87:5773–5777. doi: 10.1073/pnas.87.15.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Andrews DW, Subramani S, Rachubinski R. Mutagenesis of the amino targeting signal of Saccharomyces cerevisiae3-ketoacyl-CoA thiolase reveals conserved amino acids required for import into peroxisomes in vivo. J Biol Chem. 1994;269:7558–7563. [PubMed] [Google Scholar]
- Gorgas, K. 1987. Morphogenesis of peroxisomes in lipid synthesizing epithelia. In Peroxisomes in Biology and Medicine. H.D. Fahimi and H. Sies, editor. Springer-Verlag, Heidelberg. 3–17.
- Gould SJ, Keller GA, Hosken N, Wilkinson J, Subramani S. A conserved tripeptide sorts proteins to peroxisomes. J Cell Biol. 1989;108:1657–1664. doi: 10.1083/jcb.108.5.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser JE. Mechanisms behind the organization of membranous organelles in cells. Curr Opin Cell Biol. 1989;1:98–102. doi: 10.1016/s0955-0674(89)80044-4. [DOI] [PubMed] [Google Scholar]
- Ho WC, Allan VJ, van Meer G, Berger EG, Kreis TE. Reclustering of scattered Golgi elements occurs along microtubules. Eur J Cell Biol. 1989;48:250–263. [PubMed] [Google Scholar]
- Hollenbeck PJ, Swanson JA. Radial extension of macrophage tubular lysosomes supported by kinesin. Nature (Lond) 1990;346:864–866. doi: 10.1038/346864a0. [DOI] [PubMed] [Google Scholar]
- Kamijo K, Taketani S, Yokota S, Osumi T, Hashimoto T. The 70kDa peroxisomal membrane protein is a member of the Mdr (P-glycoprotein)-related ATP-binding protein superfamily. J Biol Chem. 1990;265:4534–4540. [PubMed] [Google Scholar]
- Keller G-A, Gould S, DeLuca M, Subramani S. Firefly luciferase is targeted to peroxisomes in mammalian cells. Proc Natl Acad Sci USA. 1987;84:3264–3268. doi: 10.1073/pnas.84.10.3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller G-A, Krisans S, Gould SJ, Sommer JM, Wang CC, Schliebs W, Kunau W, Brody S, Subramani S. Evolutionary conservation of a microbody targeting signal that targets proteins to peroxisomes, glyoxysomes, and glycosomes. J Cell Biol. 1991;114:893–904. doi: 10.1083/jcb.114.5.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly RB. Microtubules, membrane traffic, and cell organization. Cell. 1990;61:5–7. doi: 10.1016/0092-8674(90)90206-t. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Lazarow, P.B., and H.W. Moser. 1994. Disorders of peroxisome biogenesis. In The Metabolic Basis of Inherited Disease. Seventh edition. C.R. Scriver, A.L. Beaudet, W.S. Sly, and A.D. Valle, editors. McGraw-Hill Inc., New York. 2287–2324.
- McDonald HB, Stewart RJ, Goldstein LS. The kinesin-like ncd protein of Drosophilais a minus end-directed microtubule motor. Cell. 1990;63:1159–1165. doi: 10.1016/0092-8674(90)90412-8. [DOI] [PubMed] [Google Scholar]
- McNew JA, Goodman JM. The targeting and assembly of peroxisomal proteins: some old rules do not apply. Trends Biochem Sci. 1996;21:54–58. [PubMed] [Google Scholar]
- Monosov E, Wenzel T, Lüers G, Heyman JA, Subramani S. Labeling of peroxisomes with green fluorescent protein in living Pichia pastoriscells. J Histochem Cytochem. 1996;44:581–589. doi: 10.1177/44.6.8666743. [DOI] [PubMed] [Google Scholar]
- Morris RL, Hollenbeck PJ. Axonal transport of mitochondria along microtubules and F-actin in living vertebrate neurons. J Cell Biol. 1995;131:1315–1326. doi: 10.1083/jcb.131.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi T, Tsukamoto T, Hata S, Yokota S, Miura S, Fujiki Y, Hijikata M, Miyazawa S, Hashimoto T. Amino-terminal presequence of the precursor of peroxisomal 3-ketoacyl-CoA thiolase is a cleavable signal peptide for peroxisomal targeting. Biochem Biophys Res Commun. 1991;181:947–954. doi: 10.1016/0006-291x(91)92028-i. [DOI] [PubMed] [Google Scholar]
- Rapp S, Saffrich R, Anton M, Jäkle U, Ansorge W, Gorgas K, Just W. Microtubule-based peroxisome movement. J Cell Sci. 1996;109:837–849. doi: 10.1242/jcs.109.4.837. [DOI] [PubMed] [Google Scholar]
- Rickard JE, Kreis TE. CLIPs for organelle-microtubule interactions. Trends Cell Biol. 1996;6:1278–1283. doi: 10.1016/0962-8924(96)10017-9. [DOI] [PubMed] [Google Scholar]
- Rizzuto R, Brini M, Pizzo P, Murgia M, Pozzan T. Chimeric green fluorescent protein as a tool for visualizing subcellular organelles in living cells. Curr Biol. 1995;5:635–642. doi: 10.1016/s0960-9822(95)00128-x. [DOI] [PubMed] [Google Scholar]
- Schliwa, M. 1984. Intracellular organelle transport. In Cell and Muscle Motility. J.W. Shay, editor. Plenum Press, New York. 1–82. [DOI] [PubMed]
- Schrader M, Burkhardt JK, Baumgart E, Lüers G, Spring H, Völkl A, Fahimi HD. Interaction of microtubules with peroxisomes. Tubular and spherical peroxisomes in HepG2 cells and their alterations induced by microtubule-active drugs. Eur J Cell Biol. 1996;69:24–35. [PubMed] [Google Scholar]
- Simon VR, Swayne TC, Pon LA. Actin-dependent mitochondrial motility in mitotic yeast and cell-free systems: identification of a motor activity on the mitochondrial surface. J Cell Biol. 1995;130:345–354. doi: 10.1083/jcb.130.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto GE, Young SJ, Martone ME, Deerinck TJ, Lamont S, Carragher BO, Hama K, Ellisman MH. Serial section electron tomography: a method for three-dimensional reconstruction of large structures. Neuroimage. 1993;1:230–243. doi: 10.1006/nimg.1994.1008. [DOI] [PubMed] [Google Scholar]
- Subramani S. Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- Swinkels BW, Gould SJ, Bodnar AG, Rachubinski RA, Subramani S. A novel, cleavable peroxisomal targeting signal at the amino-terminus of the rat 3-ketoacyl-CoA thiolase. EMBO (Eur Mol Biol Organ) J. 1991;10:3255–3262. doi: 10.1002/j.1460-2075.1991.tb04889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaki M, Chen LB, Fujiwara K. Microtubules and the endoplasmic reticulum are highly interdependent structures. J Cell Biol. 1986;103:1557–1568. doi: 10.1083/jcb.103.4.1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaler CD, Haimo LT. Microtubules and microtubule motors: mechanisms of regulation. Int Rev Cytol. 1996;164:269–327. doi: 10.1016/s0074-7696(08)62388-8. [DOI] [PubMed] [Google Scholar]
- Vale RD, Reese TS, Sheetz MP. Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell. 1985;42:39–50. doi: 10.1016/s0092-8674(85)80099-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch H, Schutgens RBH, Wanders RJA, Tager JM. Biochemistry of peroxisomes. Annu Rev Biochem. 1992;61:157–197. doi: 10.1146/annurev.bi.61.070192.001105. [DOI] [PubMed] [Google Scholar]
- Wang E, Goldman RD. Functions of cytoplasmic fibers in intracellular movements in BHK-21 cells. J Cell Biol. 1978;79:708–726. doi: 10.1083/jcb.79.3.708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G, Wickner W. Organelle inheritance. Cell. 1996;84:395–400. doi: 10.1016/s0092-8674(00)81284-2. [DOI] [PubMed] [Google Scholar]
- Weisman LS, Emr SD, Wickner WT. Mutants of Saccharomyces cerevisiaethat block intervacuole vesicular traffic and vacuole division and segregation. Proc Natl Acad Sci USA. 1990;87:1076–1080. doi: 10.1073/pnas.87.3.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemer EAC, Lüers G, Faber KN, Wenzel T, Veenhuis M, Subramani S. Isolation and characterization of Pas2p, a peroxisomal membrane protein essential for peroxisome biogenesis in the methylotrophic yeast Pichia pastoris. . J Biol Chem. 1996;271:18973–18980. doi: 10.1074/jbc.271.31.18973. [DOI] [PubMed] [Google Scholar]
- Yamamoto K, Fahimi HD. Biogenesis of peroxisomes in regenerating rat liver. I. Sequential changes of catalase and urate oxidase detected by ultrastructural cytochemistry. Eur J Cell Biol. 1987;43:293–300. [PubMed] [Google Scholar]