Abstract
The replication licensing factor (RLF) is an essential initiation factor that is involved in preventing re-replication of chromosomal DNA in a single cell cycle. In Xenopus egg extracts, it can be separated into two components: RLF-M, a complex of MCM/P1 polypeptides, and RLF-B, which is currently unpurified. In this paper we investigate variations in RLF activity throughout the cell cycle. Total RLF activity is low in metaphase, due to a lack of RLF-B activity and the presence of an RLF inhibitor. RLF-B is rapidly activated on exit from metaphase, and then declines during interphase. The RLF inhibitor present in metaphase extracts is dependent on the activity of cyclin-dependent kinases (Cdks). Affinity depletion of Cdks from metaphase extracts removed the RLF inhibitor, while Cdc2/cyclin B directly inhibited RLF activity. In metaphase extracts treated with the protein kinase inhibitor 6-dimethylaminopurine (6-DMAP), both cyclin B and the RLF inhibitor were stabilized although the extracts morphologically entered interphase. These results are consistent with studies in other organisms that invoke a key role for Cdks in preventing re-replication of DNA in a single cell cycle.
During each S phase of the eukaryotic cell division cycle, the entire genome is precisely duplicated. To achieve this, many hundreds or thousands of replication origins must each fire once and only once in each S phase. Studies using cell-free systems derived from Xenopus eggs have revealed that re-replication of chromosomal DNA in a single cell cycle is prevented by the action of two distinct replication signals (Blow and Laskey, 1988; Chong et al., 1996). The first signal, replication licensing factor (RLF)1, “licenses” chromosomal DNA by putting replication origins into an initiation-competent state. The second signal, S-phase promoting factor (SPF), induces licensed origins to initiate and, in doing so, removes the licence. To achieve precise duplication of chromosomal DNA, the licensing and initiation signals must act on the DNA sequentially, and never at the same time. This is achieved in two different ways. First, active RLF cannot cross the nuclear envelope, so it can only license DNA when the nuclear envelope has broken down in mitosis (Blow and Laskey, 1988; Leno et al., 1992; Coverley et al., 1993; Blow, 1993); in contrast, SPF can only initiate DNA replication on licensed DNA within an intact nucleus (Blow and Watson, 1987; Newport, 1987; Sheehan et al., 1988; Blow and Sleeman, 1990). Secondly, both activities are periodic in the cell cycle: RLF is abruptly activated after the metaphase–anaphase transition and decays during interphase (Blow, 1993), while SPF activity can only be detected during interphase (Blow and Nurse, 1990). The spatial separation of RLF and SPF activities is thereby enhanced by a temporal regulation.
The activation of RLF that occurs at the metaphase– anaphase transition in Xenopus can be blocked by certain protein kinase inhibitors, such as 6-dimethylaminopurine (6-DMAP), that are known to inhibit cyclin-dependent kinases (Blow, 1993; Kubota and Takisawa, 1993; Vesely et al., 1994). The licensing system has been subjected to biochemical and immunological analysis using extracts treated with these kinase inhibitors (Chong et al., 1995; Kubota et al., 1995). RLF activity can be separated chromatographically into two essential components, RLF-M and RLF-B, both of which are required for licensing (Chong et al., 1995). RLF-M has been purified to apparent homogeneity, and it consists of a complex of all six currently identified members of the Xenopus MCM/P1 family, XMcm2– XMcm7 (Chong et al., 1995, 1996; Thömmes, P., Y. Kubota, H. Takisawa, and J.J. Blow, manuscript submitted for publication). Anti-XMcm3 antibodies coprecipitated all six Xenopus MCM/P1 proteins in a complex closely resembling the RLF-M complex (Kubota et al., 1995; Madine et al., 1995a ,b; Thömmes, P., Y. Kubota, H. Takisawa, and J.J. Blow, manuscript submitted for publication) and depleted the extracts of RLF-M activity (Chong et al., 1995).
MCM/P1 genes were originally identified in Saccharomyces cerevisiae in a screen for mutants unable to efficiently initiate replication at particular replication origins (Maine et al., 1984), due to a failure of the initiation process (Maiti and Sinha, 1992; Yan et al., 1993). The proteins show cell cycle–dependent changes in subnuclear localization, being observed within the nucleus only during late mitosis and G1 (Hennessy et al., 1990; Yan et al., 1993; Dalton and Whitbread, 1995). Homologous MCM/P1 genes have been identified in a wide range of eukaryotes including insects, plants, amphibians, and mammals, where they fall into six related groups designated MCM2–MCM7 (Chong et al., 1996; Kearsey et al., 1996). Consistent with their role in yeast, MCM/P1 proteins in higher eukaryotes are required for DNA replication (Kimura et al., 1994; Todorov et al., 1994; Chong et al., 1995; Kubota et al., 1995; Madine et al., 1995a ; Treisman et al., 1995) and behave in accordance with their role as a central component of the licensing system. They associate tightly with chromatin during late mitosis and G1, but are removed during replication (Kimura et al., 1994; Chong et al., 1995; Kubota et al., 1995; Madine et al., 1995a ,b; Todorov et al., 1995; Coué et al., 1996). Reassociation of XMcm3 with chromatin, which is required before an additional round of replication can take place, only occurs after permeabilization or breakdown of the nuclear envelope (Chong et al., 1995; Kubota et al., 1995; Madine et al., 1995a ,b). The licensing of chromatin by RLF-M and RLF-B in Xenopus requires the presence of the Xenopus origin recognition complex (ORC) on the chromatin (Rowles et al., 1996).
In this paper we analyze the cell cycle control of RLF activity in Xenopus egg extracts. Total RLF activity is sharply periodic in the cell cycle. An RLF inhibitor is present in metaphase extracts and appears to be directly dependent on the activity of cyclin-dependent kinases. When the inhibitor is removed or diluted away, RLF-M, but not RLF-B, activity can be detected. RLF-B, but not RLF-M, activity is unstable in interphase and decays over a period of 60–120 min. However, the decay of RLF-B activity in interphase is not associated with activation of an RLF inhibitor and does not depend on cyclin-dependent kinases.
Materials and Methods
Preparation and Use of Xenopus Egg Extract
Metaphase-arrested and interphase Xenopus extract was prepared as described (Blow, 1993) and stored in 20-μl aliquots in liquid nitrogen. Just before use, extract was thawed at room temperature and supplemented with 250 μg/ml cycloheximide, 25 mM phosphocreatine, and 15 μg/ml creatine phosphokinase. Metaphase-arrested extract was released into interphase by the addition of 0.3 mM CaCl2. To block RLF activation, metaphase-arrested extracts were supplemented with 3 mM 6-DMAP and mixed thoroughly before CaCl2 addition (6-DMAP–treated extract). For replication assays, extract was supplemented with 1/200 vol 10 mCi/ml [α32P]dATP (Amersham Intl., Little Chalfont, UK). Extract incubations were performed at 23°C.
Suc1 depletion was performed essentially as described (Blow and Nurse, 1990). Metaphase extract was mixed with an equal volume of p13suc1-coupled Sepharose (10 μg Suc1/ml beads, washed in LFB1 [40 mM Hepes KOH, pH 8.0, 20 mM K2HPO4/KH2PO4, pH 8.0, 2 mM MgCl2; 1 mM EGTA, 2 mM DTT, 10% sucrose, 1 μg/ml each of leupeptin, pepstatin, and aprotinin] and incubated on a rotating wheel at 4°C for 60 min. The supernatant was recovered by spinning the bead/extract slurry past a glass ball wedged in a sawn-off yellow Gilson tip in an Eppendorf tube.
Preparation of DNA Templates
Demembranated Xenopus sperm nuclei were prepared by lysolecithin treatment as described (Blow and Laskey, 1986). They were assembled into chromatin in 6–DMAP-treated extract, to form “6-DMAP chromatin” as follows (Chong et al., 1995, 1997): 80 μl of a metaphase-arrested extract was supplemented with phosphocreatine, creatine phosphokinase, 3 mM 6-DMAP, and CaCl2 as described above, and then supplemented with 1.3 × 106 demembranated sperm nuclei (∼50 ng DNA per μl extract). The mixture was incubated for 12 min at 23°C, and then diluted in 1 ml NIB (50 mM KCl; 50 mM Hepes KOH, pH 7.6; 5 mM MgCl2; 2 mM DTT, 0.5 mM spermidine 3HCl; 0.15 mM spermine 4HCl; 1 μg/ml each leupeptin, aprotinin, and pepstatin) and underlayered with 100 μl NIB supplemented with 15% sucrose. The chromatin was isolated by centrifugation in a swing-out rotor at 6,200 g for 5 min at 4°C. The pellet was resuspended in 45 μl NIB (to a final concentration of 80 ng DNA/per μl) and frozen in 5-μl aliquots in liquid nitrogen.
Preparation of RLF-B and RLF-M
Polyethylene glycol (PEG)–fractionated RLF-B and RLF-M were prepared as described (Chong et al., 1995, 1997). Eggs were dejellied in 2% cysteine solution, washed three times in Barth solution (88 mM NaCl, 2 mM KCl, 1 mM MgCl2, 15 mM Tris HCl, pH 7.4, 0.5 mM CaCl2), and then incubated for 5–10 min in 100 ml Barth plus 20 μl 10 μg/ml calcium ionophore A23187. The activated eggs were then washed three times in Barth solution at 23°C and three times in EB (50 mM KCl, 50 mM Hepes KOH, pH 7.6, 5 mM MgCl2, 2 mM DTT) at 4°C. All additional steps were performed at 4°C or on ice. Eggs were packed by centrifuging at 800 g in a swing-out rotor for 1 min. Excess buffer and sick eggs, which float to the surface during this treatment, were then removed. Packed eggs were spincrushed by centrifugation at 10,000 g for 10 min in a swing-out rotor. The cytoplasmic layer was taken, supplemented to a final concentration of 10 μg/ml cytochalasin B and 15% EDB-S (50 mM KCl, 50 mM Hepes KOH, pH 7.6, 10% sucrose, 2 mM DTT, 0.4 mM MgCl2, 0.4 mM EGTA, 1 μg/ml each of pepstatin, leupeptin, and aprotinin), and then diluted with 4 vol of LFB1 containing 50 mM KCl and 0.5 mM freshly added PMSF. This diluted extract was spun at 60,000 g for 20 min at 4°C in a swing-out rotor. The supernatant (“licensing factor extract”) was decanted, drop frozen in liquid nitrogen, and stored at −70°C. Since it is fivefold diluted over neat extract, licensing factor extract is designated a concentration of 0.2×.
Differential PEG precipitation was performed as follows (Chong et al., 1995, 1997): licensing factor extract was supplemented with 0.075 vol of a 50% PEG solution (50% PEG 6000 [BDH Chemicals Ltd., Poole, UK] in LFB1) to give a final concentration of 3.5% PEG. Extract was incubated on ice for 30 min, and then centrifuged at 10,000 g for 10 min in a fixed- angle rotor. The supernatant was decanted, and residual PEG solution was removed after an additional brief spin. The pellet was then resuspended in LFB1 at 1/25 of the original volume of licensing factor extract to give 5× concentrated RLF-B, and was stored in aliquots at −70°C. The supernatant was supplemented with an additional 0.11 vol of a 50% PEG solution, giving a final concentration of 9% PEG, incubated on ice, and pelleted as before. The pellet was resuspended in LFB1 at 1/25 of the original volume, to give 5× concentrated RLF-M, and was stored in aliquots at −70°C.
RLF-M was further fractionated on Q Sepharose to produce RLFMinter as described (Chong et al., 1995, 1997). RLF-M produced by PEG precipitation was diluted to 1× in LFB1 plus 100 mM KCl and was adsorbed in batch for 30 min at 4°C onto an equal volume of loose Q Sepharose (Pharmacia, Uppsala, Sweden) preequilibrated in LFB1 plus 100 mM KCl. The media was then packed into a column, washed with LFB1 plus 100 mM KCl, and eluted as a step in LFB1 plus 325 mM KCl. Eluted material was reprecipitated with PEG and resuspended in LFB1. RLF-Mmeta was produced in the same way as RLF-Minter, except that the starting material was metaphase-arrested extract.
For the maturation promoting factor (MPF) inhibition experiment (see Fig. 4 b), RLF-M from the Q Sepharose step was further purified by phenyl Sepharose chromatography and gel filtration, as previously described (Chong et al., 1995, 1997). For this experiment, the RLF-B preparation was also further purified, according to the protocol previously described for Xenopus ORC (Rowles et al., 1996; Chong et al., 1997). Briefly, PEGfractionated RLF-B was bound to phosphocellulose equilibrated in LFB1 plus 150 mM KCl, and then step-eluted in LFB1 plus 500 mM KCl. Eluted material, containing both RLF-B and Xenopus ORC activity, was precipitated with 40% ammonium sulfate, and the pellet was resuspended in LFB1 for assay.
Figure 4.
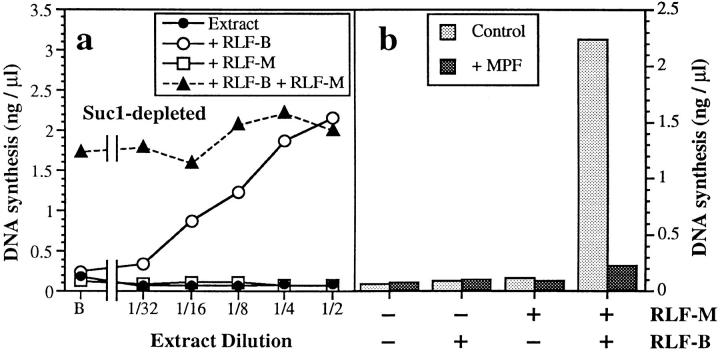
Evidence for a Cdk-dependent RLF inhibitor. (a) Metaphase-arrested extracts were depleted of Cdks with an equal volume of p13suc1-coupled Sepharose. Depleted extract was subjected to serial dilution, and then mixed with an equal volume of either buffer (○̶ ), crude RLF-B (●̶ ), crude RLF-M (□̶ ), or a mixture of crude RLF-B and crude RLF-M (▲̶ ). B, buffer in place of diluted extract. Unlicensed chromatin was incubated in the mixture (12 ng DNA/μl) for 15 min at 23°C to allow licensing, and then was transferred to 2.5 vol 6-DMAP–treated extract containing [α32P]dATP. The total DNA synthesized after further incubation for 90 min at 23°C was measured. (b) Preincubations were performed for 15 min with different combinations of partially purified RLF-B, purified RLF-M, and Cdc2/cyclin B (MPF). Unlicensed chromatin was then added, and the incubation was continued for an additional 15 min to allow licensing to occur. Samples were then transferred to 6-DMAP–treated extract containing [α32P]dATP, and the total DNA synthesized after further incubation for 90 min at 23°C was measured.
Licensing Assay
RLF was assayed in a two-step protocol as described (Chong et al., 1995, 1997). 2-μl samples to be assayed were incubated with 0.3 μl 6-DMAP chromatin (80 ng DNA/μl) for 15 min for the “licensing reaction.” In assays for individual components, the 2-μl samples were typically composed of 1-μl fraction of interest, plus 1-μl PEG-cut RLF-B and/or RLF-M diluted to 0.5× in LFB1, plus 50 mM KCl and 2.5 mM Mg-ATP. For extract dilution experiments, dilutions were performed with LFB1 plus 50 mM KCl. Once the licensing reaction was finished, 5.7 μl 6-DMAP–treated extract containing cycloheximide, phosphocreatine, creatine phosphokinase, 6-DMAP, CaCl2 and [α32P]dATP were added. The incubation was continued for an additional 90 min at 23°C (the “replication reaction”). Replication reactions were terminated by the addition of 150 μl Stop-C (20 mM Tris HCl, pH 7.5, 5 mM EDTA, 0.5% SDS) plus 0.2 μg/ml proteinase K and incubated at 37°C for 15 min. TCA precipitation was performed as described (Blow and Laskey, 1986; Chong et al., 1997), and the total DNA synthesized (in ng DNA/μl) was calculated, assuming 50 μM endogenous dATP in the extract (Blow and Laskey, 1986). Bromodeoxyuridine triphosphate (BrdUTP) density substitution was performed as described (Blow and Laskey, 1986).
Alkaline Agarose Gel Electrophoresis
Reactions for agarose gel electrophoresis were stopped by digesting with Stop-N (20 mM Tris-Cl, 200 mM NaCl, 5 mM EDTA, 0.5% SDS, pH 8.0) containing 2 μg/ml RNase A and 200 μg/ml Proteinase K for 30 min at 37°C. DNA was extracted sequentially with phenol, phenol-chloroform, and chloroform, and then precipitated with ethanol. Pellets were resuspended in alkali gel loading buffer (50 mM NaOH, 1 mM EDTA, 1.25% Ficoll, 0.0125% bromocresol green). Alkaline gels were prepared by adding the required amount of agarose to 50 mM NaCl and 1 mM EDTA. Once set, the gels were allowed to equilibrate for 1 h in alkaline gel running buffer (30 mM NaOH, 1 mM EDTA), and then were run at 2 V/cm. After electrophoresis was complete, gels were fixed in 7% TCA, dried, and autoradiographed.
Cip1 and Cdc2/Cyclin B
Glutathione-S-transferase (GST)–tagged Cip1 was prepared as described (Strausfeld et al., 1994). It was added to Xenopus extracts at 150 nM, an amount sufficient to inhibit DNA replication by >95% (Strausfeld et al., 1994). Purified Cdc2/cyclin B was purchased from Upstate Biotechnology Inc.(Lake Placid, NY) and was used in the licensing reaction at a final concentration of 1.25 ng/μl.
Antibodies and Western Blotting
For Western blotting, chromatin was isolated from extract as described for the preparation of 6-DMAP chromatin, except that the NIB buffer was supplemented with 0.1% NP-40 (Chong et al., 1995, 1997). Chromatin pellets were resuspended in gel loading buffer, electrophoresed, and Western blotted by standard techniques. Anti-XMcm3 rabbit polyclonal antibodies were raised against recombinant GST-tagged XMcm3 (amino acids 423– 798) produced from the construct described by Madine et al. (1995a).
35S Labeling and Autoradiography
200 μCi [35S]methionine (Amersham Intl.) was dried under vacuum and redissolved in 40 μl metaphase-arrested extract supplemented with phosphocreatine and creatine phosphokinase. The extract was incubated for 30 min at 23°C to label newly synthesized proteins, before the addition of 250 μg/ml cycloheximide. Extract was then supplemented with various concentrations of 6-DMAP and then with 0.3 mM CaCl2, before being incubated for an additional 30 min at 23°C to allow cyclin degradation to occur. Cyclin–cyclin-dependent kinase (Cdk) complexes were then collected on p13suc1 beads at 4°C for 45 min. The beads were washed several times, and then were resuspended in sample loading buffer for electrophoresis and autoradiography according to standard techniques.
Results
Quantification of RLF-M Required for DNA Replication
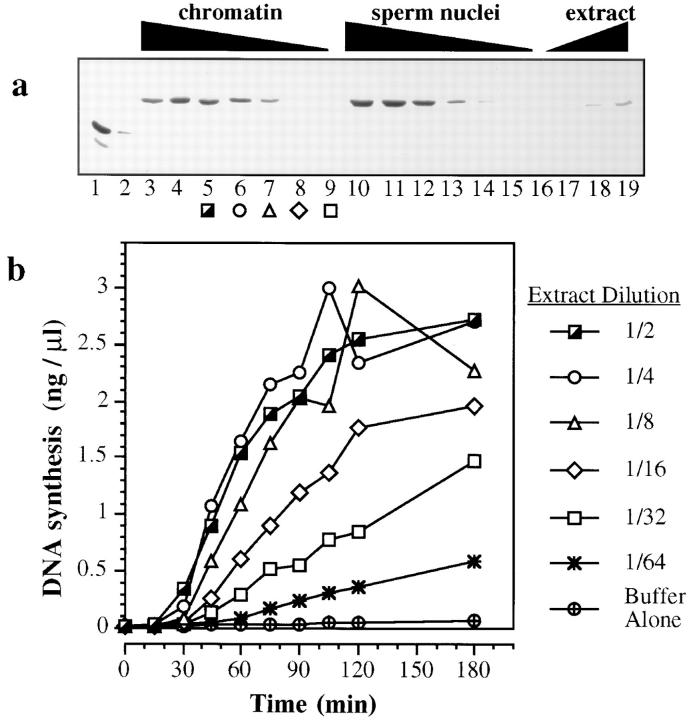
RLF-M consists of a complex of MCM/P1 proteins, including XMcm3 (Chong et al., 1995). To correlate the amount of RLF-M activity in Xenopus extracts with the amount of XMcm3 protein, we performed quantitative Western blotting on extracts and on chromatin assembled in these extracts (Fig. 1 a). An estimate of 75–150 μg XMcm3 protein per ml Xenopus extract (0.75–1.5 μM) was provided by a number of duplicate blots (e.g., Fig. 1 a, lanes 1 and 2 and 17–19). Fig. 1 a also quantifies the association of XMcm3 with chromatin. Demembranated Xenopus sperm nuclei (Fig. 1, lanes 10–16) or chromatin previously assembled in 6-DMAP–treated extract (6-DMAP chromatin; Fig. 1, lanes 3–9) were briefly incubated with serial dilutions of Xenopus egg extract. Chromatin was then isolated by centrifugation through sucrose and immunoblotted for XMcm3. As more extract was incubated with a fixed quantity of chromatin, the amount of chromatin-associated XMcm3 increased. Only at the highest concentrations of DNA (6–12 ng DNA per μl extract) was there any sign of the chromatin becoming saturated with XMcm3. Comparison with the recombinant XMcm3 standard (Fig. 1, lanes 1 and 2) suggests that the extract can assemble in excess of 30 ng XMcm3 onto 300 ng DNA, equivalent to one XMcm3 molecule bound to every 1.5 kb of DNA on average. Previous studies have estimated that the replicon size of the early Xenopus embryo is 10–20 kb (Blow and Watson, 1987; Mahbubani et al., 1992; Hyrien and Méchali, 1993), suggesting that there may be >10 XMcm3 molecules bound to each replicon.
Figure 1.
Effect of serial dilution on XMcm3 assembled onto chromatin and rate of replication. Interphase Xenopus extract was subjected to serial dilution, and 30-μl aliquots (or 60-μl for a, lanes 3 and 10) were incubated with equal quantities (360 ng DNA) of unlicensed chromatin (a, lanes 3–9; b) or sperm nuclei (a, lanes 10–16) for 15 min at 23°C. (a) Equal quantities of chromatin (∼300 ng DNA) were isolated by centrifugation through sucrose, run on a 7.5% polyacrylamide gel, and immunoblotted with an anti-XMcm3 antibody. Dilutions were: (lanes 3 and 10) undiluted (60 μl); (lanes 4 and 11) undiluted (30 μl); (lanes 5 and 12) 1/2; (lanes 6 and 13) 1/4; (lanes 7 and 14) 1/8; (lanes 8 and 15) 1/16; (lanes 9 and 16) 1/32. Samples of recombinant GST-tagged XMcm3 (lane 1, 40 ng; lane 2, 20 ng) and interphase Xenopus egg extract (lane 17, 0.125 μl; lane 18, 0.25 μl; lane 19, 0.5 μl) were blotted in parallel. (b) Samples were transferred to 6-DMAP– treated extract containing [α32P]dATP. The total DNA synthesized at different times during an incubation at 23°C was measured. Samples corresponding to the blots in a are indicated with identical symbols.
Fig. 1 b examines the RLF activity of chromatin licensed with different dilutions of interphase extract. Chromatin was licensed with serial dilutions of extract, and then transferred to 6-DMAP–treated extract (lacking RLF activity) supplemented with [α32P]dATP. At different times after transfer, the amount of DNA synthesis was measured. When incubated in extract at concentrations of above 1/4–1/8 (corresponding to 100–200 ng DNA per μl neat extract), chromatin was efficiently licensed, leading to almost complete replication after an additional 90-min incubation in 6-DMAP–treated extract. This is similar to replication rates seen in untreated extract. When licensed with lower amounts of extract (1/16 dilution and below), the chromatin replicated at a lower rate, as though only a fraction of the replication origins had initiated. Comparison of Fig. 1 a and 1 b suggests that maximal replication rates were still seen with subsaturating quantities of XMcm3 assembled onto the chromatin.
Cell Cycle Variation in RLF Activity
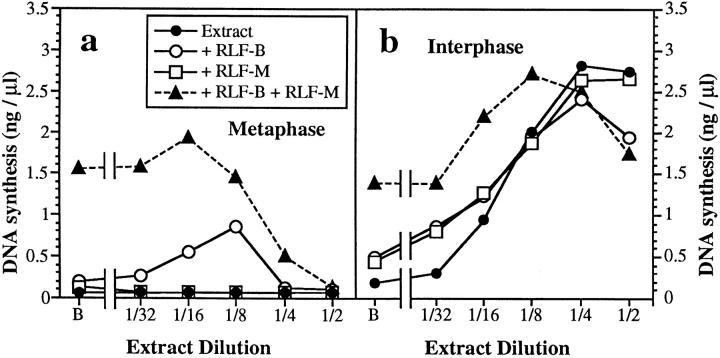
The Xenopus egg is arrested at metaphase of meiosis II. At fertilization, a calcium wave is released that overcomes the metaphase arrest, and the early embryo embarks on a series of 12 rapid cell cycles. Extracts prepared from unfertilized eggs maintain the metaphase arrest and can be released into interphase by addition of CaCl2 (Lohka and Masui, 1984). RLF activity is low in such metaphase- arrested extracts, but it rapidly increases upon CaCl2 addition (Blow, 1993). We first determined the quantity of RLF-B and RLF-M activity present in metaphase-arrested and in interphase egg extracts (Fig. 2). Chromatin was licensed with serial dilutions of extract and transferred to 6-DMAP–treated extract, and the total DNA synthesized over a 90-min incubation was measured (Fig. 2, filled circles). Consistent with the time courses shown in Fig. 1 b, interphase extract efficiently licensed DNA synthesis after dilution as low as 1/4–1/8 (Fig. 2 b). In contrast, no RLF activity was provided by the metaphase-arrested extract (Fig. 2 a).
Figure 2.
RLF activity in serial dilutions of metaphase and interphase extract. Metaphase arrested (a) or interphase (b) Xenopus extracts were subjected to serial dilution, and then mixed with an equal volume of either buffer (○̶ ), crude RLF-B (●̶ ), crude RLF-M (□̶ ), or a mixture of crude RLF-B and crude RLF-M (▲̶ ). B, buffer in place of diluted extract. Chromatin was incubated in the mixture (12 ng DNA/μl) for 15 min at 23°C to allow licensing, and then was transferred to 2.5 vol 6-DMAP–treated extract containing [α32P]dATP. The total DNA synthesized after further incubation for 90 min at 23°C was measured.
The titrations of RLF activity in metaphase and interphase extracts were repeated in the presence of exogenous RLF-B (Fig. 2, open circles) or RLF-M (Fig. 2, open squares) to respectively determine the RLF-M and RLF-B activity of the samples. Titrations of activity in interphase extract under these conditions remained virtually unchanged (Fig. 2 b), suggesting that these extracts had approximately equal activities of RLF-M and RLF-B. At high concentrations of metaphase extract, addition of exogenous RLF-B or RLF-M induced no RLF activity (Fig. 2 a). However, when metaphase extracts were diluted down to 1/8–1/16, significant RLF-M activity could be detected. No RLF-B activity was observed at any dilution of metaphase extract. The appearance of RLF-M activity after dilution of metaphase extract was unexpected and suggested the presence of an RLF inhibitor in these extracts that was being diluted away until it no longer inhibited the reaction. The presence of an RLF inhibitor was demonstrated by mixing serial dilutions of metaphase extract with a constant amount of active RLF (RLF-B plus RLF-M; Fig. 2 a, triangles). The licensing activity of exogenous RLF was inhibited by high concentrations of metaphase extract. When metaphase extracts were diluted further, the inhibitory activity was titrated away, allowing exogenous RLF to license chromatin. The disappearance of the inhibitory activity upon dilution corresponded to concentrations where RLF-M activity became assayable. In contrast, no RLF inhibitor was observed in interphase extracts (Fig. 2 b, triangles). The simplest explanation for the ability to assay RLF-M activity after dilution of metaphase extract is that the RLF inhibitor affects only RLF-B activity. Consistent with this interpretation, we could find no evidence for the inhibition of RLF-M activity in metaphase (see below).
To quantify cell cycle variations of RLF activity, extract was incubated for various times, and then diluted to 1/8, where RLF activity should be in the linear range for the assay (Fig. 3). In these extracts, chromatin decondensation and nuclear assembly are complete by ∼30–40 min after CaCl2 addition, while S phase occurs at 40–90 min (Blow, 1993). RLF levels in metaphase were low, but, within 15 min after release from the metaphase arrest by CaCl2 addition, maximum levels of total RLF activity were observed (Fig. 3 a, filled circles). Total RLF activity decayed over 1–2 h (Fig. 3 b, filled circles), although the kinetics of the decay varied from extract to extract. This decay of total RLF activity was almost entirely due to decay of RLF-B activity (Fig. 3 b, open squares), while RLF-M activity remained fairly constant throughout the cell cycle (open circles).
Figure 3.
Time course of RLF activity during the in vitro cell cycle. Interphase extract (b) or metaphase extract released into interphase with CaCl2 (a) was incubated at 23°C for various times. Extract was then diluted eightfold and mixed with an equal volume of either buffer (○̶ ), crude RLF-B (●̶ ), or crude RLF-M (□̶ ). Unlicensed chromatin was incubated in the mixture (12 ng DNA/μl) for 15 min at 23°C to allow licensing, and then was transferred to 2.5 vol 6-DMAP–treated extract containing [α32P]dATP. The total DNA synthesized after further incubation for 90 min at 23°C was measured. M, metaphase-arrested extract that was not released with CaCl2.
Cdk Dependence of the RLF Inhibitor
The above results suggest the presence of an RLF inhibitor in metaphase extracts. Interphase Xenopus extracts can be induced to enter metaphase by addition of MPF, a Cdk activity that can be provided by either Cdc2/cyclin B (Labbé et al., 1989a ; Gautier et al., 1990) or Cdc2/cyclin A (Luca et al., 1991; Roy et al., 1991). We have previously shown that the addition of high levels of cyclin A to interphase Xenopus extracts inactivates RLF (Blow, 1993) and blocks DNA replication (Strausfeld et al., 1996). We therefore examined whether the RLF inhibitor present in metaphase extracts was dependent on Cdk activity. Cyclin-dependent kinases can be affinity depleted from Xenopus extracts using p13suc1 coupled to Sepharose beads (Dunphy et al., 1988). When p13suc1 depletion is performed on metaphase-arrested Xenopus extracts, subsequent DNA synthesis is blocked (Blow and Nurse, 1990). Fig. 4 a (filled circles) shows that p13suc1 depleted metaphase extracts lack RLF activity. However, unlike the undepleted extracts (Fig. 2 a), RLF-M activity could be detected at the highest extract concentrations (Fig. 4 a, open circles). Thus, no RLF inhibitor could be detected in these extracts (Fig. 4 a, triangles), and the lack of RLF activity was apparently due only to a lack of RLF-B activity (Fig. 4 a, squares).
The Xenopus early embryo has at least three distinct cyclin-dependent kinases: Cdc2/cyclin A, Cdc2/cyclin B, and Cdk2/cyclin E. Similar to the effect of p13suc1 depletion, immunoprecipitation of Cdc2 from metaphase extracts blocked activation of RLF-B (data not shown) and prevented subsequent DNA replication (Blow and Nurse, 1990). To demonstrate a direct role for Cdc2 in inhibiting RLF, we prepared purified RLF-M (Chong et al., 1995) and partially purified RLF-B (Rowles et al., 1996; Chong et al., 1997). Incubation of recombinant Cdc2/cyclin B with these purified fractions caused strong inhibition of licensing (Fig. 4 b). This result strongly suggests that Cdc2/cyclin B can directly inhibit the RLF system. Consistent with this idea, we show below that stabilization of the RLF inhibitor correlates well with stabilization of cyclin B.
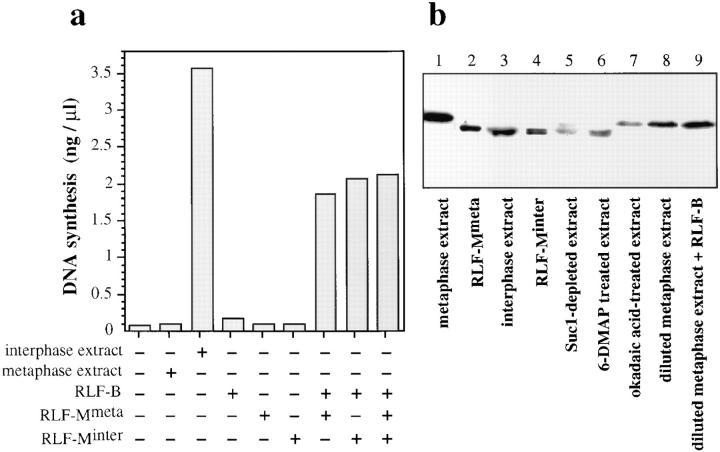
We next performed a partial (∼50-fold) purification of RLF-M from metaphase and interphase extracts to investigate whether active RLF-M can be found in a metaphase-arrested extract (Fig. 5 a). RLF-M prepared from metaphase extracts (RLF-Mmeta) showed virtually identical activity to RLF-M prepared from interphase extracts (RLF-Minter). Mixing RLF-Mmeta and RLF-Minter together showed that the RLF-M had been purified away from the metaphase-specific inhibitory activity. At this stage we cannot be completely certain that RLF-M is fully active in metaphase extracts, as it is possible that an inhibitory modification (such as phosphorylation) might be reversed during the purification protocol. In metaphase Xenopus extracts, the XMcm4 protein, a component of RLF-M (Chong et al., 1995; Thömmes, P., Y. Kubota, H. Takisawa, and J.J. Blow, manuscript submitted for publication), is phosphorylated and migrates slower on SDS gels (Fig. 5 b, lanes 1 and 3; Coué et al., 1996). This phosphorylation can be blocked with phosphatase inhibitor okadaic acid (Fig. 5 b, lane 7; Coué et al., 1996), but is lost during the purification of the RLF-Mmeta fraction (Fig. 5 b, lane 2). However, the presence of the slow migrating form of XMcm4 does not correlate with the presence of the RLF inhibitor, since XMcm4 is fast migrating in 6-DMAP– treated extract (Fig. 5, lane 6) where the RLF inhibitor is present (see below). Furthermore, XMcm4 is still slow migrating in metaphase extracts diluted to 1/8 and incubated with RLF-B (Fig. 5 b, lanes 8 and 9), under which conditions the inhibitor has been titrated away (Fig. 2 a). Conversely, most of the XMcm4 is fast migrating in Suc1- depleted extracts (Fig. 5 b, lane 5) where the inhibitor is absent. Thus, we can find no evidence that mitotic phosphorylation of RLF-M inhibits its activity.
Figure 5.
Features of RLF-M prepared from metaphase or interphase extract. (a) Unlicensed chromatin was incubated for 15 min at 23°C with interphase extract, metaphase extract, or mixtures of RLF-B and RLF-Mmeta (RLF-M prepared from metaphase extract) and RLF-Minter (RLF-M prepared from interphase extract). Samples were then transferred to 6-DMAP–treated extract containing [α32P]dATP. The total DNA synthesized after further incubation for 90 min at 23°C was measured. (b) Protein samples electrophoresed on a 7.5% polyacrylamide gel and immunoblotted with an anti-XMcm4 antibody. Samples: 1, metaphase extract; 2, RLFMmeta; 3, interphase extract; 4, RLF-Minter; 5, metaphase extract depleted with p13suc1-Sepharose; 6, metaphase extract treated with 3 mM 6-DMAP before CaCl2 release (6-DMAP–treated extract); 7, metaphase extract treated with 2 μM okadaic acid before CaCl2 release; 8, metaphase extract diluted eightfold and incubated for 15 min at 23°C; 9, metaphase extract diluted eightfold, mixed with crude RLF-B, and incubated for 15 min at 23°C.
Since the RLF inhibitor present in metaphase extracts is dependent on cyclin-dependent kinases, we next investigated whether the decline in RLF activity during interphase is also dependent on cyclin-dependent kinases. Both cyclin A and cyclin B are abruptly degraded during late mitosis, and must be resynthesized during the next cell cycle before entry into mitosis. Therefore, treatment of Xenopus egg extracts with protein synthesis inhibitors such as cycloheximide blocks the reappearance of these two cyclins. However, consistent with our previous results (Blow, 1993), cycloheximide only slightly delayed the decay of RLF that normally occurs during interphase (Fig. 6 a, halffilled diamonds). In contrast to cyclins A and B, cyclin E remains at approximately constant levels throughout the early embryonic cell cycles (Rempel et al., 1995; Chevalier et al., 1996). Treatment of Xenopus extracts with the Cdk2 inhibitor Cip1 inhibits the SPF activity of Cdk2/cyclin E and thus blocks the initiation of DNA replication (Strausfeld et al., 1994; Jackson et al., 1995). However, addition of Cip1 to cycloheximide-treated extracts did not stabilize RLF activity in the cell cycle (Fig. 6 a, filled diamonds). Similarly, affinity depletion of Cdks from interphase extract using p13suc1-coupled Sepharose did not stabilize RLF activity (data not shown). These results suggest that the decay of RLF activity during interphase is independent of Cdk activity.
Figure 6.
Effect of cycloheximide and Cip1 of RLF stability. (a) Metaphase extract was released into interphase with CaCl2 either untreated (control) or in the presence of 100 μg/ml cycloheximide (+ CHX), or 100 μg/ml cycloheximide plus 150 nM p21Cip1 (+ CHX + Cip1). At the indicated times, samples were diluted eightfold and incubated for 15 min with unlicensed chromatin. Samples were then transferred to 6-DMAP–treated extract containing [α32P]dATP. The total DNA synthesized after further incubation for 90 min at 23°C was measured. (b) Interphase extract was incubated at 23°C for 2 h to allow RLF activity to decay. The extract was subjected to serial dilution, and then mixed with an equal volume of either buffer (○̶ ), crude RLF-B (●̶ ), crude RLF-M (□̶ ), or a mixture of crude RLF-B and crude RLF-M (). B, buffer in place of diluted extract. Unlicensed chromatin was incubated in the mixture (12 ng DNA/μl) for 15 min at 23°C to allow licensing, and then was transferred to 2.5 vol 6-DMAP–treated extract containing [α32P]dATP. The total DNA synthesized after further incubation for 90 min at 23°C was measured.
Fig. 6 b shows the quantity of RLF-B and RLF-M activity in interphase extracts that had been incubated at 23°C for 2 h. Total RLF activity was reduced approximately eightfold over extract that had recently been released from metaphase (compare Figs. 2 b and 6 b, filled circles). This was almost exclusively due to a loss of RLF-B activity (open circles), while RLF-M activity remained high (squares). Despite the decline in RLF-B activity, no RLF inhibitor could be detected (Fig. 6 b, triangles). Thus, Xenopus extracts can be defective in RLF activity in at least two distinct ways. In metaphase, a potent inhibitor of RLF activity is present, and this inhibitor is directly dependent on the presence of Cdk activity. If this inhibitor is diluted away, RLF-M but not RLF-B activity can be detected. In contrast, when RLF-B activity decays during late interphase, this decay does not require Cdk activity and no RLF inhibitor becomes activated.
Role of RLF in the Initiation of Replication
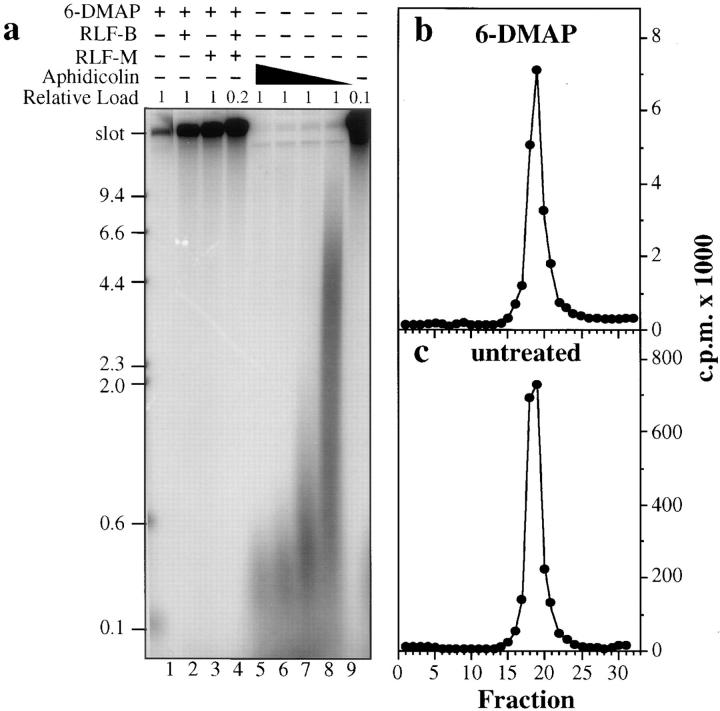
6-DMAP–treated extracts lack active RLF and therefore cannot replicate double-stranded DNA (Blow, 1993). Despite this fact, they are still competent to support complementary strand synthesis on single-stranded DNA and to elongate replication forks stalled with aphidicolin (Blow, 1993). This suggests that RLF activity is required for the initiation, but not the elongation, of replication forks. However, Yan and Newport (1995) have suggested on the basis of immunofluorescence data that 6-DMAP–treated extracts are able to support the synthesis of DNA primers at replication origins, but that these primers are not correctly processed into functional replication forks. We therefore analyzed the residual DNA synthesis seen in 6-DMAP–treated extracts by alkaline gel electrophoresis (Fig. 7 a) and BrdUTP density substitution (Fig. 7, b and c). 6-DMAP treatment of the metaphase-arrested extract inhibited subsequent DNA synthesis by 95%. However, BrdU density substitution showed that all of the residual DNA synthesized in the 6-DMAP–treated extract (Fig. 7 b) was of the same density as DNA synthesized in untreated extract (Fig. 7 c), as expected of fully replicated heavy/light DNA produced by semiconservative replication. This suggests that a small number of replicons escape the 6-DMAP inhibition and produce relatively large tracts of fully replicated DNA. This conclusion was confirmed by analyzing nascent strand length by alkaline agarose gel electrophoresis (Fig. 7 a). In both 6-DMAP–treated (lane 1) and untreated extract (lane 9), virtually all of the nascent DNA was in the form of high molecular weight DNA (>23,000 nt in size). In contrast, when replication forks are stalled close to the origin by treating extracts with aphidicolin, most of the nascent DNA is observed in a broad smear of between 100 and 300 nt in length (Fig. 7 a, lanes 5–8). These results strongly suggest that 6-DMAP blocks the initiation of DNA replication at a stage before significant primer synthesis has occurred. The few replication forks that do initiate in 6-DMAP–treated extracts then proceed to synthesize extensive stretches of nascent DNA. No low molecular weight products are observed when chromatin is preincubated with RLF-B, RLF-M, or a mixture of both before incubation in 6-DMAP–treated extract (Fig. 7 a, lanes 3–5), suggesting that both of these activities are required for efficient initiation of DNA replication in 6-DMAP– treated extracts.
Figure 7.
Analysis of the stage of replication blocked in 6-DMAP– treated extracts. (a) Alkaline agarose gel of nascent DNA. (Lanes 1–4) Chromatin was licensed for 15 min with either buffer (lane 1), RLF-B (lane 2), RLF-M (lane 3), or RLF-B plus RLF-M (lane 4), transferred to 6-DMAP–treated extract containing [α32P]dATP, and incubated for 90 min. (Lanes 5–9) Sperm nuclei was incubated for 90 min in interphase Xenopus extract containing [α32P]dATP and various concentrations of aphidicolin (lane 5, 30 μg/ml; lane 6, 20 μg/ml; lane 7, 10 μg/ml; lane 8, 5 μg/ml; lane 9, no aphidicolin). To compensate for the much higher incorporation of 32P in the reactions, the total DNA loaded in lane 4 was reduced to 20%, and the total DNA in lane 9 was reduced to 10%. (b and c) BrdUTP density substitution of nascent DNA synthesized after incubation of sperm nuclei for 90 min in untreated (c) or 6-DMAP–treated extract (b).
Effect of 6-DMAP on the Licensing System
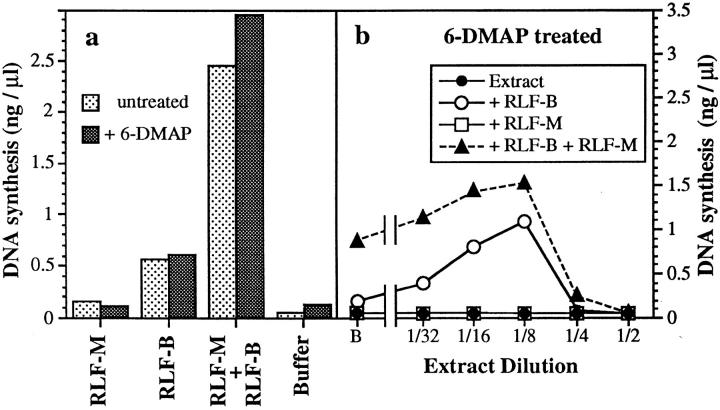
When 6-DMAP is added to metaphase-arrested Xenopus extracts, MPF activity is inhibited, and the extracts spontaneously exit from metaphase into interphase without the need for CaCl2 addition (Zhang and Masui, 1992; Blow, 1993). These 6-DMAP–treated extracts assemble DNA into normal nuclei and, with the exception of RLF, contain all the activities required for complete chromosome replication (Blow, 1993). Similar results have been reported for two other kinase inhibitors: staurosporine (Blow, 1993; Kubota and Takisawa, 1993) and olomoucine (Vesely et al., 1994). For all these inhibitors, there is a good correlation between the inhibition of MPF activity (as judged by spontaneous assembly of interphase nuclei) and the inhibition of RLF activity. To cause inhibition of the licensing system, the kinase inhibitors must be added to Xenopus extracts before exit from metaphase arrest (Blow, 1993; Kubota and Takisawa, 1993). This suggests that the protein kinase inhibitors block the RLF activation that normally occurs upon exit from metaphase, rather than inhibiting the action of active RLF. This conclusion is confirmed in Fig. 8 a. Active RLF-M and RLF-B fractions prepared from interphase Xenopus extract by differential PEG precipitation were resuspended in buffers plus or minus 3 mM 6-DMAP. This concentration of 6-DMAP is sufficient to block RLF activation in metaphase extracts (Blow, 1993). Efficient licensing of DNA replication required both RLFB and RLF-M, but it was not significantly affected by the presence of 6-DMAP.
Figure 8.
Effect of 6-DMAP on RLF activity. (a) Unlicensed chromatin was incubated for 15 min plus or minus 3 mM 6-DMAP with crude RLF-B, crude RLF-M, or a mixture of the two. Samples were then transferred to 6-DMAP–treated extract containing [α32P]dATP. The total DNA synthesized after further incubation for 90 min at 23°C was measured. (b) Metaphase-arrested Xenopus extract was treated with 3 mM 6-DMAP and 0.3 mM CaCl2, followed by serial dilution in buffer containing 3 mM 6-DMAP. Samples were then mixed with equal volumes of either buffer (○̶ ), crude RLF-B (●̶ ), crude RLF-M (□̶ ), or a mixture of crude RLF-B and crude RLF-M (▲̶ ). B, buffer in place of diluted extract. Chromatin was incubated in the mixture (12 ng DNA/μl) for 15 min at 23°C to allow licensing, and then was transferred to 2.5 vol 6-DMAP–treated extract containing [α32P]dATP. The total DNA synthesized after further incubation for 90 min at 23°C was measured.
To measure RLF-B, RLF-M, and RLF inhibitor activity when 6-DMAP is added to metaphase-arrested extracts, we performed serial dilutions of 6-DMAP–treated metaphase extract using buffer supplemented with an additional 3 mM 6-DMAP (Fig. 8 b). The results of the titration were strikingly similar to those of untreated metaphase extracts (compare Figs. 8 b and 2 a). RLF activity was not detected at any dilution (Fig. 8 b, filled circles), but an RLF inhibitor was detected at higher concentrations (triangles). Although no RLF-B could be detected at any dilution (squares), significant RLF-M activity could be detected once the inhibitor had been diluted away (open circles). It therefore appears that although the 6-DMAP–treated extracts have morphologically exited from metaphase into interphase, they still contain the RLF inhibitor normally present in metaphase extracts.
Since the RLF inhibitor is dependent on the presence of Cdk activity, we determined whether Cdc2/cyclin B is present in 6-DMAP–treated extracts. Cyclins A and B are normally degraded on exit from mitosis. Fig. 9 a shows an autoradiograph of 35S-labeled cyclin B in extracts released from metaphase by CaCl2 in the presence of different concentrations of 6-DMAP. Consistent with previous reports (Felix et al., 1989; Luca and Ruderman, 1989), 6-DMAP at concentrations of above 2 mM blocked cyclin B degradation that normally occurs after CaCl2 addition (Fig. 9 a, lanes 5–8). However, these extracts all entered interphase as judged by the assembly of DNA into interphase nuclei, despite the stability of cyclin B at higher concentrations (Fig. 9 b; +CaCl2). Above 1 mM, 6-DMAP could inhibit MPF and cause metaphase extract to spontaneously enter interphase (as judged by the assembly of DNA into interphase nuclei) in the absence of added CaCl2 (Fig. 9 b, −CaCl2; Zhang and Masui, 1992). The concentrations of 6-DMAP just sufficient to block cyclin B degradation correlated well with concentrations at which RLF became stably inhibited (Fig. 9 c). It therefore seems most likely that while high levels of Cdc2 kinase are required to trigger cyclin B degradation (Felix et al., 1989), lower levels of kinase sufficient to inhibit RLF activity are still present in extracts treated with 3 mM 6-DMAP.
Figure 9.
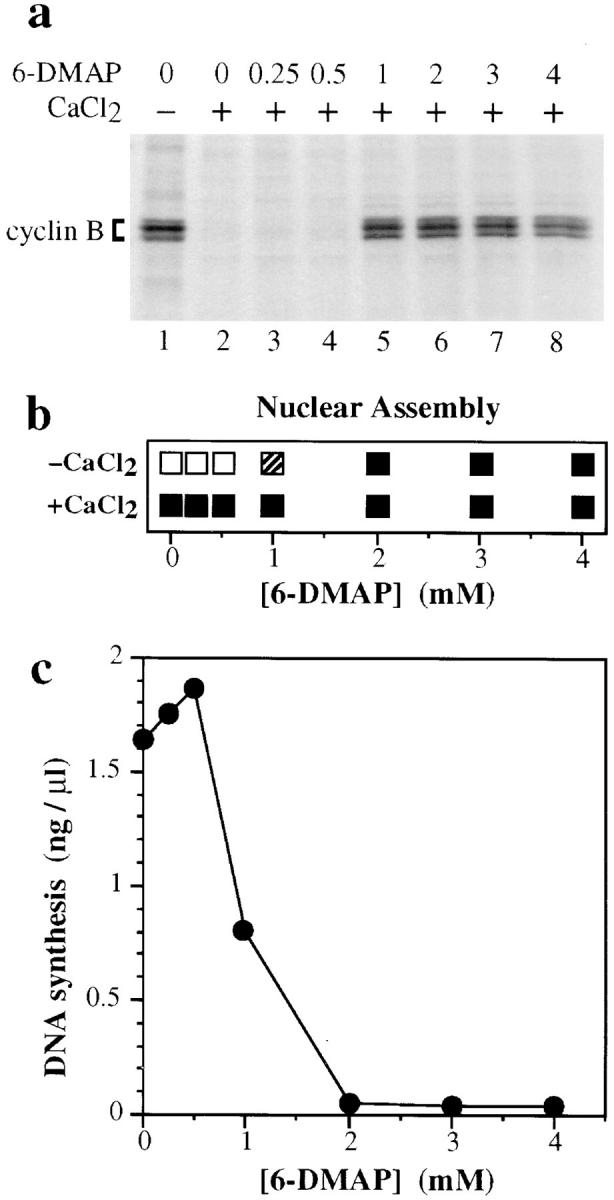
Titration of 6-DMAP into metaphase extract. (a) Metaphase extract was labeled for 20 min with [35S]methionine, and then was supplemented with 100 μg/ml cycloheximide and the indicated concentrations of 6-DMAP. Extract was then further supplemented plus (lanes 2–8) or minus (lane 1) 0.3 mM CaCl2. After incubation at 23°C for 30 min, cyclin–Cdk complexes were collected on p13suc1 beads, electrophoresed on polyacrylamide gels, and autoradiographed. The migration of cyclin B is indicated. (b) Metaphase extract was supplemented with sperm nuclei (3 ng DNA/μl) and various concentrations of 6-DMAP, plus (lower panel) or minus (upper panel) 0.3 mM CaCl2. The morphology of chromatin after 1.5 h at 23°C was assessed by microscopy: (□), condensed chromosomes; (▪), interphase nuclei; (▨ ), partial nuclear assembly. (c) Metaphase extract was supplemented with sperm nuclei (3 ng DNA/μl), 0.3 mM CaCl2, [α32P]dATP, and various concentrations of 6-DMAP. The total DNA synthesized after 3 h at 23°C was measured.
Discussion
RLF is an essential initiation factor that ensures that chromosomal DNA is not re-replicated in a single cell cycle. In this paper we measure the RLF activity present in Xenopus extracts and show that the ability of extracts to license chromatin varies throughout the cell cycle. In metaphase extract, RLF-M but not RLF-B activity can be assayed, and, in addition, an RLF inhibitor is present. This RLF inhibitor is dependent on the presence of cyclin-dependent kinases and is stabilized when cyclin B is stabilized by the protein kinase inhibitor 6-DMAP. RLF activity is rapidly activated upon exit from metaphase, and then decays during late interphase. The decay of RLF activity in late interphase is mainly due to decay of RLF-B activity and is not associated with reactivation of the RLF inhibitor.
RLF Activity of Xenopus Egg Extracts
Interphase Xenopus extract can license ∼100–200 ng DNA per μl extract for efficient replication. Titration of RLF-M and RLF-B activity suggested that these two activities were present at approximately equal levels. Chromatin licensed at 100 ng DNA per μl extract could replicate at rates typical of Xenopus egg extracts. Western blotting of XMcm3, a component of RLF-M (Chong et al., 1995), suggests that, at 100 ng DNA per μl extract, one XMcm3 molecule is assembled on average onto every 5–10 kb DNA. This represents on average one to three XMcm3 molecules per replicon. However, at lower concentrations of DNA, more XMcm3 can be assembled onto chromatin (up to one XMcm3 molecule per 1.5 kb, or 10 XMcm3 molecules per replicon) without significantly affecting the overall replication rate. This higher density of XMcm3 is more likely to reflect conditions in vivo, since up to the midblastula transition (when zygotic transcription starts), the concentration of chromosomal DNA is <25 ng/μl. A similar excess of Mcm3 over replication origins has been reported in human cells (Burkhart et al., 1995) and budding yeast (Lei et al., 1996).
Cell Cycle Variation of RLF Activity
RLF activity varies throughout the in vitro cell cycle. In metaphase, RLF is largely inactive but is rapidly activated after the metaphase–anaphase transition; during interphase, activity then gradually declines (Blow, 1993). We have analyzed some of the basic causes of this variation. In metaphase an RLF inhibitor was present that inhibited licensing by exogenous RLF. When this metaphase extract was diluted so that the inhibitor no longer interfered with the licensing assay, levels of RLF-M were detected comparable with those seen in interphase extract. In contrast, no RLF-B activity was detected. Partially purified RLF-M prepared from metaphase extracts (RLF-Mmeta) was also fully active. XMcm4, a component of RLF-M (Thömmes, P., Y. Kubota, H. Takisawa, and J.J. Blow, manuscript submitted for publication), was phosphorylated during metaphase (Coué et al., 1996), but this phosphorylated XMcm4 was still associated with active RLF-M. The simplest explanation for these results is that the target of the RLF inhibitor is RLF-B, not RLF-M, although we cannot completely rule out an inhibitory modification to RLF-M that we have been unable to stabilize.
The decline of RLF activity that occurs during interphase causes a different defect than is seen in metaphase extracts. RLF-M activity remains high, but RLF-B activity declines. Unlike metaphase extracts, the loss of RLF-B activity during interphase occurs without the appearance of an RLF inhibitor. At present we cannot be certain whether the decline of RLF-B activity during interphase is of physiological relevance or whether it is an artifact of the cell-free system. However, the decline of RLF-B activity is fairly specific, since extracts retain RLF-M activity and remain competent to replicate previously licensed chromatin (Blow, 1993).
Role of RLF in the Initiation of Replication
The initiation of chromosomal DNA replication is likely to consist of several steps, including the unwinding of origin DNA, the synthesis of short DNA primers, and the establishment of a mature replication fork. On the basis of immunofluorescence data showing that 6-DMAP–treated extracts are able to support low levels of DNA synthesis, it has been suggested that RLF is not required for the synthesis of DNA primers at replication origins, but it is required for correct assembly of replication forks (Yan and Newport, 1995). We have examined this point in detail to determine whether RLF is required for the synthesis of DNA primers early in initiation. Alkaline gel electrophoresis of nascent strands showed that in 6-DMAP– treated extracts lacking RLF, no short DNA primers could be observed. In contrast, short nascent strands could be readily detected when replication forks were blocked by the polymerase inhibitor aphidicolin. Both RLF-B and RLF-M were required for efficient DNA replication, but in the presence of both these activities, nascent strands were rapidly extended into high molecular weight DNA. We therefore conclude that RLF is required for the initiation of DNA replication before the synthesis of DNA primers.
The RLF Inhibitor Depends on Cdk Activity
Cdks are key regulators of cell cycle progression (for review see Nigg, 1995). MPF is an activity present in metaphase cells capable of inducing entry into mitosis. It consists of a Cdc2/cyclin B heterodimer (Dunphy et al., 1988; Gautier et al., 1988, 1990; Labbé et al., 1989a ,b). Cdc2/cyclin A, which is also active at this stage of the cell cycle, can also generate MPF activity (Luca et al., 1991; Roy et al., 1991; Strausfeld et al., 1996). The RLF inhibitor present in metaphase extracts appears to be directly dependent on Cdks active at this time. When Cdks were affinity depleted from metaphase extracts using p13suc1, the RLF inhibitor was removed. Inhibition of RLF function by kinase inhibitors such as 6-DMAP caused persistence into interphase state of the RLF inhibitor normally only present during metaphase. This also correlated with inhibition of MPF activity (Blow, 1993; Kubota and Takisawa, 1993; Vesely et al., 1994) and stabilization of cyclin B. Active RLF was also inhibited by the activation of Cdks. When recombinant cyclin A was added to interphase extract, RLF activity abruptly decayed (Blow, 1993). Furthermore, purified Cdc2/cyclin B directly inhibited licensing activity provided by purified RLF-M and partially purified RLF-B. These results strongly suggest a direct inhibition of the licensing system by Cdc2 kinase. Although MCM/P1 polypeptides are expected to be good substrates for this kinase (Coué et al., 1996), the results discussed above suggest that inhibition of licensing is unlikely to be mediated by inhibition of RLF-M function. Instead, RLF-B appears to be a more likely target of Cdk inhibition.
Inhibition of RLF function by Cdks is consistent with experiments in other organisms that show, when Cdk activity is inhibited in G2 cells, re-replication of chromosomal DNA can occur without cells passing into mitosis. This effect is plausibly caused by reactivation of RLF and relicensing of replicated DNA. Re-replication of chromosomal DNA is seen in Saccharomyces pombe cells containing a temperature-sensitive cdc2 gene when they are shifted to the nonpermissive temperature during G2 (Broek et al., 1991). Repeated rounds of DNA synthesis are seen in S. pombe cells lacking the cdc13 (cyclin B) gene (Hayles et al., 1994) or cells that overexpress the Cdc2 inhibitor Rum1 (Moreno and Nurse, 1994). In S. cerevisiae, the establishment of a pre-replicative footprint over origins of replication that probably corresponds to the licensed state can only occur in late mitosis and G1 when Cdk levels are low (Diffley et al., 1994; Piatti et al., 1996). Furthermore, inhibition of Cdks in G2/M-phase S. cerevisiae cells is sufficient to induce a pre-replicative footprint over origins of replication (Dahmann et al., 1995). Treatment of mammalian cells with certain protein kinases that inhibit Cdks was sufficient to allow re-replication of DNA in the absence of mitosis (Usui et al., 1991). Similar treatments allow mammalian G2 nuclei to become relicensed for replication in Xenopus egg extracts (Coverley et al., 1996). A requirement for Cdks to prevent re-replication of DNA late in the cell cycle therefore appears widespread throughout the eukaryotic kingdom and is plausibly mediated by an RLF inhibitor that prevents relicensing of replicated DNA. This is consistent with the results presented here demonstrating a Cdk-dependent RLF inhibitor present in metaphase extract.
However, RLF regulation in the Xenopus system appears somewhat different from that in the other cell types. First, loss of cytoplasmic RLF during interphase in Xenopus is not dependent on Cdk activity and is not associated with an RLF inhibitor. Secondly, although relicensing of DNA is dependent on nuclear envelope permeabilization in both Xenopus and Drosophila cell-free systems (Blow and Laskey, 1988; Crevel and Cotterill, 1991), this does not seem necessary in other cell types. One attractive explanation for these apparent differences is that in Xenopus, relicensing of DNA in replicated nuclei is prevented by a Cdk-dependent inhibitor that is active only within the nucleus. Permeabilization of the nuclear envelope would then release the inhibitor and permit relicensing of the DNA by RLF-B and RLF-M.
Abbreviations used in this paper
- BrdUTP
bromodeoxyuridine triphosphate
- Cdk
cyclin-dependent kinase
- 6-DMAP
6-dimethyl aminopurine
- GST
glutathione-S-transferase
- MPF
maturation promoting factor
- ORC
origin recognition complex
- PEG
polyethylene glycol
- RLF
replication licensing factor
- SPF
S-phase promoting factor
Footnotes
J.J. Blow is a Lister Institute Research fellow. We thank Noel Lowndes and John Diffley for comments on the manuscript.
Received for publication 16 August 1996 and in revised form 23 October 1996.
Address all correspondence to J. Julian Blow, Imperial Cancer Research Fund, Clare Hall Laboratories, Blanche Lane, South Mimms, Potters Bar, Herts EN6 3LD, United Kingdom. Tel.: (44) 171-269-3923. Fax: (44) 171269-3801. e-mail: blow@icrf.icnet.uk
References
- Blow JJ. Preventing re-replication of DNA in a single cell cycle: evidence for a Replication Licensing Factor. J Cell Biol. 1993;122:993–1002. doi: 10.1083/jcb.122.5.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopuseggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature (Lond) 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Nurse P. A cdc2-like protein is involved in the initiation of DNA replication in Xenopusegg extracts. Cell. 1990;62:855–862. doi: 10.1016/0092-8674(90)90261-c. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Sleeman AM. Replication of purified DNA in Xenopusegg extract is dependent on nuclear assembly. J Cell Sci. 1990;95:383–391. doi: 10.1242/jcs.95.3.383. [DOI] [PubMed] [Google Scholar]
- Blow JJ, Watson JV. Nuclei act as independent and integrated units of replication in a Xenopuscell-free system. EMBO (Eur Mol Biol Organ) J. 1987;6:1997–2002. doi: 10.1002/j.1460-2075.1987.tb02463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broek D, Bartlett R, Crawford K, Nurse P. Involvement of p34cdc2 in establishing the dependency of S phase on mitosis. Nature (Lond) 1991;349:388–393. doi: 10.1038/349388a0. [DOI] [PubMed] [Google Scholar]
- Burkhart R, Schulte D, Hu D, Musahl C, Gohring F, Knippers R. Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur J Biochem. 1995;228:431–438. [PubMed] [Google Scholar]
- Chevalier S, Couturier A, Chartrain I, Le Guellec R, Beckhelling C, Le Guellec K, Philippe M, Ford CC. Xenopuscyclin E, a nuclear phosphoprotein, accumulates when oocytes gain the ability to initiate DNA replication. J Cell Sci. 1996;109:1173–1184. doi: 10.1242/jcs.109.6.1173. [DOI] [PubMed] [Google Scholar]
- Chong JPJ, Mahbubani MH, Khoo C-Y, Blow JJ. Purification of an Mcm-containing complex as a component of the DNA replication licensing system. Nature (Lond) 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- Chong JPJ, Thömmes P, Blow JJ. The role of MCM/P1 proteins in the licensing of DNA replication. Trends Biochem Sci. 1996;21:102–106. [PubMed] [Google Scholar]
- Chong, J.P.J., P. Thömmes, A. Rowles, H.M. Mahbubani, and J.J. Blow. 1997. Characterisation of the Xenopus replication licensing system. Methods Enzymol. In press. [DOI] [PubMed]
- Coué M, Kearsey SE, Méchali M. Chromatin binding, nuclear localization and phosphorylation of Xenopuscdc21 are cell-cycle dependent and associated with the control of initiation of DNA replication. EMBO (Eur Mol Biol Organ) J. 1996;15:1085–1097. [PMC free article] [PubMed] [Google Scholar]
- Coverley D, Downes CS, Romanowski P, Laskey RA. Reversible effects of nuclear membrane permeabilization on DNA replication: evidence for a positive licensing factor. J Cell Biol. 1993;122:985–992. doi: 10.1083/jcb.122.5.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coverley D, Wilkinson HR, Downes CS. A protein kinase-dependent block to reinitiation of DNA replication in G2 phase in mammalian cells. Exp Cell Res. 1996;225:294–300. doi: 10.1006/excr.1996.0179. [DOI] [PubMed] [Google Scholar]
- Crevel G, Cotterill S. DNA replication in cell-free extracts from Drosophila melanogaster. . EMBO (Eur Mol Biol Organ) J. 1991;10:4361–4369. doi: 10.1002/j.1460-2075.1991.tb05014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmann C, Diffley JF, Nasmyth KA. S-phase-promoting cyclindependent kinases prevent re-replication by inhibiting the transition of replication origins to a pre-replicative state. Curr Biol. 1995;5:1257–1269. doi: 10.1016/s0960-9822(95)00252-1. [DOI] [PubMed] [Google Scholar]
- Dalton S, Whitbread L. Cell cycle-regulated nuclear import and export of Cdc47, a protein essential for initiation of DNA replication in budding yeast. Proc Natl Acad Sci USA. 1995;92:2514–2518. doi: 10.1073/pnas.92.7.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffley JF, Cocker JH, Dowell SJ, Rowley A. Two steps in the assembly of complexes at yeast replication origins in vivo. Cell. 1994;78:303–316. doi: 10.1016/0092-8674(94)90299-2. [DOI] [PubMed] [Google Scholar]
- Dunphy WG, Brizuela L, Beach D, Newport J. The Xenopuscdc2 protein is a component of MPF, a cytoplasmic regulator of mitosis. Cell. 1988;54:423–431. doi: 10.1016/0092-8674(88)90205-x. [DOI] [PubMed] [Google Scholar]
- Felix MA, Pines J, Hunt T, Karsenti E. A post-ribosomal supernatant from activated Xenopuseggs that displays post-translationally regulated oscillation of its cdc2+ mitotic kinase activity. EMBO (Eur Mol Biol Organ) J. 1989;8:3059–3069. doi: 10.1002/j.1460-2075.1989.tb08457.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier J, Norbury C, Lohka M, Nurse P, Maller J. Purified maturation-promoting factor contains the product of a Xenopushomolog of the fission yeast cell cycle control gene cdc2+ Cell. 1988;54:433–439. doi: 10.1016/0092-8674(88)90206-1. [DOI] [PubMed] [Google Scholar]
- Gautier J, Minshull J, Lohka M, Glotzer M, Hunt T, Maller JL. Cyclin is a component of maturation-promoting factor from Xenopus. . Cell. 1990;60:487–494. doi: 10.1016/0092-8674(90)90599-a. [DOI] [PubMed] [Google Scholar]
- Hayles J, Fisher D, Woollard A, Nurse P. Temporal order of S phase and mitosis in fission yeast is determined by the state of the p34cdc2– mitotic B cyclin complex. Cell. 1994;78:813–822. doi: 10.1016/s0092-8674(94)90542-8. [DOI] [PubMed] [Google Scholar]
- Hennessy KM, Clark CD, Botstein D. Subcellular localization of yeast CDC46 varies with the cell cycle. Genes & Dev. 1990;4:2252–2263. doi: 10.1101/gad.4.12b.2252. [DOI] [PubMed] [Google Scholar]
- Hyrien O, Méchali M. Chromosomal replication initiates and terminates at random sequences but at regular intervals in the ribosomal DNA of Xenopusearly embryos. EMBO (Eur Mol Biol Organ) J. 1993;12:4511–4520. doi: 10.1002/j.1460-2075.1993.tb06140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson PK, Chevalier S, Philippe M, Kirschner MW. Early events in DNA replication require cyclin E and are blocked by p21CIP1 . J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearsey SE, Maiorano D, Holmes EC, Todorov IT. The role of MCM proteins in the cell cycle control of genome duplication. Bioessays. 1996;18:183–190. doi: 10.1002/bies.950180305. [DOI] [PubMed] [Google Scholar]
- Kimura H, Nozaki N, Sugimoto K. DNA polymerase alpha associated protein P1, a murine homolog of yeast MCM3, changes its intranuclear distribution during the DNA synthetic period. EMBO (Eur Mol Biol Organ) J. 1994;13:4311–4320. doi: 10.1002/j.1460-2075.1994.tb06751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Takisawa H. Determination of initiation of DNA replication before and after nuclear formation in Xenopusegg cell-free extracts. J Cell Biol. 1993;123:1321–1331. doi: 10.1083/jcb.123.6.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of XenopusDNA Replication Licensing Factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- Labbé JC, Capony JP, Caput D, Cavadore JC, Derancourt J, Kaghad M, Lelias JM, Picard A, Doree M. MPF from starfish oocytes at first meiotic metaphase is a heterodimer containing one molecule of cdc2 and one molecule of cyclin B. EMBO (Eur Mol Biol Organ) J. 1989a;8:3053–3058. doi: 10.1002/j.1460-2075.1989.tb08456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbé JC, Picard A, Peaucellier G, Cavadore JC, Nurse P, Doree M. Purification of MPF from starfish: identification as the H1 histone kinase p34cdc2 and a possible mechanism for its periodic activation. Cell. 1989b;57:253–263. doi: 10.1016/0092-8674(89)90963-x. [DOI] [PubMed] [Google Scholar]
- Lei M, Kawasaki Y, Tye BK. Physical interactions among Mcm proteins and effects of Mcm dosage on DNA replication in Saccharomyces cerevisiae. . Mol Cell Biol. 1996;16:5081–5090. doi: 10.1128/mcb.16.9.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leno GH, Downes CS, Laskey RA. The nuclear membrane prevents replication of human G2 nuclei but not G1 nuclei in Xenopusegg extract. Cell. 1992;69:151–158. doi: 10.1016/0092-8674(92)90126-w. [DOI] [PubMed] [Google Scholar]
- Lohka MJ, Masui Y. Effects of Ca2+ions on the formation of metaphase chromosomes and sperm pronuclei in cell-free preparations from unactivated Rana pipiens eggs. Dev Biol. 1984;103:434–442. doi: 10.1016/0012-1606(84)90331-2. [DOI] [PubMed] [Google Scholar]
- Luca FC, Ruderman JV. Control of programmed cyclin destruction in a cell-free system. J Cell Biol. 1989;109:1895–1909. doi: 10.1083/jcb.109.5.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca FC, Shibuya EK, Dohrmann CE, Ruderman JV. Both cyclin A delta 60 and B delta 97 are stable and arrest cells in M-phase, but only cyclin B delta 97 turns on cyclin destruction. EMBO (Eur Mol Biol Organ) J. 1991;10:4311–4320. doi: 10.1002/j.1460-2075.1991.tb05009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madine MA, Khoo C-Y, Mills AD, Laskey RA. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature (Lond) 1995a;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- Madine MA, Khoo C-Y, Mills AD, Musahl C, Laskey RA. Nuclear envelope prevents re-replication by restricting binding of MCM3 to chromatin. Curr Biol. 1995b;5:1270–1279. doi: 10.1016/s0960-9822(95)00253-3. [DOI] [PubMed] [Google Scholar]
- Mahbubani HM, Paull T, Elder JK, Blow JJ. DNA replication initiates at multiple sites on plasmid DNA in Xenopusegg extracts. Nucleic Acids Res. 1992;20:1457–1462. doi: 10.1093/nar/20.7.1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine GT, Sinha P, Tye BK. Mutants of S. cerevisiaedefective in the maintenance of minichromosomes. Genetics. 1984;106:365–385. doi: 10.1093/genetics/106.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti AK, Sinha P. The mcm2 mutation of yeast affects replication, rather than segregation or amplification of the two micron plasmid. J Mol Biol. 1992;224:545–558. doi: 10.1016/0022-2836(92)90543-s. [DOI] [PubMed] [Google Scholar]
- Moreno S, Nurse P. Regulation of progression through the G1 phase of the cell cycle by the rum1+ gene. Nature (Lond) 1994;367:236–242. doi: 10.1038/367236a0. [DOI] [PubMed] [Google Scholar]
- Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Cyclin-dependent protein kinases: key regulators of the eukaryotic cell cycle. Bioessays. 1995;17:471–480. doi: 10.1002/bies.950170603. [DOI] [PubMed] [Google Scholar]
- Piatti S, Bohm T, Cocker JH, Diffley JFX, Nasmyth K. Activation of S-phase-promoting CDKs in late G1 defines a “point of no return” after which Cdc6 synthesis cannot promote DNA replication in yeast. Genes & Dev. 1996;10:1516–1531. doi: 10.1101/gad.10.12.1516. [DOI] [PubMed] [Google Scholar]
- Rempel RE, Sleight SB, Maller JL. Maternal XenopusCdk2-cyclin E complexes function during meiotic and early embryonic cell cycles that lack a G1 phase. J Biol Chem. 1995;270:6843–6855. doi: 10.1074/jbc.270.12.6843. [DOI] [PubMed] [Google Scholar]
- Rowles A, Chong JPJ, Brown L, Howell M, Evan GI, Blow JJ. Interaction between the Origin Recognition Complex and the replication licensing system in Xenopus. . Cell. 1996;87:287–296. doi: 10.1016/s0092-8674(00)81346-x. [DOI] [PubMed] [Google Scholar]
- Roy LM, Swenson KI, Walker DH, Gabrielli BG, Li RS, Piwnica H, Worms, Maller JL. Activation of p34cdc2 kinase by cyclin A. J Cell Biol. 1991;113:507–514. doi: 10.1083/jcb.113.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan MA, Mills AD, Sleeman AM, Laskey RA, Blow JJ. Steps in the assembly of replication-competent nuclei in a cell-free system from Xenopuseggs. J Cell Biol. 1988;106:1–12. doi: 10.1083/jcb.106.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strausfeld UP, Howell M, Rempel R, Maller JL, Hunt T, Blow JJ. Cip1 blocks the initiation of DNA replication in Xenopusextracts by inhibition of cyclin-dependent kinases. Curr Biol. 1994;4:876–883. doi: 10.1016/s0960-9822(00)00196-2. [DOI] [PubMed] [Google Scholar]
- Strausfeld UP, Howell M, Descombes P, Chevalier S, Rempel RE, Adamczewski J, Maller JL, Hunt T, Blow JJ. Both cyclin A and cyclin E have S-phase promoting (SPF) activity in Xenopusegg extracts. J Cell Sci. 1996;109:1555–1563. doi: 10.1242/jcs.109.6.1555. [DOI] [PubMed] [Google Scholar]
- Todorov IT, Pepperkok R, Philipova RN, Kearsey SE, Ansorge W, Werner D. A human nuclear protein with sequence homology to a family of early S phase proteins is required for entry into S phase and for cell division. J Cell Sci. 1994;107:253–265. doi: 10.1242/jcs.107.1.253. [DOI] [PubMed] [Google Scholar]
- Todorov IT, Attaran A, Kearsey SE. BM28, a human member of the MCM2-3-5 family, is displaced from chromatin during DNA replication. J Cell Biol. 1995;129:1433–1445. doi: 10.1083/jcb.129.6.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treisman JE, Follette PJ, O'Farrell PH, Rubin GM. Cell proliferation and DNA replication defects in a DrosophilaMCM2 mutant. Genes & Dev. 1995;9:1709–1715. doi: 10.1101/gad.9.14.1709. [DOI] [PubMed] [Google Scholar]
- Usui T, Yoshida M, Abe K, Osada H, Isono K, Beppu T. Uncoupled cell cycle without mitosis induced by a protein kinase inhibitor, K-252a. J Cell Biol. 1991;115:1275–1282. doi: 10.1083/jcb.115.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vesely J, Havlicek L, Strand M, Blow JJ, Donnella-Deana A, Pinna L, Letham DS, Kato J, Detivaud L, Leclerc S, et al. Inhibition of cyclin- dependent kinases by purine analogues. Eur J Biochem. 1994;224:771–786. doi: 10.1111/j.1432-1033.1994.00771.x. [DOI] [PubMed] [Google Scholar]
- Yan H, Newport J. An analysis of the regulation of DNA synthesis by cdk2, Cip1, and licensing factor. J Cell Biol. 1995;129:1–15. doi: 10.1083/jcb.129.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Merchant AM, Tye BK. Cell cycle-regulated nuclear localization of MCM2 and MCM3, which are required for the initiation of DNA synthesis at chromosomal replication origins in yeast. Genes & Dev. 1993;7:2149–2160. doi: 10.1101/gad.7.11.2149. [DOI] [PubMed] [Google Scholar]
- Zhang SC, Masui Y. Activation of Xenopus laeviseggs in the absence of intracellular Ca activity by the protein phosphorylation inhibitor, 6-dimethylaminopurine (6-DMAP) J Exp Zool. 1992;262:317–329. doi: 10.1002/jez.1402620312. [DOI] [PubMed] [Google Scholar]