Abstract
The carboxy-terminal domain (CTD) of the largest subunit of RNA polymerase II (Pol II) contains multiple tandem copies of the consensus heptapeptide, TyrSerProThrSerProSer. Concomitant with transcription initiation the CTD is phosphorylated. Elongating polymerase has a hyperphosphorylated CTD, but the role of this modification is poorly understood. A recent study revealed that some hyperphosphorylated polymerase molecules (Pol IIo) are nonchromosomal, and hence transcriptionally unengaged (Bregman, D.B., L. Du, S. van der Zee, S.L. Warren. 1995. J. Cell Biol. 129: 287–298). Pol IIo was concentrated in discrete splicing factor domains, suggesting a possible relationship between CTD phosphorylation and splicing factors, but no evidence beyond immunolocalization data was provided to support this idea. Here, we show that Pol IIo co-immunoprecipitates with members of two classes of splicing factors, the Sm snRNPs and non-snRNP SerArg (SR) family proteins. Significantly, Pol IIo's association with splicing factors is maintained in the absence of pre-mRNA, and the polymerase need not be transcriptionally engaged. We also provide definitive evidence that hyperphosphorylation of Pol II's CTD is poorly correlated with its transcriptional activity. Using monoclonal antibodies (mAbs) H5 and H14, which are shown here to recognize phosphoepitopes on Pol II's CTD, we have quantitated the level of Pol IIo at different stages of the cell cycle. The level of Pol IIo is similar in interphase and mitotic cells, which are transcriptionally active and inactive, respectively. Finally, complexes containing Pol IIo and splicing factors can be prepared from mitotic as well as interphase cells. The experiments reported here establish that hyperphosphorylation of the CTD is a good indicator of polymerase's association with snRNP and SR splicing factors, but not of its transcriptional activity. Most importantly, the present study suggests that splicing factors may associate with the polymerase via the hyperphosphorylated CTD.
The largest subunit of RNA polymerase II (Pol II LS)1 is part of the catalytic core of the enzyme that synthesizes pre-mRNAs in all eukaryotic cells (for reviews see Young, 1991; Corden, 1993; Dahmus, 1996). The carboxy-terminal domain (CTD) of Pol II LS is comprised of tandemly repeated heptapeptides with the consensus sequence TyrSerProThrSerProSer (Corden et al., 1985). The CTD of mammals, D. melanogaster and S. cerevisiae have 52, 44, and 26 heptapeptide repeats, respectively (Corden et al., 1985; Allison et al., 1985; Zehring et al., 1988). In mammalian cells, Pol II LS exists as an unphosphorylated form (Pol IIa; M r = 220 kD), or one of a variety of hyperphosphorylated forms migrating at ∼240 kD (collectively called “Pol IIo”) (Cadena and Dahmus, 1987; Zhang and Corden, 1991).
The CTD harbors all known phosphorylation sites on eukaryotic Pol II LS molecules (reviewed in Corden, 1993; Dahmus, 1996). It has been repeatedly shown that Pol IIa is more efficiently recruited to in vitro pre-initiation complexes than Pol IIo (Lu et al., 1991; Chesnut et al., 1992), and that elongating Pol II is hyperphosphorylated on the CTD (Cadena and Dahmus, 1987; Payne et al., 1989; Laybourn and Dahmus, 1990; Chesnut et al., 1992; Weeks et al., 1993; O'Brien et al., 1994). CTD phosphorylation occurs concomitant with the initiation of transcription (Kang and Dahmus, 1993; Lu et al., 1991; Payne et al., 1989; Laybourn and Dahmus, 1990; Chesnut et al., 1992; Weeks et al., 1993). However, the bulk of evidence indicates that CTD phosphorylation is not needed for transcription. In the presence of CTD kinase inhibitors RNA polymerase II can synthesize RNA in vitro (Serizawa et al., 1993). In fact, Pol II molecules lacking a CTD can transcribe certain genes in vitro (Kim and Dahmus, 1989; Buratowski and Sharp, 1990; Zehring and Greenleaf, 1990). Although transcription from certain promoters requires a TFIIHassociated CTD kinase (Akoulitchev et al., 1995), an absolute requirement for CTD phosphorylation has yet to be demonstrated, since this kinase may phosphorylate transcriptional proteins besides Pol II's CTD. Indeed, basal and activated in vitro transcription were clearly shown to take place without CTD phosphorylation in another system (Mäkela et al., 1995). Therefore, despite the repeated observation that the CTD becomes phosphorylated as the polymerase begins to transcribe, it remains unclear whether CTD phosphorylation regulates Pol II's transcription cycle, or whether this modification merely coincides with the initiation of transcription.
Several putative CTD-binding proteins have been reported. Usheva and colleagues reported that Pol IIa, but not Pol IIo, interacts directly with TATA-binding protein (TBP), a key subunit of TFIID (Usheva et al., 1992). This finding, together with the timing of CTD phosphorylation, is consistent with the “promoter clearance” hypothesis, whereby phosphorylation of the CTD releases the polymerase from the promoter (discussed in Dahmus, 1996). In addition, cross-linking studies indicate that unphosphorylated CTD peptides directly interact with other transcription factors, viz., subunits of TFIIE and TFIIF (Kang and Dahmus, 1996). The CTD has also been shown to associate with a “mediator” complex containing multiple basal transcription factors and suppressers of RNA polymerase B (SRB) proteins (Thompson et al., 1993; Kim et al., 1994; Koleske and Young, 1994; for review see Koleske and Young, 1995). Pre-assembled “holoenzymes” containing an unphosphorylated CTD can be recruited to preinitiation complexes. The “mediator” promotes phosphorylation of the CTD via TFIIH-associated kinase(s), and it bestows upon polymerase the ability to respond to transactivator proteins in vitro. Thus, in agreement with the promoter clearance model of CTD function, transactivating proteins may stimulate the mediator to activate CTD phosphorylation coincident with transcriptional initiation. The mediator protein(s) that directly bind to the unphosphorylated CTD have yet to be identified. In addition, at least one other CTD kinase, apparently not part of the mediator, has been shown to be required for normal CTD phosphorylation in vivo (Sterner et al., 1995; Lee and Greenleaf, 1991). Finally, Yuryev and colleagues used a yeast two-hybrid screen to identify CTD interacting proteins in rat cells (Yuryev et al., 1996). Four proteins were identified, each containing repetitive SerArg dipeptide (SR) motifs characteristic of the SR superfamily of proteins. Among the putative CTD-binding proteins reported to date, rA1 is the only one that interacts with Pol II molecules containing a hyperphosphorylated CTD.
Many studies indicate a close correlation between CTD phosphorylation and the initiation of transcription, but little is known about the phosphorylation state or fate of Pol II molecules that have disengaged from the chromatin. Recently, a substantial fraction of Pol IIo was shown to be concentrated in discrete nuclear domains that overlap with 20–50 “speckle” domains, which are enriched with splicing factors (Bregman et al., 1994, 1995). Studies from other labs indicated that the bulk of Pol II transcription takes place in the nucleoplasm outside of these domains (Wansink et al., 1993; Jackson et al., 1993), so it was proposed that much of the Pol IIo in these domains is probably not engaged in transcription (Bregman et al., 1995). Since that time, a handful of specific Pol II transcripts have been localized to the periphery of the speckle domains, and some transcripts (e.g., collagen Iα1) overlap extensively with the speckle domains. Thus, the precise spatial relationship between Pol II transcription and the speckle domains remains poorly understood, and the interested reader is referred to several recent reviews of this topic (Fakan, 1994; Wansink et al., 1994; Bazett-Jones, 1995; Moen et al., 1995; Spector et al., 1995; van Driel et al., 1995). Nevertheless, our finding that transcriptional inhibitors and heat shock induce multiple SR proteins, Sm snRNPs, and Pol IIo to synchronously reorganize into enlarged, round speckles, suggests that some Pol IIo molecules in these domains are not engaged in transcription. A similar, but more striking reorganization takes place at the onset of mitosis when transcription abruptly ceases: Pol IIo, multiple SR proteins and Sm snRNPs become sequestered in mitotic interchromatin granule clusters (MIGs), dot-like regions (∼0.1 μm – ∼1 μm in diameter) scattered throughout the dividing cell, far removed from the condensed chromatin (Warren et al., 1992; Bregman et al., 1994).
Our previous immunolocalization studies indicated that splicing factors and transcriptionally unengaged Pol IIo molecules are sequestered in the same discrete nonchromosomal compartment when transcription diminishes or ceases (Bregman et al., 1995). Here, we show that Pol IIo co-immunoprecipitates with splicing factors of the Sm snRNP and non-snRNP SerArg (RS) families. We also report two novel and intriguing findings. First, splicing factors are tightly associated with Pol IIo in the absence of pre-mRNA; and second, the hyperphosphorylated polymerase need not be transcriptionally engaged to associate with splicing factors. Perhaps the most important implication of these findings is that the splicing factors associate with polymerase via the phosphorylated CTD. In agreement with the above findings, we also provide definitive evidence that hyperphosphorylation of the CTD correlates poorly with Pol II's transcriptional activity in vivo. In summary, we have established for the first time that CTD phosphorylation is a better indicator of Pol II's association with Sm snRNPs and SR proteins than its transcriptional activity. The outcome of the experiments described here prompted us to ask whether a functional relationship exists between Pol II's CTD and pre-mRNA splicing, an idea explored in an accompanying paper (Du and Warren, 1997).
Materials and Methods
Antibodies
Anti-Pol II LS mAbs (H5, H14 and 8WG16) and control IgM (mAb H22) are described elsewhere (Bregman et al., 1995; Thompson et al., 1989). Anti-RPII is a rabbit polyclonal antibody directed at Pol II LS (Kim and Dahmus, 1986). Anti-NuMa is a polyclonal rabbit antibody directed at the 236-kD nuclear mitotic apparatus protein (Yang et al., 1992). The following mAbs are directed towards Sm antigens and SR family proteins: mAb Y12 (Lerner et al., 1981; Pinto and Steitz, 1989) recognizes the Sm snRNP B/B′ and D, as well as a 70-kD proteolytic fragment of intron-binding protein (IBP). mAb 7.13 specifically recognizes Sm snRNP protein D (Bloom et al., 1993). mAb 104 (Roth et al., 1990) and mAb 3C5 (Turner and Franchi, 1987) are IgMs that recognize several members of the SR family. mAb SC35 is an IgG directed against spliceosome assembly factor, a 35-kD SR family protein (Fu and Maniatis, 1990). mAb B4A11 is a mouse IgM that intensely stains nuclear speckle domains (Blencowe et al., 1994), and mAb 3A7/64 is a mouse IgG directed at the 64-kD subunit of the cleavage stimulation factor (CstF) involved in 3′ polyadenylation and processing of premRNAs (Takagaki et al., 1990). mAb M2 (Kodak) binds to the FlagR peptide, AspTyrLysAspAspAspAspLys.
Cell Culture, Immunofluorescence Microscopy, and Image Analysis
Cells were maintained as described (Bregman et al., 1994). Immunostaining was performed as described (Bregman et al., 1994). To visualize mitotic chromosomes, cells were stained with 4′, 6-diamidino-2-phenylindole (DAPI) as described (Warren et al., 1992). Images were captured using a Photometrics CH250 CCD camera (CE200A/LC200 liquid cooling), mounted on an Axioskop (Zeiss). Image analysis software included NIHImageR, Registration v1.1d2R, and Adobe Photoshop 2.0R.
CTD Peptide Expression Plasmids and Transient Transfection
Complete descriptions of pF-CTD52 and pF-CTDless are given in the accompanying manuscript (Du and Warren, 1997). pF-CTD52 expresses a fusion protein comprised of an NH2-terminal Flag peptideR epitope (AspTyrLysAspAspAspAspLys) attached to 636 amino acids derived from the COOH terminus of human Pol II LS, whereas “pF-CTDless” expresses a “non-CTD” segment of Pol II LS (residues 1335 to 1570) that is Flagtagged in the NH2 terminus. Cells were transfected using LipofectinR (GIBCO BRL, Gaithersburg, MD), according to the manufacturer's instructions.
Cyanogen Bromide Digestion
Pol II was cyanogen bromide (CNBr) digested as described (Cadena and Dahmus, 1987).
Centrifugal Elutriation, Preparation of Nocodazole-arrested Mitotic Cells and Flow Cytometric Cell Cycle Analysis
Performed as described (Bregman et al., 1994; Tobey et al., 1967). Nocodazole-arrested, mechanically detached mitotic cells were judged to be 99% pure by counting ∼200 DAPI-stained mitotic figures under the fluorescence microscope.
Nuclear Extracts, Immunoprecipitation, and Immunoblotting
HeLa cell nuclei (Dignam et al., 1983) were extracted with ice-cold 50 mM Tris-HCl, 250 mM NaCl, 1% Triton X-100, 1 mM PMSF, 0.2 mM NaVO4, 5 mM β-glycerophosphate, pH 7.4 (T buffer). Insoluble material was centrifuged for 10 min at 15,000 g, and the supernatant was used for immunoprecipitation. Nuclear extracts were incubated with 50 μl of staphylococcal protein G coupled–agarose beads, precharged with an excess of antibody. After incubation for 4 h at 4°C, the beads were washed three times with ice cold T buffer, boiled in SDS sample buffer and subjected to SDS-PAGE and immunoblotting. Immunoblotting was performed as described (Bregman et al., 1995).
Metabolic Labeling with [32P]Orthophosphate, Immunoprecipitation, RNase A Digestion, and Analysis of RNAs
HeLa cells were grown for 24 h in phosphate-free RPMI 1640 containing 10% dialyzed FBS, 1% undialyzed FBS, and [32P]orthophosphate (0.5 mCi/ml). Cells were washed three times in ice-cold PBS, lysed in T Buffer (see above), and centrifuged to remove insoluble material. The supernatant was used for immunoprecipitations as described above. To determine the content of 32P-labeled RNA in the complexes, immunoprecipitates were incubated with or without RNase A (5 μg/ml), phenol extracted, ethanol precipitated, rehydrated in loading buffer, and resolved on a 5% or 10% polyacrylamide/urea gel as described (Sambrook et al., 1989). The gels were dried and processed for autoradiography.
Results
mAbs H5 and H14 Bind to Different Phosphoepitopes on Pol II's CTD
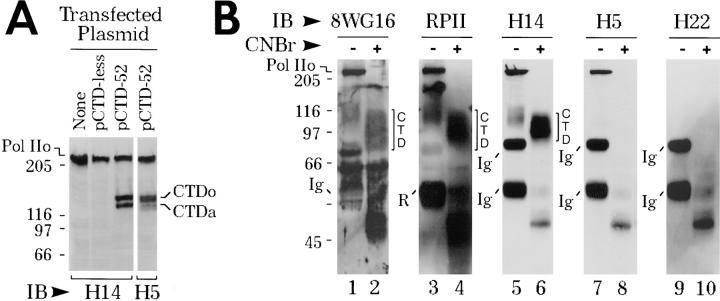
In a previous study we showed that pretreatment of Pol II LS with alkaline phosphatase abolishes binding of mAbs H5 and H14 (Bregman et al., 1995), suggesting that these mAbs may recognize phosphoepitopes on the CTD (Cadena and Dahmus, 1987; Zhang and Corden, 1991). However, we had no direct evidence to support this idea. Here, we ask whether mAbs H5 and H14 bind to recombinant human CTD-derived proteins expressed in HeLa cells (Materials and Methods). Interestingly, the recombinant CTD proteins lack a conventional nuclear localization signal, but they still accumulate in the nucleus (see Du and Warren, 1997). Furthermore, CTD proteins are apparently phosphorylated in vivo by endogenous protein kinase(s) to yield two species, “CTDa” and “CTDo” (Fig. 1 A). Both antibodies recognize the full-length CTD (52 repeats) (Fig. 1 A). Note that both mAbs bind strongly to CTDo, whereas only mAb H14 blots strongly to CTDa (Fig. 1 A). Finally, in vitro pre-incubation of these CTDs with alkaline phosphatase abolishes binding by either mAb (data not shown).
Figure 1.
mAbs H5 and H14 bind to the CTD of RNA polymerase II. (A) mAbs H5 and H14 recognize recombinant CTD proteins expressed in HeLa cells. HeLa cells were transfected with pF-CTDless or pF-CTD52 (see Materials and Methods). 24 h later, the cells were solubilized with SDS sample buffer, subjected to 7% SDS-PAGE, and immunoblotted with mAb H5 or mAb H14. (B) mAb H14 binds to CTD of native RNA polymerase II. Pol II LS was immunoprecipitated with mAb H14 and digested with CNBr (Cadena and Dahmus, 1987). CNBr digested material was solubilized in SDS sample buffer, subjected to 7% SDS-PAGE, and immunoblotted with mAb 8WG16, RPII (polyclonal rabbit anti-Pol II; Kim and Dahmus, 1986), mAb H14, mAb H5, and mAb H22 (control IgM). IB, immunoblotting antibody. Ig, mouse immunoglobulin chains; R, rabbit immunoglobulin chains. CNBr, cyanogen bromide. CNBr-released CTD indicated by brackets on the right hand side of gels. Pol IIo, hyperphosphorylated largest subunit of Pol II. Numbers at margins of panels indicate apparent molecular weights in kilodaltons.
Next, we asked whether mAbs H5 and H14 also bind to CTD epitopes on native Pol IIo molecules. Pol II was immunoprecipitated from HeLa cells with mAb H14 and digested with CNBr to prepare intact CTD fragments (Cadena and Dahmus, 1987). The immunoprecipitated, CNBr-digested Pol II molecules were subjected to SDSPAGE and blotted with two antibodies known to recognize the CTD: mAb 8WG16 (Thompson et al., 1989) and anti-RPII (Kim and Dahmus, 1986) (Fig. 1 B, lanes 2 and 4, respectively; CTD fragment is indicated by a bracket). mAb H14 also recognizes the chemically excised CTD peptide (Fig. 1 B, lane 6, bracket). However, the H5 phosphoepitope is destroyed during the CNBr procedure (Fig. 1 B, lane 8). Taken together, the above results indicate that mAbs H5 and H14 recognize distinct phosphoepitopes in Pol II's CTD.
It is well established that the CTD is phosphorylated concomitant with the initiation of transcription (see Introduction). In a study to be reported elsewhere, we have used an in situ photoaffinity labeling technique (Bartholomew et al., 1986) to show that mAbs H5 and H14 recognize a population of elongating Pol IIo molecules (Zeng et al., 1997). Indeed, multiple species of Pol II LS, which are differentially phosphorylated on the CTD, are transcriptionally active under the conditions of the nuclear run-on assay (Zeng et al., 1997). These findings are in agreement with previous transcription studies performed in vitro (Kang and Dahmus, 1993; Lu et al., 1991; Payne et al., 1989; Laybourn and Dahmus, 1990; Chesnut et al., 1992) and in vivo (O'Brien et al., 1994; Weeks et al., 1993).
The Steady-state Level of CTD Phosphorylation Is Relatively Constant during the Cell Cycle
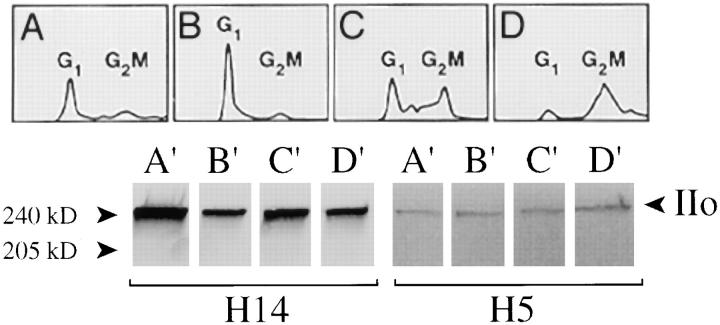
The overall level of Pol II transcription fluctuates widely during the cell cycle (Taylor, 1960; Prescott and Bender, 1962; Shermoen and O'Farrell, 1991). An abrupt cessation of transcription takes place at the onset of mitosis (Shermoen and O'Farrell, 1991; Ferreira et al., 1994), and abundant nonchromosomal Pol II is present during mitosis (Warren et al., 1992; Bregman et al., 1994). Pol II transcription resumes during late telophase and continues until G2/M (Ferreira et al., 1994). To better understand the functional significance of CTD phosphorylation, we asked whether the level of Pol IIo fluctuates in parallel with marked changes in transcriptional activity that takes place during the cell cycle (Fig. 2). HeLa cells were separated by centrifugal elutriation into ∼10 fractions, which were analyzed by flow cytometry. Four representative cell cycle– enriched fractions are presented in Fig. 2, A–D. An equal number of cells from each fraction was solubilized in SDS sample buffer, subjected to SDS-PAGE and immunoblotted with mAb H14 or mAb H5 (Fig. 2, A′–D′, bottom panels). Quantitative scanning of these blots reveals less than a twofold fluctuation in the level of Pol IIo as determined by blotting with mAb H14, and less than a 5% fluctuation with mAb H5. Significantly, the G2/M-enriched cells have an overall level of Pol IIo that is very similar to the G1 enriched cells (Fig. 2, compare B′ and D′). Similar results have been obtained using pharmacologically synchronized G1, S, and M phase cell populations (Kim, E., L. Du, D.B. Bregman, and S.L. Warren, unpublished results). We conclude that phosphorylation of the CTD does not correlate well with Pol II's transcriptional activity.
Figure 2.
Overall CTD phosphorylation remains relatively constant during the cell cycle. S3 HeLa cells were separated by centrifugal elutriation as described (Bregman et al., 1994), and 10 fractions were analyzed by flow cytometry. An equal number of cells from each fraction was boiled in SDS sample buffer, electrophoresed, and blotted with mAb H14 or H5. Four representative fractions, each enriched for a different stage of the cell cycle, are presented in A–D (top). Lower panels A′–D′ correspond to upper panels A–D. IIo, hyperphosphorylated largest subunit of RNA polymerase II. Numbers at margins of panels indicate apparent molecular weights in kilodaltons.
Pol IIo Is Associated with Sm snRNPs and SR Proteins In Vivo
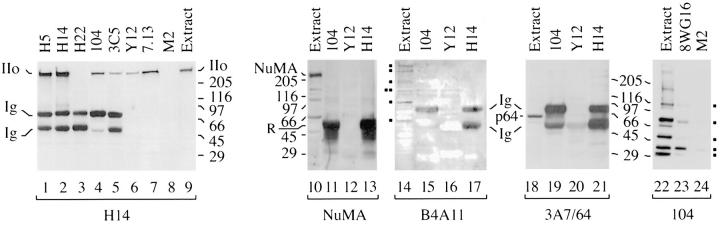
Our previous observations that Pol IIo and multiple splicing factors are colocalized in nuclear domains (Bregman et al., 1994, 1995) prompted us to ask whether Pol IIo can be co-immunoprecipitated with splicing factors. Antibodies that bind to a variety of snRNPs and SR family proteins were used to immunoprecipitate complexes from HeLa cell nuclear extracts under nondenaturing conditions (Fig. 3). First, the presence of SR proteins and Sm snRNPs in the complexes was confirmed by immunoblotting; mAb Y12 immunoprecipitates the B/B′ Sm snRNPs, a ∼70-kD proteolytic fragment of the intron-binding protein (Pinto and Steitz, 1989), and several SR family proteins recognized by mAb 104 (data not shown). In addition, complexes prepared with mAbs 104 and 3C5 contain multiple SR family proteins as shown previously (Roth et al., 1991; Turner and Franchi, 1987; Blencowe et al., 1995; data not shown). To ascertain whether these previously reported immunoprecipitates also contain Pol IIo, additional samples were electrophoresed and immunoblotted with mAb H14 (Fig. 3, lanes 4–7). The results demonstrate that Pol IIo is co-immunoprecipitated with two mAbs directed at SR family proteins (Fig. 3, lanes 4 and 5). Pol IIo is also co-immunoprecipitated with mAb 7.13, which binds to the Sm snRNP D (Bloom et al., 1993) (Fig. 3, lane 7), and mAb Y12, which binds to Sm snRNPs B and B′ (Fig. 3, lane 6). We have compared the amount of Pol IIo immunoprecipitated by an excess of the following mAbs: 104, 3C5, Y12, 7.13, and H14. Quantitative scanning of three immunoblots similar to Fig. 3 indicates that the anti-Sm snRNP and anti-SR antibodies immunoprecipitate 5–20% as much Pol IIo from nuclear extracts as does anti-Pol IIo mAb H14 (Kim, E., L. Du, D.B. Bregman, and S.L. Warren, unpublished results).
Figure 3.
The largest subunit of RNA polymerase II is associated with SR family proteins and Sm snRNPs. HeLa cell nuclear extracts were used for immunoprecipitations as described in the Materials and Methods. Immunoprecipitations with mAbs Y12 (anti-Sm), H5 (antiPol II CTD), H14 (anti-Pol II CTD), H22 (control IgM), 104 (anti-SR family proteins), and 3C5 (anti-SR family proteins), 7.13 (antiSm) and M2 (control IgG) were washed, boiled in SDS sample buffer, electrophoresed, and immunoblotted with mAb H14 (lanes 1–9), anti-NuMA (lanes 10–13), mAb B4A11 (lanes 14–17), mAb 3A7/64 (lanes 18–21), or mAb 104 (lanes 14–16). Blocks at left of lane 14, proteins recognized by nuclear matrix antibody B4A11. Blocks at right margin of lane 24, mAb 104-reactive SR proteins. Ig, mouse immunoglobulin μ chains, present only in samples immunoprecipitated with IgM antibodies; R, rabbit anti-mouse immunoglobulin added to immunoprecipitates and detected only in anti-NuMA blot. IIo, hyperphosphorylated form(s) of the largest subunit of RNA polymerase II. p64, cleavage stimulation factor (64 kD subunit). NuMA, nuclear mitotic apparatus protein, 236 kD. Numbers at margins of panels indicate apparent molecular weights in kilodaltons.
Several control immunoprecipitations were performed to determine whether or not other nuclear proteins are present in these complexes (Fig. 3, lanes 10–21). One control is the nuclear mitotic apparatus protein (NuMA), a 236-kD protein, which has been reported to exist in a diffuse nucleoplasmic distribution (Yang et al., 1992; Compton et al., 1992) and in nuclear speckle domains during interphase (Zeng et al., 1994). NuMa is abundant in whole nuclear extracts (Fig. 3, lane 10), but it is not detected in immunoprecipitates prepared with mAbs 104, Y12, or H14 (Fig. 3, lanes 11–13). Another control, mAb B4A11, recognizes a ∼300-kD nuclear matrix antigen and intensely stains speckle domains (Blencowe et al., 1994). Neither the ∼300-kD protein, or the other proteins recognized by mAb B4A11 were found to co-immunoprecipitate with mAbs 104, Y12, or H14 (Fig. 3, lanes 14–17). A third control, mAb 3A7/64, recognizes the 64-kD subunit of cleavage stimulation factor (CstF), which participates in the processing of 3′ ends of pre-mRNAs (Takagaki et al., 1990). The p64 protein is readily detected in nuclear extracts, but not in immunoprecipitates prepared with mAbs 104, Y12, or H14 (Fig. 3, lanes 19–21). Finally, antiNuMA, mAb B4A11, mAb 3A7/64, and several additional control mAbs directed at nuclear proteins failed to co-immunoprecipitate Pol IIo (Kim, E., L. Du, D.B. Bregman, and S.L. Warren, unpublished results). The above controls clearly show the specificity of Pol IIo's association with the splicing factors (Fig. 3, lanes 4–7).
Repeated attempts to co-immunoprecipitate Sm snRNPs and SR proteins with mAbs H5 and H14 failed. In fact, in the presence of mAb H5 or mAb H14 (IgMs), Pol IIo is not co-immunoprecipitated with mAb Y12 (IgG) implying that these CTD-specific mAbs cause Pol IIo to be released from the complexes (unpublished results). In an attempt to co-immunoprecipitate SR proteins with a different antibody directed at Pol II, we chose mAb 8WG16. In contrast to mAbs H5 and H14, which are multivalent IgMs directed at phosphoepitopes on the CTD, mAb 8WG16 is a bivalent IgG directed against unphosphorylated epitope(s) on the CTD. To interpret such an experiment, it is essential to remember that even “hyperphosphorylated” Pol II molecules (i.e., Pol IIo species) contain many unphosphorylated serine and threonine residues on the CTD (Zhang and Corden, 1991). Thus, although mAb 8WG16 immunoprecipitates and blots predominantly hypophosphorylated Pol IIa molecules (∼220 kD), it also immunoprecipitates and blots Pol IIo molecules (240 kD), albeit weakly (see Bregman et al., 1995). In contrast to mAbs H5 and H14, mAb 8WG16 co-immunoprecipitates several mAb 104-reactive SR proteins (Fig. 3, lane 23).
Anti-SR Specific mAb 104 Preferentially Co-immunoprecipitates Pol IIo
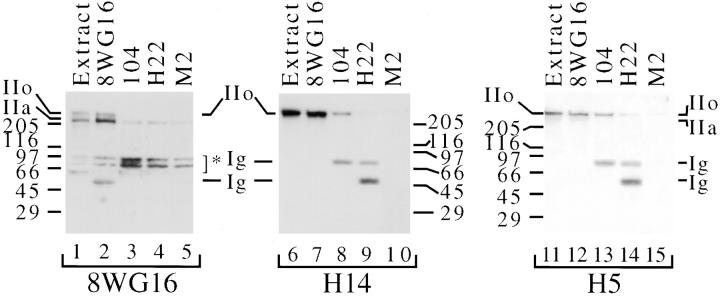
Next, we asked whether the Pol II LS molecules that are associated with SR proteins have a hyperphosphorylated or hypophosphorylated CTD (Fig. 4). To answer this question, we have again taken advantage of mAbs H5, H14, and 8WG16, which recognized different phosphorylated forms of Pol II LS (Bregman et al., 1995). mAb H5 binds specifically to a subset of Pol IIo molecules, which migrate at ∼240 kD (Bregman et al., 1995), whereas mAb H14 recognizes various phosphorylated forms that migrate between 220 and 240 kD (Bregman et al., 1995). mAb 8WG16 binds to unphosphorylated CTD epitopes (Thompson et al., 1989; Bregman et al., 1995), so it preferentially binds to Pol IIa (∼220 kD). Recall that although mAb 8WG16 can recognize Pol IIo, mAbs H5 and H14 are much more sensitive probes for Pol IIo.
Figure 4.
Anti-SR specific mAb 104 preferentially co-immunoprecipitates Pol IIo. Nuclear extracts and immunoprecipitations were prepared as in Fig. 3. Immunoprecipitations were performed using mAbs indicated at the top of the panels and immunoblotting with mAbs at the bottom. Extract, HeLa cell nuclear extract. Ig, mouse immunoglobulin chains; IIo, hyperphosphorylated largest subunit of RNA polymerase II. IIa, hypophosphorylated largest subunit of RNA polymerase II. Numbers at margins of panels indicate apparent molecular weights in kilodaltons. Asterisks indicate mAb 8WG16 cross-reactive proteins at ∼100 kD and ∼85 kD.
HeLa cell nuclear extracts were incubated with mAb 8WG16, mAb 104, mAb H22 (IgM control), or mAb M2 (IgG control). Immune complexes were captured on agarose beads, washed, boiled in SDS sample buffer, electrophoresed, and immunoblotted with mAb 8WG16, mAb H14, or mAb H5 (Fig. 4). The results show that mAb 104 co-immunoprecipitates Pol IIo (Fig. 4, lanes 8 and 13). Immunoblotting with mAb 8WG16 reveals only background levels of Pol IIa in the complexes (Fig. 4, compare lane 3 to lanes 4 and 5). The Pol IIo molecules which are co-immunoprecipitated with mAb 104 can be easily detected by mAbs H5 and H14, but mAb 8WG16 does not detect this subset of Pol IIo. The amount of Pol IIo in these complexes may be insufficient to be detected by mAb 8WG16. Alternatively, mAb 104 may co-immunoprecipitate a subset of Pol IIo molecules that has few unphosphorylated serine and threonine residues in the CTD.
Pol IIo's Association with Sm snRNPs and Certain SR Proteins Resists Exhaustive RNase Digestion
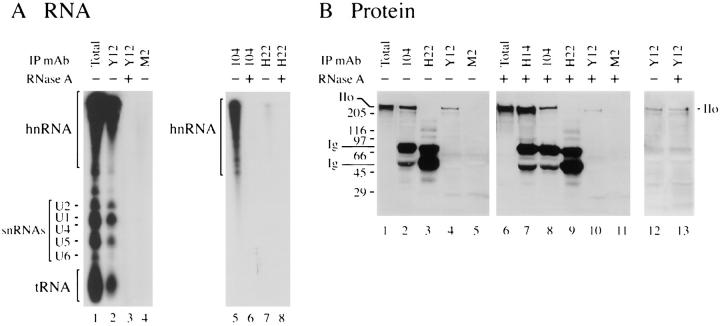
One possible explanation for the above results is that the Pol IIo/SR protein/Sm snRNP “complexes” are transcriptionally engaged Pol IIo molecules connected to snRNPs and SR proteins via partially transcribed pre-mRNAs. If this is true, then RNase digestion should release Pol IIo from the immunoprecipitated complexes. To test this idea, we first analyzed the RNA content in the immunoprecipitated material (Fig. 5 A). HeLa cells were metabolically labeled with [32P]orthophosphate, washed in PBS and extracted with T buffer (see Materials and Methods). The supernatant was used for immunoprecipitation with mAb Y12 (directed at Sm snRNPs) and mAb 104 (directed at SR family proteins), under nondenaturing conditions as in Figs. 3 and 4.
Figure 5.
Pol IIo's association with SR proteins and Sm snRNPs resists RNase digestion. HeLa cells were grown overnight in low phosphate medium supplemented with [32P]orthophosphate (0.5 mCi/ml). The cells were extracted for immunoprecipitation as in Figs. 3 and 4. Immunoprecipitating antibodies include Y12 (anti-Sm snRNP), 104 (anti-SR family proteins), H14 (anti-Pol II), M2 (control IgG), and H22 (control IgM). Immunoprecipitates were washed extensively, and then incubated with RNase (+) or without RNase (−) as described in Materials and Methods. (A) RNA content of immunoprecipitates before and after RNase treatment. Immunoprecipitates prepared from 32P-labeled cells were phenol extracted, ethanol precipitated, rehydrated in loading buffer, and resolved on a 5% (left panel) or 10% (right panel) polyacrylamide/urea gel. The gels were dried and processed for autoradiography. hnRNA, heterogeneous RNAs. U1, U2, U4, U5, and U6, U-rich Sm snRNAs. tRNA, transfer RNA. (B) Proteins in immunoprecipitates before and after RNase treatment. Immunoprecipitates prepared under nonradioactive, but otherwise identical conditions, to those in A. After the RNase digestion step, samples were boiled in SDS sample buffer, subjected to 7% SDS-PAGE electrophoresis, transferred to nitrocellulose, and immunoblotted with anti-Pol II mAb H14. Ig, mouse immunoglobulin chains; IIo, hyperphosphorylated Pol II. Numbers at margins of panels indicate apparent molecular weights in kilodaltons.
The results show that mAb Y12 co-immunoprecipitates multiple RNA species, including hnRNA and Sm snRNAs, whereas mAb 104 co-immunoprecipitates predominantly hnRNA (Fig. 5 A, lanes 2 and 5, respectively). Significantly, all 32P-labeled RNA species are destroyed by incubating the immunoprecipitated complexes with RNAase A (Fig. 5 A, lanes 3 and 6, respectively). Despite the exhaustive RNase A treatment, the amount of Pol IIo which co-immunoprecipitate with the SR proteins is not diminished significantly (Fig. 5 B, compare lanes 2 and 8). RNAase treatment reduced the amount of Pol IIo co-immunoprecipitating with Sm snRNPs in some experiments (Fig. 5 B, compare lanes 4 and 10), while minimal changes were detected in other experiments (Fig. 5 B, compare lanes 12 and 13).
A Hyperphosphorylated Form of Pol II LS Is Associated with Sm snRNPs and SR Family Proteins during Mitosis
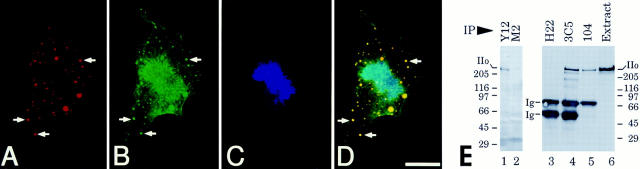
The above results argue that Pol IIo's association with mAb 104 reactive SR proteins and Sm snRNPs may not be dependent on pre-mRNA. To further explore this idea, we have taken advantage of mitotic cells, which do not synthesize RNA (Taylor, 1960; Prescott and Bender, 1962; Ferreira et al., 1994). In addition, nascent Pol II transcripts are aborted during mitosis (Shermoen and O'Farrell, 1991) and Pol II accumulates in nonchromosomal MIGs (Warren et al., 1992; Bregman et al., 1994). First, we compared the subcellular distributions of condensed mitotic chromosomes, Pol IIo and SC35 (a representative SR family splicing protein in MIGs) using imaging techniques that are superior to our earlier studies (Fig. 6, A–D). Prometaphase chromosomes are clearly visible as indicated by blue pseudocolor (Fig. 6 C). As shown by red pseudocolor in Fig. 6 A, SC35 is distributed exclusively in nonchromosomal dot-like MIGs, shown previously to harbor SR family proteins and Sm snRNPs (Spector et al., 1991; Ferreira et al., 1994). The distribution of mAb H5 reactive Pol IIo is more complex (Fig. 6 B, green pseudocolor). Note that SC35 and Pol IIo are precisely colocalized in the dot-like MIGs as indicated in the merged image (Fig. 6 D, yellow). This population of Pol IIo molecules is not associated with chromosomes. Another population of Pol IIo is distributed in a perichromosomal pattern (Fig. 6, B and D). Neither population of the polymerase can be transcriptionally engaged at this stage of the cell cycle, but all of the Pol II visualized in Fig. 6 is hyperphosphorylated on the CTD (mAb H5 recognizes only Pol IIo). Significantly, there is a striking colocalization of SC35 and Pol IIo in the MIGs.
Figure 6.
Pol IIo is associated with splicing factors in nonchromosomal domains of mitotic cells. (A–D) Colocalization of Pol IIo and SC35 in mitotic interchromatin granule clusters (MIGs). Mitotic MDCK cells were fixed with 1.75% paraformaldehyde, permeabilized with 0.5% Triton X-100 and triple stained using anti-spliceosome assembly factor, mAb SC35 (A), anti-Pol IIo (B), and DNA-binding dye, 4′, 6-diamidino-2-phenylindole (DAPI) (C). Digital images were pseudocolored and merged using Registration v1.1d2R image analysis software (see Materials and Methods). Arrows, mitotic interchromatin granule clusters (MIGs). Note that the SC35/Pol IIo enriched MIGs are clearly separate from the DAPI-stained prometaphase chromosomes. Another population of Pol IIo is distributed in a stocking-like pattern which surrounds the individual chromosomes (B and D). Similar results were obtained using HeLa and many other types of mammalian cells (Kim, E., L. Du, D.B. Bregman, and S.L. Warren, unpublished results). (E) Co-immunoprecipitation of Pol IIo, Sm snRNPs, and SR family proteins from mitotic cell extracts. Pure mitotic HeLa cell extracts were prepared as described (see Materials and Methods). Immunoprecipitations were performed using the mAbs listed at top of panel: Y12 (anti-Sm snRNP), 3C5 and 104 (anti-SR family proteins), M2 (control IgG), and H22 (control IgM). Extract, HeLa cell mitotic extract. Immunoprecipitated material was boiled in SDS sample buffer, electrophoresed, and immunoblotted with anti-Pol II mAb H14. Ig, mouse immunoglobulin μ chains; IIo, hyperphosphorylated form(s) of the largest subunit of RNA polymerase II. Numbers at margins of panels indicate apparent molecular weights in kilodaltons.
The above results prompted us to ask whether Pol IIo can be co-immunoprecipitated with SR proteins and Sm snRNPs from mitotic cells. Thus, we prepared 99% pure mitotic HeLa cell extracts (Materials and Methods), which were used for immunoprecipitation with mAbs Y12, 3C5, 104, M2 (control IgG), and H22 (control IgM). The immunoprecipitates were washed extensively, boiled in SDS sample buffer, subjected to SDS-PAGE, and blotted with mAb H14 (Fig. 6 E). The results show that Pol IIo can be co-immunoprecipitated with Sm snRNPs and SR proteins during mitosis (Fig. 6, lanes 1, 4, and 5). Similar results were obtained by blotting with mAb H5 (data not shown). Note that in pure mitotic cell extracts Pol IIo migrates as a thin ∼240-kD band (Fig. 6 E, lane 6), in agreement with the results obtained with G2/M-enriched cells prepared by elutriation (Fig. 2 D′). These experiments, together with previous studies, indicate that “megacomplexes” comprised of SR family proteins, Sm snRNPs, and hyperphosphorylated Pol II molecules are sequestered in discrete nonchromosomal domains (i.e., MIGs in mitotic cells and speckle domains in the interphase nucleus).
Discussion
In the present study, we first established that mAbs H5 and H14 bind to phosphoepitopes on Pol II's CTD (see Fig. 1). As expected from previous studies, these phosphoepitopes are present on transcriptionally engaged polymerase molecules (Zeng et al., 1997). But hyperphosphorylation of the CTD does not necessarily indicate that Pol II is transcriptionally engaged, since interphase cells and transcriptionally inactive mitotic cells have similar levels of Pol IIo (see Fig. 2). Taken together with the immunolocalization results on mitotic (Fig. 6) and interphase cells (Bregman et al., 1995), these data indicate that one fraction of Pol IIo molecules is transcriptionally engaged, while another fraction is not. The latter conclusion is further supported by recent immunolocalization studies using Triton X-100–permeabilized, transcriptionally active cells in which the majority of nascent, BrU-labeled transcripts are located in the nucleoplasm outside (or at the periphery) of the large Pol IIo-rich domains (Zeng et al., 1997). Thus, hyperphosphorylation of the CTD is poorly correlated with transcriptional activity, even though CTD phosphorylation takes place at the time when polymerase initiates transcription.
The discrete Pol IIo-rich domains are highly enriched with splicing factors (Bregman et al., 1995). These morphological observations led us to ask whether Pol IIo can be co-immunoprecipitated with Sm snRNPs and/or SR proteins. The results of the present study clearly show that Pol II molecules containing a hyperphosphorylated CTD associate with two classes of splicing factors, the Sm snRNPs and non-snRNP SR proteins (Figs. 3 and 4). Significantly, these associations resist exhaustive in vitro digestion with RNase A (Fig. 5), and Pol IIo co-immunoprecipitates with Sm snRNPs and SR proteins during mitosis, when transcription is inactive (Fig. 6). Taken together, these results indicate that transcriptionally unengaged, hyperphosphorylated Pol II molecules are associated with Sm snRNPs and certain SR proteins without the involvement of premRNA. Sm snRNPs and SR proteins may also be associated with transcriptionally engaged Pol IIo molecules, but this hypothesis cannot be tested directly at the present time since our reagents cannot distinguish transcriptionally active from unengaged Pol II molecules. Interestingly, a correlation between hyperphosphorylation of the CTD and in vivo splicing was made previously in a study of polytene nuclei in Drosophila, where a splicing factor was localized only at chromosomal sites containing Pol IIo (Weeks et al., 1993).
How can the results of the present study be reconciled with the mounting evidence that the CTD has a transcriptional function? First, the majority of studies indicating a transcriptional role for the CTD have focused on early steps of transcription; indeed, interactions involving the CTD during elongation and after Pol II terminates transcription have yet to be reported. Furthermore, with the exception of the SR-like proteins described by Corden and colleagues (Yuryev et al., 1996), all proteins reported to interact with the CTD recognize this repetitive domain in its unphosphorylated state (see Introduction). These reported interactions are consistent with promoter clearance models of CTD function, whereby unphosphorylated CTD heptapeptides tether Pol II complexes to promoter-associated transcription factors at the pre-initiation stage of the transcription cycle (see Introduction). But none of the reported interactions involving the unphosphorylated CTD preclude the possibility that the phosphorylated CTD may mediate Pol II's association with Sm snRNPs, SR proteins, or other molecules. Similarly, our data indicating that hyperphosphorylated Pol II LS molecules associate with Sm snRNPs and SR proteins do not contradict the possibility that the unphosphorylated CTD regulates early step(s) in Pol II's transcription cycle. Thus, after the CTD is phosphorylated, the promoter may “release” the polymerase complex, which initiates RNA synthesis and becomes competent to associate with Sm snRNPs and SR proteins (see below).
Genetic analyses of CTD function in S. cerevisiae have revealed no connections between the CTD and splicing proteins (see Introduction of the following paper). Inactivation of a temperature sensitive mutant CTD kinase (KIN28) promptly shuts down RNA synthesis (Cismowski et al., 1995), and partial truncation of Pol II's CTD leads to diminished transcription of specific genes (reviewed in Young, 1991; Gerber et al., 1995). While both observations strongly suggest a transcriptional role for the CTD, the potential effects of CTD kinases or CTD truncations on cotranscriptional pre-mRNA splicing would not manifest themselves in the absence of pre-mRNA synthesis. Finally, mutant yeast SRB genes were genetically selected for their ability to suppress CTD truncation mutants, and in agreement with the CTD's probable role in transcriptional initiation, they encode proteins which associate with basal transcription factors in the Pol II holoenzyme (for review see Koleske and Young, 1995). But these results do not contradict the possibility that the hyperphosphorylated CTD interacts with splicing factors.
In summary, we have provided strong evidence indicating that Pol IIo associates with splicing factors of two major families, the Sm snRNPs and the non-snRNP SR proteins. The splicing factors associate with Pol IIo, apparently without the direct involvement of pre-mRNA and at times when the polymerase is not transcribing. Our observation that certain SR proteins can be co-immunoprecipitated with mAb 8WG16, but not with mAbs H5 and H14, suggests that Pol IIo's association with SR proteins may involve CTD phosphoepitopes. In agreement with the data presented here, Pol IIo was recently shown to be associated with active spliceosomes assembled on exogenous RNA templates in vitro (Blencowe et al., 1996). The present study, in conjunction with other recent studies (Blencowe et al., 1996; Yuryev et al., 1996), suggests that a phosphorylated form of the CTD may mediate Pol II'o association with splicing factors. In an accompanying study, we pursue this idea by asking whether CTD-derived proteins affect splicing in vivo (Du and Warren, 1997).
Acknowledgments
We are grateful to Phillip Sharp, Ben Blencowe, and Michael Dahmus for helpful discussions and suggestions throughout these studies. We are also grateful to Michael Dahmus for advice about the CNBr cleavage of Pol II, and for providing anti-RPII antibodies. We thank David Ward for the use of his CCD camera. We thank Jeffery Nickerson for mAbs B1C8 and B4A11, Philip Cohen for mAb 7.13, Clint MacDonald for mAb 3A7/64, Joan Steitz for mAb Y12, Mark Roth for mAb 104, Bryan Turner for mAb 3C5, Xiang-Dong Fu for mAb SC35, and Michael Snyder for antiNuMA. We are grateful for the excellent technical assistance of Xun Sun.
Abbreviations used in this paper
- CTD
carboxy-terminal domain of RNA polymerase II
- MIG
mitotic interchromatin granule cluster
- NuMA
nuclear mitotic apparatus protein
- Pol IIa
unphosphorylated largest RNA polymerase II subunit
- Pol II LS
largest subunit of RNA polymerase II
- Pol IIo
hyperphosphorylated largest RNA polymerase II subunit
- Sm sn RNP
Smith antigen-containing small nuclear ribonucleoprotein
- SR
serine-arginine dipeptide repeat motif
Footnotes
The work was supported by the Council for Tobacco Research (#3881) and National Institutes of Health (NIH) (K08-CA01339) to S.L. Warren and by the NIH (F32 CA09281) to D.B. Bregman.
Please address all correspondence to S.L Warren, Brady Memorial Laboratories, Room B117, Department of Pathology, Yale University School of Medicine, P.O. Box 208023, New Haven, CT 06520-8023. Tel.: (203) 737-2247. Fax: (203) 785-7303. E-mail: stephen.warren@yale.edu
References
- Akoulitchev S, Mäkelä T, Weinberg RA, Reinberg D. Requirement for TFIIH kinase activity in transcription by RNA polymerase. Nature (Lond) 1995;377:557–560. doi: 10.1038/377557a0. [DOI] [PubMed] [Google Scholar]
- Allison LA, Wong JK, Fitzpatrick VD, Moyle M, Ingles CJ. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogasterand mammals: a conserved structure with an essential function. Mol Cell Biol. 1985;8:321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomew B, Dahmus ME, Meares CF. RNA contacts subunits IIo and IIc in HeLa RNA Polymerase II transcription complexes. J Biol Chem. 1986;261:14226–14231. [PubMed] [Google Scholar]
- Bazett-Jones DP. RNA polymerase II transcription and the functional organization of the mammalian cell nucleus. Chromosoma. 1995;103:509–516. doi: 10.1007/BF00355315. [DOI] [PubMed] [Google Scholar]
- Blencowe BJ, Nickerson JA, Issner R, Penman S, Sharp PA. Association of nuclear matrix antigens with exon-containing splicing complexes. J Cell Biol. 1994;127:593–607. doi: 10.1083/jcb.127.3.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Issner R, Kim J, McCaw P, Sharp PA. New proteins related to the SerArg family of splicing factors. RNA. 1995;1:852–865. [PMC free article] [PubMed] [Google Scholar]
- Blencowe BJ, Mortillaro MJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R. A hyperphosphorylated form of the largest subunit of RNA polymerase II is associated with splicing complexes. Proc Natl Acad Sci USA. 1996;93:8253–8257. doi: 10.1073/pnas.93.16.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloom DD, Davignon J-L, Cohen PL, Eisenberg RA, Clarke SH. Overlap of the anti-Sm and Anti-DNA Responses of MLR/Mp-lpr/lpr Mice. J Immunol. 1993;150:1579–1590. [PubMed] [Google Scholar]
- Bregman DB, Du L, Ribisi S, Warren SL. Cytostellin distributes to nuclear regions enriched with splicing factors. J Cell Sci. 1994;10:387–396. doi: 10.1242/jcs.107.3.387. [DOI] [PubMed] [Google Scholar]
- Bregman DB, Du L, van der Zee S, Warren SL. Transcriptiondependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratowski S, Sharp PA. Transcription initiation complexes and upstream activation with RNA polymerase II lacking the C-terminal domain of the largest subunit. Mol Cell Biol. 1990;10:5562–5564. doi: 10.1128/mcb.10.10.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadena DL, Dahmus ME. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J Biol Chem. 1987;262:12468–12474. [PubMed] [Google Scholar]
- Chesnut JD, Stephens JH, Dahmus ME. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J Biol Chem. 1992;267:10500–10506. [Google Scholar]
- Cismowski MJ, Laff GM, Solomon MJ, Reed SI. KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiaebut lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol Cell Biol. 1995;15:2983–2992. doi: 10.1128/mcb.15.6.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton DA, Szilak I, Cleveland DW. Primary structure of NuMA, an intranuclear protein that defines a novel pathway for segregation of proteins at mitosis. J Cell Biol. 1992;116:1395–1408. doi: 10.1083/jcb.116.6.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corden JL. RNA polymerase II transcription cycles. Curr Opin Gen Dev. 1993;3:213–218. doi: 10.1016/0959-437x(93)90025-k. [DOI] [PubMed] [Google Scholar]
- Corden JL, Cadena DL, Ahearn JM, Dahmus ME. A unique structure of the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci USA. 1985;82:7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus ME. Reversible phosphorylation of the C-terminal domain of RNA polymerase II. J Biol Chem. 1996;271:19009–19012. doi: 10.1074/jbc.271.32.19009. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du, L., and S. Warren. 1997. A functional interaction between the carboxy terminal domain of RNA polymerase II and Pre-mRNA splicing. J. Cell Biol. In press. [DOI] [PMC free article] [PubMed]
- Fakan S. Perichromatin fibrils are in situ forms of nascent transcripts. Trends Cell Biol. 1994;4:86–90. doi: 10.1016/0962-8924(94)90180-5. [DOI] [PubMed] [Google Scholar]
- Ferreira JA, Carmo-Fonseca M, Lamond AI. Differential interaction of splicing snRNPs with coiled bodies and interchromatin granules during mitosis and assembly of daughter cell nuclei. J Cell Biol. 1994;126:11–23. doi: 10.1083/jcb.126.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X-D, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature (Lond) 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Gerber HP, Hagmann M, Seipel K, Georgiev O, West LA, Litingtung Y, Schaffner W, Corden JL. RNA polymerase II C-terminal domain required for enhancer-driven transcription. Nature (Lond) 1995;374:660–662. doi: 10.1038/374660a0. [DOI] [PubMed] [Google Scholar]
- Jackson DA, Hassan AB, Errington RJ, Cook PR. Visualization of focal sites of transcription within human nuclei. EMBO (Eur Mol Biol Organ) J. 1993;12:1059–1065. doi: 10.1002/j.1460-2075.1993.tb05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang ME, Dahmus ME. RNA polymerases IIA and IIO have distinct roles during transcription from TATA-less murine dihydrofolate reductase promoter. J Biol Chem. 1993;268:25033–25040. [PubMed] [Google Scholar]
- Kang ME, Dahmus ME. The photoactivated cross-linking of recombinant C-terminal domain to proteins in a HeLa cell transcription extract that comigrate with transcription factors IIE and IIF. J Biol Chem. 1995;270:23390–23397. doi: 10.1074/jbc.270.40.23390. [DOI] [PubMed] [Google Scholar]
- Kim WY, Dahmus ME. Immunochemical analysis of mammalian RNA polymerase II subspecies. Stability and relative in vivoconcentration. J Biol Chem. 1986;261:14219–14225. [PubMed] [Google Scholar]
- Kim WY, Dahmus ME. The major late promoter of adenovirus-2 is accurately transcribed by RNA polymerases IIO, IIA, and IIB. J Biol Chem. 1989;264:3169–3176. [PubMed] [Google Scholar]
- Koleske AJ, Young RA. An RNA polymerase II holoenzyme responsive to activators. Nature (Lond) 1994;368:466–469. doi: 10.1038/368466a0. [DOI] [PubMed] [Google Scholar]
- Koleske AJ, Young RA. The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Bjorklund S, Li Y, Sayre MH, Kornberg RD. A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell. 1994;77:599–608. doi: 10.1016/0092-8674(94)90221-6. [DOI] [PubMed] [Google Scholar]
- Laybourn PJ, Dahmus ME. Phosphorylation of RNA polymerase IIA occurs subsequent to interaction with the promoter and before the initiation of transcription. J Biol Chem. 1990;265:13165–13173. [PubMed] [Google Scholar]
- Lee JM, Greenleaf AL. CTD kinase large subunit is encoded by CTK1, a gene required for normal growth of Saccharomyces cerevisiae. . Gene Expression. 1991;1:149–167. [PMC free article] [PubMed] [Google Scholar]
- Lerner EA, Lerner MR, Janeway C, Steitz JA. Monoclonal antibodies to nucleic acid containing cellular constituents: probes for molecular biology and autoimmune disease. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Flores O, Weinmann R, Reinberg D. The nonphosphorylated form of RNA polymerase II preferentially associates with the preinitiation complex. Proc Natl Acad Sci USA. 1991;88:10004–10008. doi: 10.1073/pnas.88.22.10004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makela TP, Parvin JD, Kim J, Huber LJ, Sharp PA, Weinberg RA. A kinase-deficient transcription factor TFIIH is functional in basal and activated transcription. Proc Natl Acad Sci USA. 1995;92:5174–5178. doi: 10.1073/pnas.92.11.5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moen PT, Smith KP, Lawrence JB. Compartmentalization of specific pre-mRNA metabolism: an emerging view. Hum Mol Genetics. 1995;4:1779–1789. doi: 10.1093/hmg/4.suppl_1.1779. [DOI] [PubMed] [Google Scholar]
- O'Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature (Lond) 1994;370:75–77. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- Payne JM, Laybourn PJ, Dahmus ME. The transition of RNA polymerase II from initiation to elongation is associated with phosphorylation of the carboxyl-terminal domain of subunit IIa. J Biol Chem. 1989;264:19621–19629. [PubMed] [Google Scholar]
- Pinto A, Steitz JA. The mammalian analogue of the yeast PRP8 splicing protein is present in the U4/5/6 small nuclear ribonucleoprotein particle and the spliceosome. PNAS. 1989;86:8742–8746. doi: 10.1073/pnas.86.22.8742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott D, Bender M. Synthesis of RNA and protein during mitosis in mammalian tissue culture cells. Exp Cell Res. 1962;26:260–268. doi: 10.1016/0014-4827(62)90176-3. [DOI] [PubMed] [Google Scholar]
- Roth MB, Murphy C, Gall JG. A monoclonal antibody that recognizes a phosphorylated epitope stains lampbrush chromosome loops and small granules in the amphibian germinal vesicle. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., E.F. Fritsch, and T. Maniatis. 1989. Molecular Cloning-A Laboratory Manual. 2nd Ed. Cold Spring Harbor Press, Cold Spring Harbor, NY. pp. 6.39–6.45.
- Serizawa H, Conaway JW, Conaway RC. Phosphorylation of C-terminal domain of RNA polymerase II is not required in basal transcription. Nature (Lond) 1993;363:371–374. doi: 10.1038/363371a0. [DOI] [PubMed] [Google Scholar]
- Shermoen AW, O'Farrell PH. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Fu X-D, Maniatis T. Associations between distinct pre-mRNA splicing components and the cell nucleus. EMBO (Eur Mol Biol Organ) J. 1991;10:3467–3481. doi: 10.1002/j.1460-2075.1991.tb04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Landon S, O'Keefe RT. Organization of RNA polymerase II transcription and pre-mRNA splicing within the mammalian cell nucleus. Biochem Soc Trans. 1995;21:918–920. doi: 10.1042/bst0210918. [DOI] [PubMed] [Google Scholar]
- Sterner D, Lee JM, Hardin SE, Greenleaf AL. Yeast carboxylterminal repeat domain kinase CTDK-1 is a divergent cyclin-cyclin dependent kinase complex. Mol Cell Biol. 1995;15:5716–5724. doi: 10.1128/mcb.15.10.5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. doi: 10.1101/gad.4.12a.2112. [DOI] [PubMed] [Google Scholar]
- Taylor J. Nucleic acid synthesis in relation to the cell division cycle. Ann Rev NY Acad Sci. 1960;90:409–421. doi: 10.1111/j.1749-6632.1960.tb23259.x. [DOI] [PubMed] [Google Scholar]
- Thompson NE, Steinberg TH, Aronson DB, Burgess RR. Inhibition of in vivo and in vitrotranscription by monoclonal antibodies prepared against wheat germ RNA polymerase II that react with the heptapeptide repeat of eukaryotic RNA polymerase II. J Biol Chem. 1989;264:11511–11520. [PubMed] [Google Scholar]
- Thompson CM, Koleske AJ, Chao DM, Young RA. A multisubunit complex associated with the RNA polymerase II CTD and TATAbinding protein in yeast. Cell. 1993;73:1361–1375. doi: 10.1016/0092-8674(93)90362-t. [DOI] [PubMed] [Google Scholar]
- Tobey RA, Anderson EC, Peterson DG. Properties of mitotic cells prepared by mechanically shaking monolayer cultures of Chinese hamster cells. J Cell Physiol. 1967;70:63–68. doi: 10.1002/jcp.1040700109. [DOI] [PubMed] [Google Scholar]
- Turner BM, Franchi L. Identification of protein antigens associated with the nuclear matrix and with clusters of interchromatin granules in both interphase and mitotic cells. J Cell Sci. 1987;87:269–282. doi: 10.1242/jcs.87.2.269. [DOI] [PubMed] [Google Scholar]
- Usheva A, Maldonado E, Goldring A, Lu H, Houbavi C, Reinberg D, Aloni Y. Specific interaction between the non-phosphorylated form of RNA polymerase II and the TATA-binding protein. Cell. 1992;69:871–881. doi: 10.1016/0092-8674(92)90297-p. [DOI] [PubMed] [Google Scholar]
- van Driel R, Wansink DG, van Steensel B, Grande MA, Schul W, de Jong L. Nuclear domains and the nuclear matrix. Int Rev Cytol. 1995;162A:151–189. doi: 10.1016/s0074-7696(08)61231-0. [DOI] [PubMed] [Google Scholar]
- Wansink DG, van Driel R, de Jong L. Organization of (pre-) mRNA metabolism in the cell nucleus. Mol Biol Rep. 1994;20:45–55. doi: 10.1007/BF00996353. [DOI] [PubMed] [Google Scholar]
- Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L. Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol. 1993;122:283–293. doi: 10.1083/jcb.122.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren SL, Landolfi AS, Curtis C, Morrow JS. Cytostellin: a novel, highly conserved protein that undergoes continuous redistribution during the cell cycle. J Cell Sci. 1992;103:381–388. doi: 10.1242/jcs.103.2.381. [DOI] [PubMed] [Google Scholar]
- Weeks JR, Hardin SE, Shen J, Lee JM, Greenleaf AL. Locusspecific variation in phosphorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- Yang CH, Lambie EJ, Snyder M. NuMA: an unusually long coiled-coil related protein in the mammalian nucleus. J Cell Biol. 1992;116:1303–1317. doi: 10.1083/jcb.116.6.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA. RNA polymerase II. Annu Rev Biochem. 1991;60:689–715. doi: 10.1146/annurev.bi.60.070191.003353. [DOI] [PubMed] [Google Scholar]
- Yuryev A, Patturajan M, Litingtung Y, Joshi RV, Gentile C, Gebara M, Corden JL. The CTD of RNA polymerase II interacts with a novel set of SR-like proteins. Proc Natl Acad Sci USA. 1996;93:6975–6980. doi: 10.1073/pnas.93.14.6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehring WA, Lee JM, Weeks JR, Jokerst RS, Greenleaf AL. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. . Proc Natl Acad Sci USA. 1988;85:3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehring WA, Greenleaf A. The carboxyl-terminal repeat domain of RNA polymerase II is not required for transcription factor Sp1 to function in vitro. . J Biol Chem. 1990;265:8351–8353. [PubMed] [Google Scholar]
- Zeng, C, E. Kim, S.L. Warren, and S.M. Berget. 1997. Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO (Eur. Mol. Biol. Organ.) J. In press. [DOI] [PMC free article] [PubMed]
- Zeng C, He D, Berget SM, Brinkley BR. Nuclear-mitotic apparatus protein: a structural protein interface between the nucleoskeleton and RNA splicing. Proc Natl Acad Sci USA. 1994;91:1505–1509. doi: 10.1073/pnas.91.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Corden JL. Identification of phosphorylation sites in the repetitive carboxy-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991;266:2297–2302. [PubMed] [Google Scholar]