Abstract
Transforming growth factor-β (TGF-β) is secreted by many cell types as part of a large latent complex composed of three subunits: TGF-β, the TGF-β propeptide, and the latent TGF-β binding protein (LTBP). To interact with its cell surface receptors, TGF-β must be released from the latent complex by disrupting noncovalent interactions between mature TGF-β and its propeptide. Previously, we identified LTBP-1 and transglutaminase, a cross-linking enzyme, as reactants involved in the formation of TGF-β. In this study, we demonstrate that LTBP-1 and large latent complex are substrates for transglutaminase. Furthermore, we show that the covalent association between LTBP-1 and the extracellular matrix is transglutaminase dependent, as little LTBP-1 is recovered from matrix digests prepared from cultures treated with transglutaminase inhibitors. Three polyclonal antisera to glutathione S–transferase fusion proteins containing amino, middle, or carboxyl regions of LTBP-1S were used to identify domains of LTBP-1 involved in crosslinking and formation of TGF-β by transglutaminase. Antibodies to the amino and carboxyl regions of LTBP-1S abrogate TGF-β generation by vascular cell cocultures or macrophages. However, only antibodies to the amino-terminal region of LTBP-1 block transglutaminase-dependent cross-linking of large latent complex or LTBP-1. To further identify transglutaminase-reactive domains within the amino-terminal region of LTBP-1S, mutants of LTBP-1S with deletions of either the amino-terminal 293 (ΔN293) or 441 (ΔN441) amino acids were expressed transiently in CHO cells. Analysis of the LTBP-1S content in matrices of transfected CHO cultures revealed that ΔN293 LTBP-1S was matrix associated via a transglutaminasedependent reaction, whereas ΔN441 LTBP-1S was not. This suggests that residues 294–441 are critical to the transglutaminase reactivity of LTBP-1S.
Most cell types secrete transforming growth factor-β1 (TGF-β)1 in a biologically inactive form (42). Mature TGF-β is a homodimer composed of two 12.5-kD polypeptides joined by a disulfide bond at cysteine 77 (14). The monomeric subunits are produced by intercellular cleavage of a higher mol wt precursor at a dibasic site immediately preceding Ala-279 (17, 21). However, after secretion the propeptides remain associated with TGF-β through noncovalent interactions, rendering TGF-β inactive (20). TGF-β with its propeptide, also known as the latency associated peptide (LAP), is referred to as the small latent complex. Both in vitro and in vivo, latent TGF-β is secreted as part of a large latent complex in which a second gene product, the latent TGF-β binding protein (LTBP), is disulfide-linked to LAP (42). The dissociation of TGF-β from LAP is required for TGF-β to bind to its receptors and exert its effects on cell proliferation, extracellular matrix (ECM) deposition, cell migration, and differentiation (34, 38, 58). Latent TGF-β is activated by heat, acid or alkaline treatment, binding to thrombospondin, deglycosylation, proteolysis, or irradiation (5, 9, 34, 35, 56). The most extensively studied process for activating large latent complex is a plasmin-dependent mechanism observed in several tissue culture systems including bovine aortic endothelial (BAE) cells treated with retinoids, cocultures of endothelial cells and either smooth muscle cells or pericytes, and lipopolysaccharide (LPS)– stimulated, thioglycollate-elicited peritoneal macrophages (30, 31, 44, 54). Components of this activation mechanism include the serine protease plasmin, the cross-linking enzyme transglutaminase, LTBP-1, and the mannose 6-phosphate/insulin-like growth factor type II receptor, which appears to bind to mannose 6-phosphate residues in LAP (16, 19, 31, 54, 55).
Interactions of the proteins involved in the activation of large latent complex are not well understood. Plasmin can release TGF-β from large latent complex under cell-free conditions (36). The mannose 6-phosphate/insulin-like growth factor type II receptor binds forms of latent TGF-β, but the role of this interaction is not clear (32). The role of LTBP-1 or tissue transglutaminase in large latent complex activation is not known. As part of an effort to characterize the activation process of large latent complex, we have initiated studies to examine the potential interactions of LTBP-1 and tissue transglutaminase.
LTBP consists of a family of glycoproteins of ∼120–210 kD that contain a central core of EGF-like repeats and multiple unique eight-cysteine repeats (22, 28, 43, 46, 64). LTBPs are structurally similar to the microfibrillar proteins fibrillin-1 and -2 (48, 50, 65). Defects in fibrillins are responsible for the matrix fragility observed in patients with Marfan syndrome and congenital contractural arachnodactyly (29, 47). The best characterized member of the family is LTBP-1, which can exist as either short (LTBP1S) or long (LTBP-1L) forms (46). A number of cell types secrete LTBP-1 as a higher order complex in which the third eight-cysteine repeat in LTBP-1 is disulfide-linked to the cysteine at position 33 of LAP (22a, 49). It is unknown whether the short and long forms of LTBP-1 are expressed differentially. LTBP facilitates the secretion of small latent complex, participates in the activation of large latent complex, targets large latent complex to the ECM of fetal rat calvarial cells, fibroblasts, epithelial cells, and endothelial cells, and contributes to the formation of fibrillar structures (13, 19, 41, 44, 59–61). LTBP-1S and -1L associate differentially with the matrix, with LTBP-1L having a greater affinity (46). Matrix association of LTBP-1 appears to be covalent, as LTBP in the matrix is deoxycholate insoluble but is released upon proteolysis (Taipale, J., J. Saharinen, K. Hedman, and J. Keski-Oja. 1994. Mol. Biol. Cell. 5[Suppl.]:311a). However, the mechanism of covalent association between LTBP-1 and the ECM is unknown.
Transglutaminases are a family of structurally and functionally related calcium-dependent enzymes that catalyze the formation of isopeptide bonds between γ-carboxamide groups of glutamine residues and ε-amino groups of lysine residues (24, 33). The family is comprised of five members: plasma Factor XIII, keratinocyte transglutaminase, tissue/ endothelial transglutaminase, epidermal transglutaminase, and prostate transglutaminase. Although transglutaminase does not have a signal sequence, the enzyme can be localized to cell surfaces as well as the ECM and reacts to stabilize interactions between extracellular substrates such as plasminogen, fibronectin, nidogen, and vitronectin (3, 6, 37, 51, 63). Pericellular transglutaminase may play a role in stabilizing cell and ECM interactions (24, 37). Transglutaminase is required for the conversion of large latent complex to TGF-β in several systems, including retinoid-treated BAE cells, LPS-stimulated thioglycollate-elicited peritoneal macrophages, and cocultures of BAE and bovine smooth muscle cells (BSM) (30, 31, 44).
Because LTBP-1 is structurally homologous to microfibrillar proteins that are stabilized by transglutaminasedependent cross-linking (10) and LTBP-2 has been localized to elastin-associated microfibrils (22), we explored the possibility that the covalent association between LTBP-1 and ECM proteins is catalyzed by transglutaminase. Here we show that LTBP-1 and large latent complex are substrates for transglutaminase both as isolated molecules and in cell cultures. We demonstrate that matrix incorporation of LTBP-1 is transglutaminase-dependent and requires the amino-terminal region of LTBP-1 and that matrix association is an intermediate step in the activation mechanism used by cells to generate TGF-β.
Materials and Methods
Materials
Human plasmin (10 U/ml), isopropyl-β-d-thiogalactopyranoside (IPTG), 1-o-n-octyl-β-d-glucopyranoside (n-octylglucoside), Pefabloc SC, and protein A–agarose were purchased from Boehringer Mannheim Corp. (Indianapolis, IN). Cystamine, glutathione-agarose, guinea pig liver transglutaminase, monodansylcadaverine (MDC), and nonimmune rabbit serum were purchased from Sigma Chemical Co. (St. Louis, MO). Pyrogen-poor BSA was purchased from Pierce (Rockford, IL). l-[35S]cysteine, 35S-labeled express translabel mix, and 125I-Na (pH 12–14, high concentration) were obtained from Du Pont Company Biotechnology Systems (Wilmington, DE). Recombinant human TGF-β1 was a gift from Berlex Biosciences (South San Francisco, CA). Pan-neutralizing monoclonal mouse anti–TGF-β IgG1 was either purchased from Genzyme (Cambridge, MA) or donated by Celtrix (Santa Clara, CA) (15). Recombinant human LTBP-1S, human large latent complex, and small latent complex were provided by Drs. Hideya Ohashi and Haruhiko Tsumura (KIRIN Brewery Co., Ltd., Pharmaceutical Division, Gunma, Japan). Recombinant Factor XIII (166 kD) was provided by Dr. P.D. Bishop (ZymoGenetics Inc., Seattle, WA) (7). The human fibroblast LTBP-1S cDNA (pSV7d-BP13) was a gift from Drs. K. Miyazono and C. Heldin (Ludwig Institute for Cancer Research, Uppsala, Sweden) (28, 46). Ab 39, a polyclonal rabbit anti-LTBP IgG, was a gift from Dr. K. Miyazono (40). Ab 39 was generated against human LTBP purified from human platelets and recognizes LTBP-1 but not LTBP-2 (28, 43).
Cell Culture
Human fibrosarcoma cells (HT 1080; American Type Culture Collection, Rockville, MD) were grown in DME containing 10% (heat-inactivated) FCS, 100 U/ml penicillin G, 100 μg/ml streptomycin sulfate, and 2 mM l-glutamine (P/S/Q). Rat osteosarcoma cells (UMR-106; American Type Culture Collection) were cultured in DME containing 10% (heat-inactivated) FCS and P/S/Q. Chinese hamster ovary cells (CHO K1) were cultured in α-MEM containing 10% (heat-inactivated) FCS and P/S/Q.
Primary BAE and BSM cells were isolated and cultured in α-MEM and DME containing 10% (non–heat inactivated) calf serum plus P/S/Q, respectively (53). Stimulated peritoneal macrophages were harvested from Swiss Webster mice, which had been injected i.p. with 4% thioglycollate broth and activated by LPS in vitro as previously described (44).
Expression and Purification of Glutathione S–Transferase–LTBP-1S Fusion Proteins
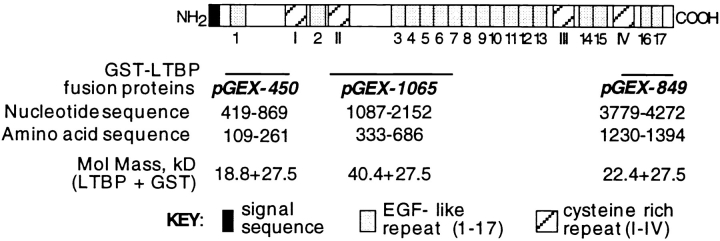
Three fusion proteins, each consisting of glutathione S–transferase (GST) fused to a region of human fibroblast LTBP-1S, were generated by expressing pGEX1n constructs encoding LTBP-1S fragments in HB101 Escherichia coli followed by purification using glutathione-agarose (57). Fusion proteins GST-450, GST-1065, and GST-849 contain sequences from the amino, middle, and carboxyl regions of human fibroblast LTBP-1S, respectively (Fig. 1) (28, 46).
Figure 1.
Regions of LTBP-1S expressed in bacteria as GST– LTBP-1S fusion proteins. Three GST–LTBP-1S fusion proteins (GST-450, GST-1065, and GST-849) were generated by expressing regions of the human LTBP-1S cDNA using pGEX1n in E. coli. Representation of LTBP-1S was adapted from Kanzaki et al. (28).
pGEX-450 was constructed by cloning a 450-bp BanI fragment of the LTBP-1S cDNA BP13 (nucleotides 419–869) (28) into a SmaI site of pGEX1n. The pGEX-1065 construct was prepared by ligating a 1065-bp BamHI-EcoRI fragment of the LTBP-1S cDNA BP13 (nucleotides 1087– 2152) and BamHI-EcoRI–digested pGEX1n. The pGEX-849 construct was generated by cloning an 849-bp StuI-SSpI fragment of the LTBP-1S cDNA BP13 (nucleotides 3779–4628) in the SmaI site of pGEX1n.
Once the insert orientation and sequence were verified for each construct, HB101 E. coli were transformed with the pGEX1n plasmid (control) or a pGEX–LTBP-1S expression construct by electroporation (Cell Porator; GIBCO BRL, Gaithersburg, MD). Fusion proteins were purified from IPTG-induced bacteria using glutathione-agarose according to the manufacturer's instructions (Pharmacia LKB Biotechnology, Inc., Piscataway, NJ). The integrity and apparent mol wts of the fusion proteins were verified by reducing SDS-PAGE followed by Coomassie brilliant blue R staining (26).
Preparation and Characterization of Antibodies to GST–LTBP-1S Fusion Proteins
Polyclonal rabbit antisera raised against purified GST-450, GST-1065, or GST-849 fusion proteins were prepared by Cocalico Biologicals, Inc. (Reamstown, PA). Affinity-purified anti-450, anti-1065, and anti-849 IgG were prepared by incubation of antisera with tosyl-activated agarose (Pierce), to which fusion proteins had been coupled as recommended by the manufacturer. Antibodies were characterized by Western blotting and immunoprecipitation of recombinant large latent complex, recombinant LTBP-1, or HT 1080 conditioned medium (CM).
Western blotting was performed using nonreducing or reducing conditions. Proteins separated by SDS-PAGE were transferred to Immobilon-P membrane (Millipore Corp., Bedford, MA) (62). Membranes were blocked with PBS containing 5% Carnation nonfat milk (Nestle Food Co., Glendale, CA), incubated with either Ab 39 serum or serum raised against GST–LTBP fusion proteins in blocking buffer, washed with Tris-buffered saline (150 mM NaCl, 50 mM Tris-HCl, pH 7.4) and 1% Triton X-100, incubated with either goat anti–rabbit IgG conjugated to alkaline phosphatase (Promega Corp., Madison, WI) or donkey anti–rabbit IgG conjugated to horseradish peroxidase (Amersham Corp., Arlington Heights, IL), and washed as described above. Alkaline phosphate– and horseradish peroxidase–containing immune complexes were revealed by incubating the membrane with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium phosphatase substrate (Kirkegaard and Perry Laboratories, Inc., Gaithersburg, MD) and enhanced chemiluminescence detection reagents (ECL; Amersham Corp.), respectively. The immunoreactivity of antisera to GST–LTBP-1S fusion proteins was compared to that of Ab 39, a polyclonal antiserum raised against intact LTBP-1 purified from human platelets (40). The mol wts of large latent complex and LTBP-1 observed in our experiments are consistent with those reported by others (data not shown) (28, 40).
Immunoprecipitations were performed using large latent complex (2–5 μg) or LTBP-1 (2–5 μg) iodinated using 250 μCi 125I-Na and 20 μg/ml chloramine T as described by McConahey et al. (39). 35S-labeled HT 1080 CM was prepared by incubating confluent cultures overnight with DME deficient in cysteine and methionine (Gibco Laboratories, Grand Island, NY), 100 μCi/ml 35S-express translabel mix, and 1% normal DME culture medium. Iodinated proteins and metabolically labeled CM were immunoprecipitated with Ab 39 serum or antiserum generated against one of the three fusion proteins. Radiolabeled proteins were incubated with antiserum for 1–1.5 h at room temperature. Immune complexes were precipitated using protein A–agarose, washed with Tris-buffered saline and 1% Triton X-100, and transferred to a 1.5-ml test tube followed by a water wash. Protein A–precipitated complexes were solubilized in nonreducing or reducing Laemmli sample buffer and separated by SDS-PAGE. Iodinated proteins were visualized by autoradiography, and metabolically labeled proteins were visualized by fluorography of gels treated with 1 M sodium salicylate, dried, and exposed to film (X-OMAT; Eastman Kodak Co., Rochester, NY) at −80°C for 7–10 d. Anti-450 and anti-849 antisera immunoprecipitated radiolabeled forms of LTBP-1 but were not as effective as Ab 39 (data not shown). Anti-1065 antisera failed to recognize any of the radiolabeled preparations containing LTBP-1 (data not shown). Anti-1065 antiserum was used as a negative control because its reactivity with nondenatured LTBP-1 was negligible. These results indicated that the antibodies to the amino- and carboxyl-terminal regions could be used for immunoprecipitation of LTBP-1 and in experiments examining the role of the amino and carboxyl termini of LTBP-1 in the cross-linking and activation of large latent complex.
Cross-Linking Reactions by Transglutaminase
Guinea pig liver transglutaminase was used at an enzyme/substrate molar ratio of 1:5. Substrates for cross-linking reactions were iodinated using 250 μCi 125I-Na and 20 μg/ml chloramine T (39). Rabbit nonimmune IgG (1 mg/ml) was added as a carrier protein to all enzymatic assays, as IgG is not a substrate for tissue transglutaminase (8). Human fibronectin (Collaborative Biomedical Products, Bedford, MA), recombinant large latent complex, and recombinant LTBP-1, all used at 167 nM, were incubated at 37°C for 1 h in 1.5-ml siliconized test tubes containing transglutaminase, 10 mM Tris-HCl, pH 8.0, 0.5 mM DTT, and 15 mM CaCl2. Specificity of cross-linking reactions was verified by including either 100 μM MDC, a competitive transglutaminase inhibitor, or 30 mM EDTA (33). Reactions were stopped by addition of reducing SDS-PAGE Laemmli sample buffer. Cross-linked proteins were separated by reducing SDS-PAGE, and gels were examined by PhosphorImager scanning analysis (Molecular Dynamics, Sunnyvale, CA).
To identify regions of LTBP-1 that contain transglutaminase-reactive sites, cross-linking reactions were performed as described above in the presence of either affinity-purified antibodies generated against GST– LTBP-1S fusion proteins or protein A–purified Ab 39, as well as 0.1 mg/ ml l-cystine, which was needed to minimize disulfide exchange within and between proteins.
Immunoprecipitation of Matrix Digests Prepared from Metabolically Labeled Cells
Subconfluent cultures were incubated in DME deficient in cysteine and methionine for 1 h at 37°C. Cultures were treated with 0.06% DMSO (control), 50 μM MDC, 100 μM cystamine, or affinity-purified antibodies during the starvation and subsequent pulse-chase period. After the cells were metabolically labeled for 3 h with 100 μCi/ml of l-[35S]cysteine in cysteine, methionine-deficient DME supplemented with 2% Optimem (Gibco Laboratories), and 150 μg/ml of l-methionine, they were transferred to Optimem medium for an overnight incubation at 37°C in a 5% CO2 atmosphere. ECMs were prepared from metabolically labeled cells after lysis with 0.5% sodium deoxycholate in Tris-buffered saline (10 mM Tris-HCl, pH 8.0, and 150 mM NaCl) (59). Matrices were digested with 0.3 U/ml plasmin in 0.1% n-octylglucoside, 3 mM MgCl2, 3 mM CaCl2, 10 mM TrisHCl, pH 8.0, and 150 mM NaCl for 1 h at 37°C. Plasmin was inhibited by 8 mM Pefabloc SC and 1 μg/ml aprotinin. Digests normalized to total TCA-precipitable cpm as described by Harlow and Lane (26) were incubated with either nonimmune or Ab 39 serum, and immune complexes were precipitated using protein A–agarose. Washed immunoprecipitates were separated by reducing SDS-PAGE and visualized by fluorography.
Immunoprecipitation of Matrix Digests Prepared from UMR-106 Matrices
35S-labeled CM were prepared using BAE or UMR-106 confluent cultures that were metabolically labeled for 20 h with 100 μCi/ml of l-[35S]cysteine. Metabolically labeled CM were concentrated fivefold using Centricon-30 ultrafilters (Amicon, Beverly, MA) and immediately added to UMR-106 ECMs prepared using 0.5% sodium deoxycholate in Tris-buffered saline (59). Additions of antibodies or 50 μM MDC were done concurrently. To catalyze the cross-linking of large latent complex present in radiolabeled CM and UMR-106 matrix proteins, thrombin-activated Factor XIII (Factor XIIIa) was added (7). Each matrix incorporation reaction was done using 1 ml of concentrated radiolabeled CM, 3 nmol of DTT-pretreated Factor XIIIa, and 4 mM CaCl2 in Tris-buffered saline on 78.5 cm2 of UMR-106 ECM for 1 h at 37°C. The LTBP-1 content of UMR-106 matrices was analyzed by immunoprecipitating plasmin digests of matrices with Ab 39 followed by SDS-PAGE and fluorography of immunoprecipitates as described in this manuscript.
Transient Transfection of CHO Cells with LTBP-1S cDNA Constructs
Three LTBP-1S cDNA constructs were generated using the pcDNA3 (InVitrogen, San Diego, CA) expression vector for transient expression of CHO cells. The three constructs are pcDNA3–wild type (WT), pcDNA-ΔN293, and pcDNA-ΔN441 (see Fig. 8 A). To generate pcDNA3-WT LTBP-1S, the fragment 68–4543 (DraI-DraI) of the human LTBP-1S cDNA BP13 (28) was subcloned into pcDNA3 downstream of the human cytomegalovirus immediate early gene promoter. pcDNA3-ΔN293 and pcDNA3ΔN441 were obtained by digesting BP13 with HpaI and DraI (nucleotides 1414–4543), and ScaI and DraI (nucleotides 970–4543), respectively; these fragments were fused in frame with the baculovirus glycoprotein GP67 signal sequence as described elsewhere (22a) and subcloned into pcDNA3.
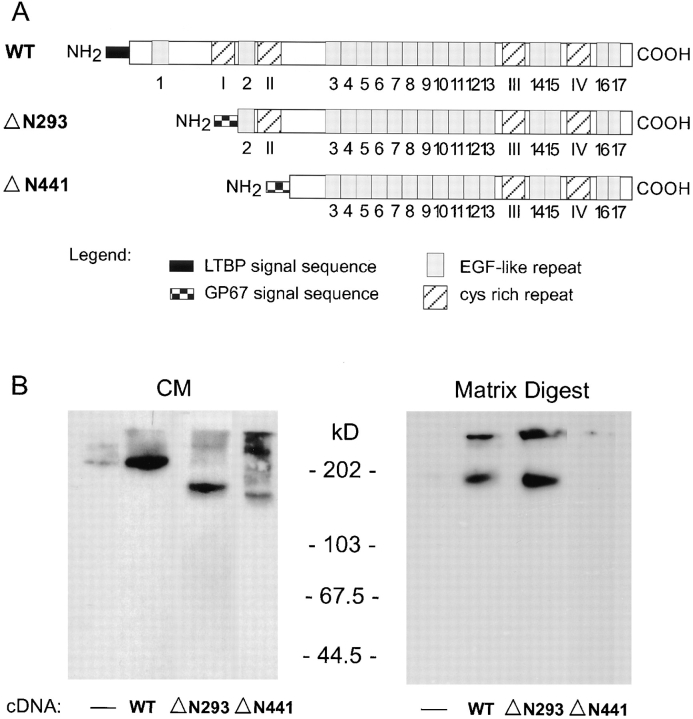
Figure 8.
Expression and matrix incorporation of amino-truncated LTBP-1S mutants by CHO cells. (A) LTBP-1S cDNA constructs were generated by subcloning LTBP-1S cDNAs into pcDNA3 for transient transfection of CHO cells. pcDNA3-WT LTBP-1S encodes the entire coding sequence of LTBP-1S. pcDNA3-ΔN293 and -ΔN441 LTBP-1S encode LTBP-1S truncated at the amino terminus by 293 and 441 amino acids, respectively. (B) CM from untransfected (−) and LTBP-1S cDNA– transfected CHO cells were immunoblotted using Ab 39 serum. The LTBP-1S content of matrix digests from corresponding cultures was analyzed by immunoblotting digests using Ab 39 serum.
CHO cells were transiently transfected using lipofectamine (Gibco Laboratories) according to the manufacturer's instructions. Briefly, cells plated in 35-mm dishes received 1–4 μg of plasmid DNA premixed with lipofectamine in Optimem. After 24 h, media were harvested and replaced with fresh Optimem. Transfected cultures were incubated for an additional 24 h, at which time media were collected and matrix digests were prepared.
Western Blotting CM and Matrix Digests of Transfected CHO Cultures
CM and matrix digests were prepared from transfected CHO cultures for Western blotting analysis using Ab 39. The CM collected after the first and second 24 h of transfection were pooled and concentrated 10-fold. The concentrated CM and matrix digests were mixed with nonreducing sample buffer for SDS-PAGE. Separated proteins were transferred electrophoretically to Immobilon-P for immunoblotting with Ab 39 serum, and immune complexes were visualized using chemiluminescence reagents as described earlier.
Preparation of CM from Cultures of BAE and BSM Cells and Stimulated Macrophages
Serum-free CM were prepared from homo- and heterotypic cultures of BAE and BSM cells and from LPS-stimulated thioglycollate-elicited macrophages as described in previous reports (44, 53, 54). CM collected after 14 h were immediately bioassayed for TGF-β activity (see below). TGFβ–dependent activity was verified by adding neutralizing anti–TGF-β IgG to parallel test samples before assay. Total TGF-β, which consists of cellactivated plus latent TGF-β, was quantitated after heating CM at 85°C for 12 min, a procedure known to convert latent TGF-β to TGF-β, before assay (9).
Mink Lung Epithelial Cells–Luciferase Assay for TGF-β Activity
This quantitative bioassay for TGF-β is based on the ability of TGF-β to stimulate plasminogen activator inhibitor-1 (PAI-1) expression (1). Mink lung epithelial cells (MLEC), stably transfected with an expression construct containing a truncated PAI-1 promoter fused to the firefly luciferase reporter gene, were incubated overnight with CM harvested from either stimulated macrophages or cultures of BAE and BSM cells. Recombinant TGF-β1 in α-MEM or DME with 0.1% BSA was used to generate a standard curve of TGF-β activity. Luciferase activity was quantitated using a luciferin substrate buffer in a ML3000 Microtiter Plate Luminometer (Dynatech Laboratories Inc., Chantilly, VA) (1).
Wound Migration Assay for TGF-β Activity
To measure TGF-β present in CM, wound migration assays were performed as described by Sato and Rifkin (52). This bioassay is based on the ability of TGF-β to inhibit endothelial cell migration. Confluent monolayers of BAE cells plated in a 35-mm dish were wounded with a razor blade and incubated with serum-free test samples for 18–20 h at 37°C in a 5% CO2 atmosphere. Cells were fixed and stained with 0.5% crystal violet in 20% methanol (44). BAE cells that had migrated from the edge of the wound were counted in successive 125-μm increments at 100× using a light microscope with an ocular grid. The number of cells migrating beyond 125 μm from seven different fields was averaged. Data are presented as a percentage of control, where the control sample consists of homotypic BAE and BSM media mixed 4:1.
Results
Transglutaminase Reactivity of LTBP and Large Latent Complex
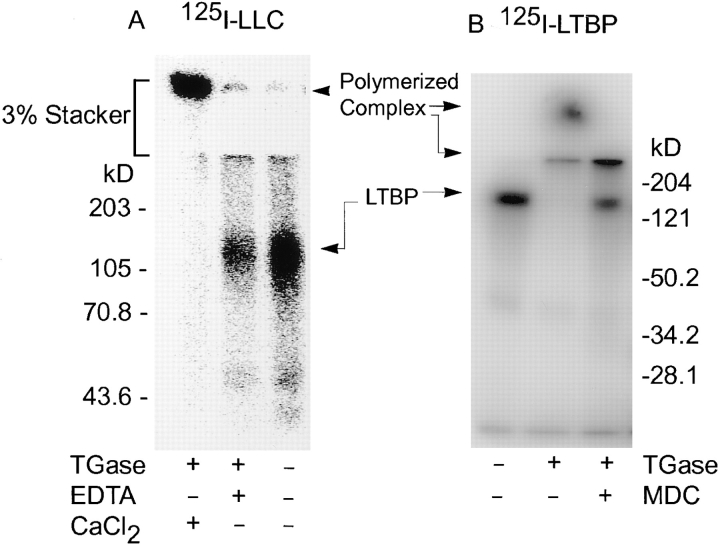
The apparent covalent interaction of LTBP-1 and ECM (33, 36) as well as the requirement for tissue transglutaminase for activation of large latent complex described for various culture systems suggested (30, 31, 44) that LTBP-1 and large latent complex might be substrates for transglutaminase. Therefore, iodinated recombinant LTBP-1 or large latent complex was incubated with guinea pig liver transglutaminase and analyzed by reducing SDS-PAGE. Samples incubated with transglutaminase revealed high mol wt complexes visible at the top of the 3% stacking polyacrylamide gel as well as intermediate-sized complexes at the interface between the 3% stacking and 7% resolving gels, indicating that both LTBP-1 and the large latent complex were polymerized by transglutaminase (Fig. 2). The efficiency of cross-linking of either LTBP-1 or large latent complex by transglutaminase varied among experiments as the relative amounts of polymerized and unreacted protein differed somewhat between reactions (data not shown). This may reflect differences in the specific activity of different transglutaminase preparations. The observed polymerization of LTBP-1 and large latent complex implies that LTBP-1 and the large latent complex each contain both acyl donor and acyl acceptor sites necessary for formation of isopeptide bonds. Cross-linking LTBP-1 or large latent complex was Ca2+ dependent and transglutaminase specific as generation of high mol wt complexes was attenuated by the addition of EDTA or MDC (Fig. 2) (33). MDC acts as a competitive inhibitor of transglutaminase by competing for reactive glutamines (33). As large latent complex consists of small latent complex and LTBP-1, transglutaminase-dependent cross-linking of large latent complex to itself could involve either LTBP-1, small latent complex, or both. Cross-linking reactions using tissue transglutaminase and recombinant small latent complex also yielded high mol wt complexes (data not shown), indicating it also contains acyl donor and acceptor sites. However, this reaction is not responsible for the matrix incorporation of large latent complex as shown by antibody inhibition studies (see below). Iodination of either LTBP-1 or large latent complex did not generate reactive sites absent in nonradiolabeled proteins because transglutaminase-catalyzed incorporation of MDC into nonradiolabeled LTBP-1 or large latent complex was observed using black light to visualize MDC-containing proteins resolved by SDS-PAGE (data not shown) (6).
Figure 2.
SDS-PAGE analysis of cross-linking large latent complex or LTBP-1 by transglutaminase. (A) Iodinated large latent complex (LLC) was incubated with guinea pig liver transglutaminase (TGase) in the presence or absence of EDTA and CaCl2. Proteins were separated by reducing SDS-PAGE using a 3% stacking and 7% resolving gel and were revealed by PhosphorImager scanning. (B) Iodinated LTBP-1 was incubated with guinea pig liver transglutaminase in the presence or absence of MDC. Proteins were separated by reducing SDS-PAGE using a 3% stacking and 12% resolving gel and were revealed by PhosphorImager scanning.
Transglutaminase-dependent Cross-Linking of LTBP-1 and ECM by Cells
Both free LTBP-1 and large latent complex have been described to associate covalently with ECMs generated by fibroblasts, endothelial cells, and epithelial cells (59–61). The results described above suggested the hypothesis that matrix incorporation of LTBP-1 and large latent complex occurs through transglutaminase-catalyzed cross-linking of LTBP-1 and matrix proteins. To test this hypothesis, we first confirmed that homotypic cultures of HT 1080 and BAE cells as well as heterotypic cultures of BAE and BSM cells generate matrix-associated LTBP-1. ECM-bound LTBP-1 was analyzed by experiments in which matrix digests prepared from metabolically labeled cultures were immunoprecipitated using Ab 39. Immunoprecipitated matrix digests prepared from HT 1080 and BAE cells, as well as from cocultures of BAE and BSM cells, revealed fragments of 120–140 and 180–210 kD, respectively (Fig. 3). These matrix fragments specifically reacted with Ab 39 because they were not observed in control immunoprecipitation experiments with nonimmune serum (data not shown). The difference in mol wt of matrix fragments immunoprecipitated with Ab 39 obtained from HT 1080 and vascular cells is consistent with a report that LTBP-1 recovered from matrix digests prepared from different cell types varies in mol wt (61). The differences in size of the LTBP-1 fragments recovered from the ECM may reflect variations in matrix composition, in differential expression of LTBP-1 isoforms incorporated into the ECM (26), or in endogenous proteolytic activities in the cell culture.
Figure 3.
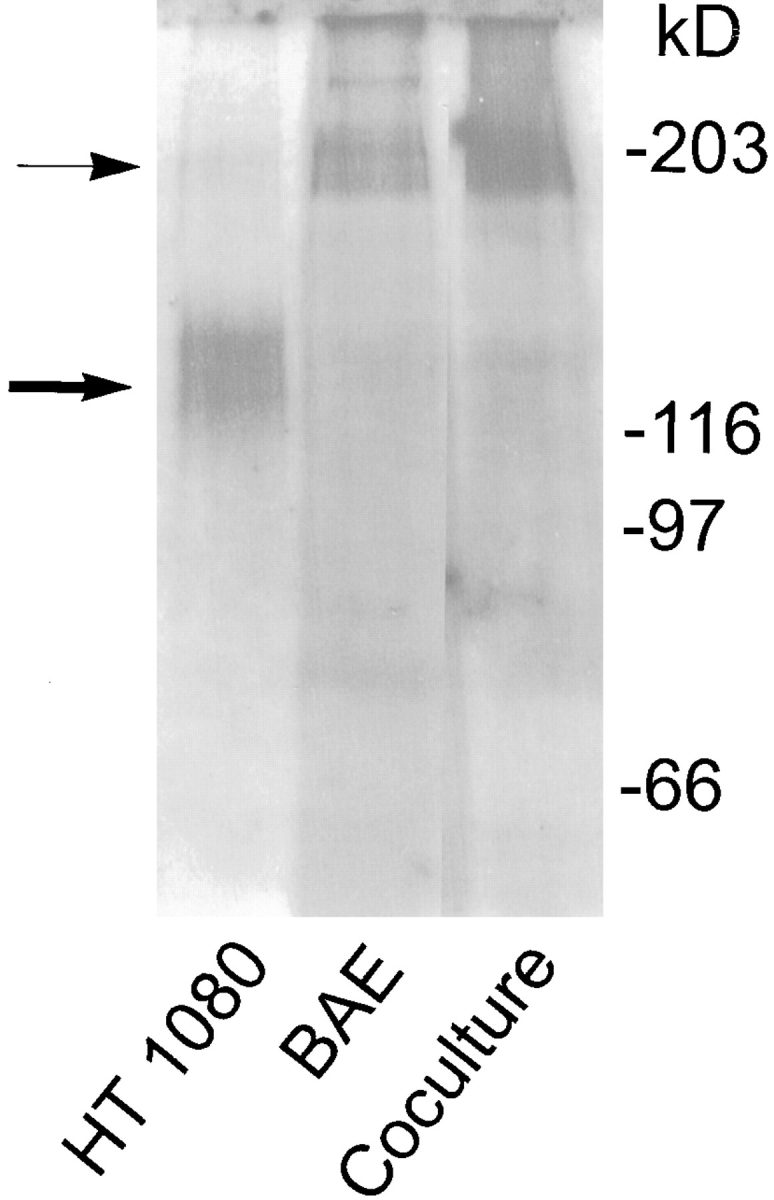
Presence of LTBP-1 in matrix digests prepared from HT 1080, BAE, and cocultures of BAE and BSM cells. Subconfluent cultures of HT 1080 and BAE cells and cocultures of BAE and BSM cells were pulsed with [35S]cysteine for 3 h and chased overnight. Matrices were prepared, followed by digestion with 0.3 U/ml of plasmin. Digests were immunoprecipitated using Ab 39 serum followed by protein A–agarose. Immunoprecipitates were analyzed by SDSPAGE followed by fluorography. ➞ , 120–140-kD LTBP-1 matrix fragments. → , 180– 210-kD LTBP-1 matrix fragments.
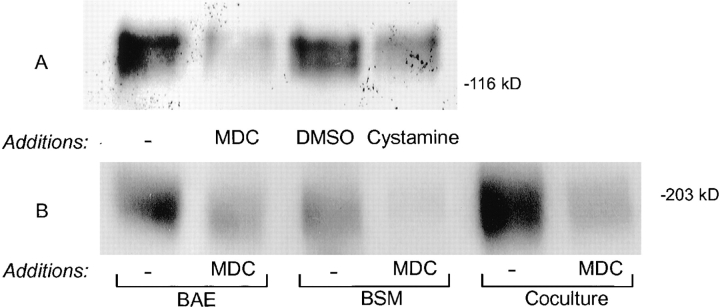
Having confirmed that LTBP-1 was present in the ECM, we next examined whether the incorporation of endogenous LTBP-1 into the ECM by cells was transglutaminase dependent. Matrices were prepared from cultures metabolically labeled in the presence of the transglutaminase competitive inhibitors MDC or cystamine (33), and the LTBP-1 content of the ECM was examined by digesting matrices with plasmin and immunoprecipitating the digests with Ab 39 followed by SDS-PAGE analysis. Significantly less LTBP-1 was recovered from matrices prepared from cultures of HT 1080, BAE, and BSM cells treated with MDC or cystamine than from control cultures (Fig. 4, A and B). Matrix digests from cultures incubated with transglutaminase inhibitors contained 2–10-fold less LTBP-1 than respective control cultures as determined by laser scanning densitometry. Cells treated with MDC incorporated less LTBP-1 into the matrix than did cystaminetreated cells (Fig. 4 A, second and fourth lanes), probably because the concentration of cystamine used in cultures was below the saturating concentration needed to completely inhibit pericellular transglutaminase activity. Levels of MDC or cystamine greater than those used in our experiments may have been necessary to completely inhibit matrix incorporation of LTBP-1. However, higher levels of these transglutaminase inhibitors were toxic to cells. To insure that results obtained using transglutaminase inhibitors did not result from effects on total protein synthesis and secretion, all matrix digests were normalized to total TCA precipitable cpms before immunoprecipitating with Ab 39 serum (11). In addition, the effect of MDC on protein synthesis and secretion was monitored by immunoprecipitating fibronectin from metabolically labeled CM harvested from untreated and MDC-treated HT 1080 cells. Fibronectin levels in treated and untreated HT 1080 CM were found to be equivalent (data not shown). Assuming that the transglutaminase inhibitors did not selectively affect LTBP-1 expression, the decreased amount of LTBP-1 observed in matrix digests prepared from cultures treated with inhibitors suggests that the matrix incorporation of LTBP-1 is transglutaminase dependent.
Figure 4.
LTBP-1 content of matrix digests prepared from cultures treated with transglutaminase inhibitors. (A) Subconfluent cultures of HT 1080 cells were either untreated or treated with 50 μM MDC, 0.06% DMSO, or 100 μM cystamine during the 3-h pulse with [35S]cysteine and overnight chase. (B) Subconfluent homotypic and heterotypic cultures of BAE and BSM cells were either untreated or MDC-treated (50 μM) during the pulse-chase period as described for HT 1080 cells. Matrix digests were prepared from all cultures and immunoprecipitated using Ab 39 serum. Immunoprecipitates were analyzed by SDS-PAGE followed by fluorography.
In similar experiments, twice as much LTBP-1 was recovered from BAE matrix digests than from BSM matrix digests as determined by immunoprecipitating plasmin digests of metabolically labeled ECM using Ab 39 (Fig. 4 B). This may reflect higher levels of tissue transglutaminase expression by BAE cells compared to BSM cells rather than differences in LTBP-1 expression, as BSM cells have been reported to produce more LTBP-1 than BAE cells (19, 31).
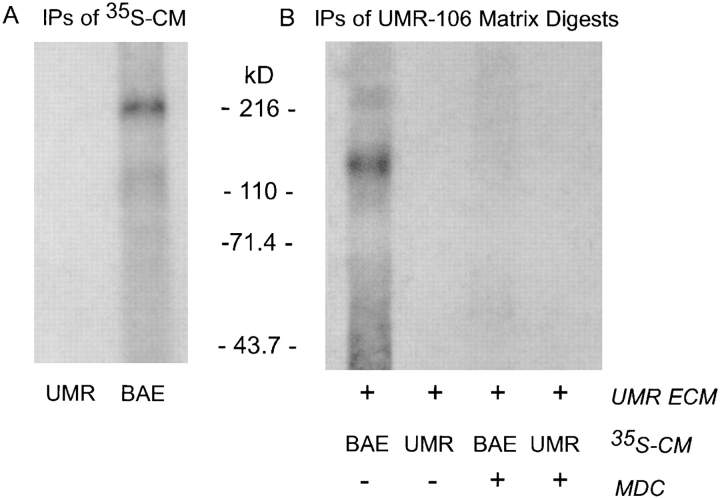
To establish whether the effect of transglutaminase inhibitors on the matrix incorporation of LTBP-1 as observed in culture was a consequence of altered matrix assembly and stabilization, matrix incorporation reactions were done using matrices from untreated UMR-106 cells, 35S-labeled BAE CM as an exogenous source of large latent complex, and the plasma transglutaminase Factor XIIIa. Matrices from UMR-106 cells were used because UMR106 cells produce latent TGF-β only as the small latent complex (12). We confirm that large latent complex was not generated by our UMR-106 cells, as no LTBP-1 was detected in Ab 39 immunoprecipitates of 35S-metabolically labeled CM (Fig. 5 A, first lane). Metabolically labeled CM from BAE cells was used as an exogenous source of large latent complex, as we have previously observed (19), and confirmed that they produce LTBP-1 as large latent complex (220 kD; Fig. 5 A, second lane ). The LTBP-1 content of UMR-106 matrices incubated with radiolabeled BAE CM in the absence or presence of MDC was analyzed by digesting matrices with plasmin and immunoprecipitating digests with Ab 39 (Fig. 5 B). Visualization of immunoprecipitates by fluorography revealed that matrices reacted with transglutaminase and BAE CM contained crosslinked LTBP-1 (Fig. 5 B, first lane), whereas digests from a parallel matrix reaction containing MDC did not (Fig. 5 B, third lane). Control matrix incorporation reactions of UMR-106 CM and UMR-106 matrices confirmed that the ECM of UMR-106 cells does not contain endogenous LTBP-1, as no LTBP-1 was recovered from matrix digests. Results from matrix incorporation experiments indicate that the decreased levels of matrix-associated LTBP-1 observed in cultures treated with MDC were due to the inhibition of cross-linking by transglutaminase rather than effects on matrix assembly and stability.
Figure 5.
Cross-linking LTBP-1 in BAE CM to UMR-106 ECM by Factor XIIIa. (A) UMR-106 and BAE cultures were metabolically labeled using [35S]cysteine and CM were immunoprecipitated using Ab 39 serum. LTBP-1 immunoprecipitates (IPs) from CM were separated by nonreducing SDS-PAGE and visualized by fluorography. (B) Matrix incorporation reactions were done using metabolically labeled CM from BAE or UMR-106 cells and UMR-106 ECMs. The cross-linking reaction was catalyzed by Factor XIIIa in the absence (−) or presence (+) of 50 μM MDC. Matrix digests were prepared by plasmin digestion and immunoprecipitated using Ab 39 serum. Immunoprecipitates were analyzed by reducing SDS-PAGE and fluorography.
Identification of LTBP-1 Domains Involved in Cross-Linking by Transglutaminase
Results presented in Figs. 2, 4, and 5 demonstrate that LTBP-1 and large latent complex are substrates for transglutaminase and that matrix incorporation of large latent complex is transglutaminase dependent. To determine whether LTBP-1 is necessary for cross-linking of large latent complex, antibodies to intact LTBP-1 or different LTBP-1 sequences were included in cross-linking reactions of transglutaminase and iodinated large latent complex as described previously. Ab 39 inhibited transglutaminase-dependent cross-linking of large latent complex, as measured by the absence of polymerized large latent complex present as a high mol wt band in the 3% stacking polyacrylamide SDS gel (Fig. 6 A). Ab 39 did not interfere with cross-linking of small latent complex by transglutaminase (data not shown). This indicated that cross-linking of the large latent complex probably requires lysine or glutamine residues present in LTBP-1. To identify what region of LTBP-1 contains transglutaminase reactive residues, affinity-purified anti-450 and anti-849 IgG were added to transglutaminase-dependent cross-linking reactions of iodinated LTBP-1 and large latent complex. Addition of anti-450 IgG blocked cross-linking of large latent complex and LTBP-1 to itself, as measured by the decreased formation of high mol wt polymers observed in the stacking gel, whereas nonimmune, anti-849, and anti-1065 IgGs did not (Fig. 6). These findings indicate that transglutaminasereactive sites are present in the amino-terminal region of LTBP-1.
Figure 6.
Effect of antibodies to LTBP-1 on transglutaminase cross-linking of large latent complex or LTBP-1. Ab 39 IgG (0.2 mg/ml) and affinity-purified IgGs (0.8 mg/ml) generated against GST-450, GST-1065, or GST-849 were added to transglutaminase cross-linking reactions of iodinated large latent complex (LLC) (A) or LTBP-1 (B). The presence of high mol wt complexes was revealed by reducing SDS-PAGE using a 3% stacking and 7% resolving gel followed by PhosphorImager scanning.
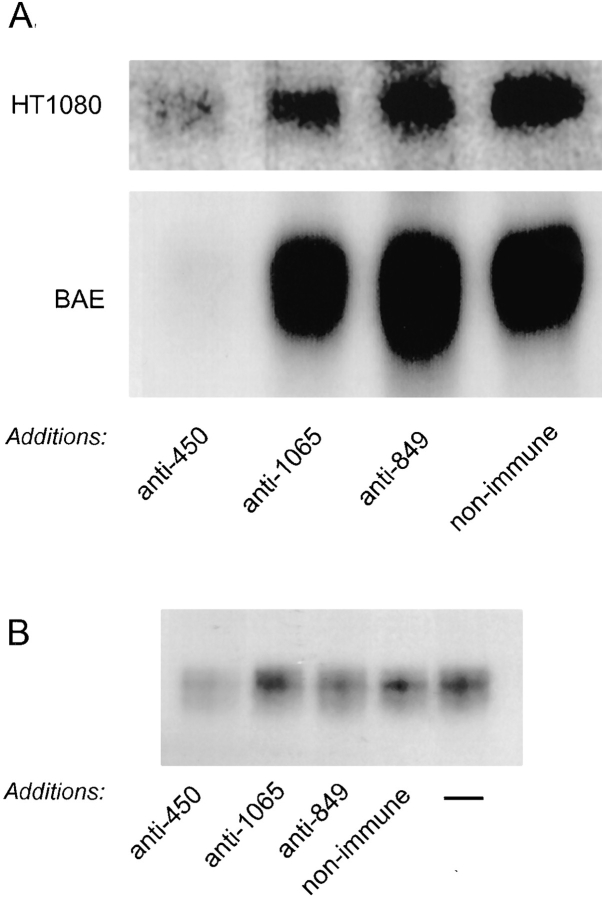
To establish whether the amino-terminal region also contained transglutaminase reactive sites involved in the matrix association of LTBP-1, affinity-purified antibodies to the three GST–LTBP-1S fusion proteins were added to cultures of HT 1080 and BAE cells throughout a pulsechase metabolic labeling period. Because HT 1080 cells are proteolytically more active than BAE cells (18), we added aprotinin, a serine protease inhibitor, to HT 1080 cultures to prevent antibodies from being degraded by endogenous proteases. The amount of LTBP-1 incorporated in the matrix by antibody-treated HT 1080 and BAE cells was assessed by immunoprecipitating matrix digests with Ab 39 followed by SDS-PAGE. The immunoprecipitation experiments revealed that the amount of LTBP-1 recovered from matrices prepared from cultures treated with anti-450 IgG was significantly decreased compared to that recovered from cultures treated with nonimmune, anti1065, or anti-849 IgG (Fig. 7 A). Laser scanning densitometry of fluorographs revealed that digests prepared from HT 1080 and BAE cultures receiving anti-450 IgG contained 5–10-fold and 70-fold less LTBP-1, respectively, than matrices prepared from cultures treated with the other antibodies. Addition of aprotinin did not affect the deposition of LTBP-1 into the matrix, as the cpms recovered by immunoprecipitating matrix digests prepared from aprotinin-treated and untreated cultures with Ab 39 were similar (data not shown). The weaker effect of anti-450 IgG on HT 1080 versus BAE cells may reflect degradation of the antibodies by metalloproteases; however, this was not verified. These results support findings reported by others that incorporation of LTBP-1 into the matrix requires its amino-terminal residues (46, 49).
Figure 7.
Effect of antibodies to LTBP-1 on matrix incorporation of LTBP-1. (A) Subconfluent cultures of HT 1080 and BAE cells were treated during the 3-h pulse and overnight chase labeling period with either protein A–purified nonimmune IgG or affinity-purified IgGs (20 μg/ml) generated against GST-450, GST1065, or GST-849. HT 1080 cultures also received 50 μg/ml aprotinin to prevent degradation of exogenously added antibodies. Matrix digests were prepared and immunoprecipitated using Ab 39 followed by SDS-PAGE and fluorography. (B) Matrix incorporation reactions of 35S-labeled BAE CM and UMR-106 ECMs were done in the absence (−) of additions or in the presence of protein A–purified non-immune rabbit IgG or affinity-purified IgGs (20 μg/ml) generated against GST-450, GST-1065, and GST-849. Matrix digests were prepared as previously described and their LTBP-1 content was analyzed by immunoblotting using Ab 39 serum as described in Materials and Methods.
To confirm the observation that addition of anti-450 IgG to cells producing large latent complex interferes with the covalent incorporation of the complex to the ECM, matrix incorporation reactions were done using UMR-106 matrices, 35S-labeled BAE CM, and Factor XIIIa, as described earlier, in the presence of antibodies to GST– LTBP-1S fusion proteins. Western blotting analysis of matrix digests with Ab 39 indicated that addition of anti-450 IgG attenuated matrix deposition of large latent complex, as less LTBP-1 was present in digests from matrix incorporation reactions containing anti-450 IgG compared to those receiving nonimmune, anti-1065, or anti-849 IgG (Fig. 7 B). Results from these matrix incorporation reactions confirm that cross-linking large latent complex to the ECM involves transglutaminase reactive residues at the amino terminus of LTBP-1.
Transglutaminase Reactivity of Amino-truncated LTBP-1S
The antibody to the amino terminus of LTBP-1S (anti-450 IgG) identified the amino-terminal region of LTBP-1 as containing transglutaminase-reactive sites. However, localization of reactive residues within the amino terminus is not possible using antibodies, as their inhibitory effect may be due to steric hindrance rather than binding to reactive residues. To identify domains within the amino-terminal region of LTBP-1S that contain the transglutaminase-reactive site(s), amino-terminal truncated mutants ΔN293 and ΔN441 LTBP-1S cDNAs (Fig. 8 A) were constructed for transient transfection of CHO cells. Control cultures were either untransfected or WT LTBP-1S cDNA–transfected cells. CHO cells transfected with WT, ΔN293, or ΔN441 LTBP-1S cDNA expressed and secreted LTBP-1S protein accordingly as determined by Western blotting CM using Ab 39 (Fig. 8 B). Untransfected (Fig. 8 B) cells, as well as mock (pcDNA3-cat) transfected cells (data not shown), produce low levels of LTBP-1 compared to cells transfected with LTBP-1S cDNA constructs. To determine if CHO cells incorporate LTBP-1S into ECM, matrix digests were prepared by plasmin treating deoxycholate-extracted matrices followed by immunoblotting using Ab 39. Matrix digests prepared from cells transfected with WT and ΔN293 LTBP-1S cDNA contained LTBP-1S, whereas little to no LTBP-1S was detected in digests from untransfected and ΔN441 LTBP-1S cDNA–transfected cells (Fig. 8 B). To determine whether matrix association of WT and ΔN293 LTBP-1S was transglutaminase dependent, MDC was added to untransfected and transfected CHO cultures. Western blotting analysis of the LTBP-1S content of matrix digests generated from MDC-treated transfected CHO cells revealed negligible levels of LTBP-1S present in digests from WT and ΔN293 LTBP-1S cDNA–transfected cells, indicating that matrix association was transglutaminase dependent (data not shown). Results from the LTBP1S transfection experiments indicate that, unlike ΔN441 LTBP-1S, ΔN293 LTBP-1S was cross-linked to matrix suggesting that residues 294–441 are critical to the transglutaminase-dependent reactivity of LTBP-1S.
Effect of Antibodies to GST–LTBP-1S Fusion Proteins on Latent TGF-β Activation
Activation of latent TGF-β by cocultures of BAE and BSM cells as well as by stimulated macrophages requires transglutaminase and LTBP-1 (19, 31, 44). Transglutaminase-dependent cross-linking of LTBP-1 into the matrix by cocultures is inhibited by anti–LTBP-1 antibodies (Fig. 4 B). Therefore, we examined whether antibodies to GST– LTBP-1S fusion proteins affected activation of latent TGF-β using cocultures of BAE and BSM cells and cultures of stimulated macrophages.
Serum-free heterotypic cultures of BAE and BSM cells were used as a cell system that generates mature TGF-β from endogenous large latent complex (14). Control cultures were serum-free homotypic cultures of BAE and BSM cells. The cocultures of BAE and BSM cells received either affinity-purified antibodies to one of the three GST–LTBP-1S fusion proteins, protein A–purified Ab 39, or nonimmune IgG, as indicated in Fig. 9. After an overnight incubation, CM from these cultures were bioassayed immediately for mature TGF-β using the MLEC-luciferase assay. These experiments revealed that CM from cocultures treated with Ab 39, anti-450, or anti-849 IgG contained less mature TGF-β than CM from untreated cocultures or cocultures treated with nonimmune or anti-1065 IgG (Fig. 9 A). We previously observed that Ab 39 IgG blocks latent TGF-β activation by cocultures of BAE and BSM cells (19). To verify that the measured luciferase activity was TGF-β dependent, neutralizing anti–TGF-β IgG was added to CM from untreated cocultures and found to inhibit 86% of the luciferase response (Fig. 9 A), indicating that most of the measured luciferase activity was TGF-β dependent. Anti-450 and anti-849 IgG blocked TGF-β generation at ID50's of 20 and 1 μg/ml, respectively. CM from cocultures treated with the anti–LTBP-1S IgGs contained levels of latent TGF-β similar to those from untreated cocultures, as determined by the MLEC-luciferase assay (data not shown). Furthermore, addition of the anti– LTBP-1S IgGs to CM from untreated cocultures did not inhibit TGF-β activity (data not shown). Therefore, the effects of the antibodies did not result from changes in total TGF-β levels.
Figure 9.
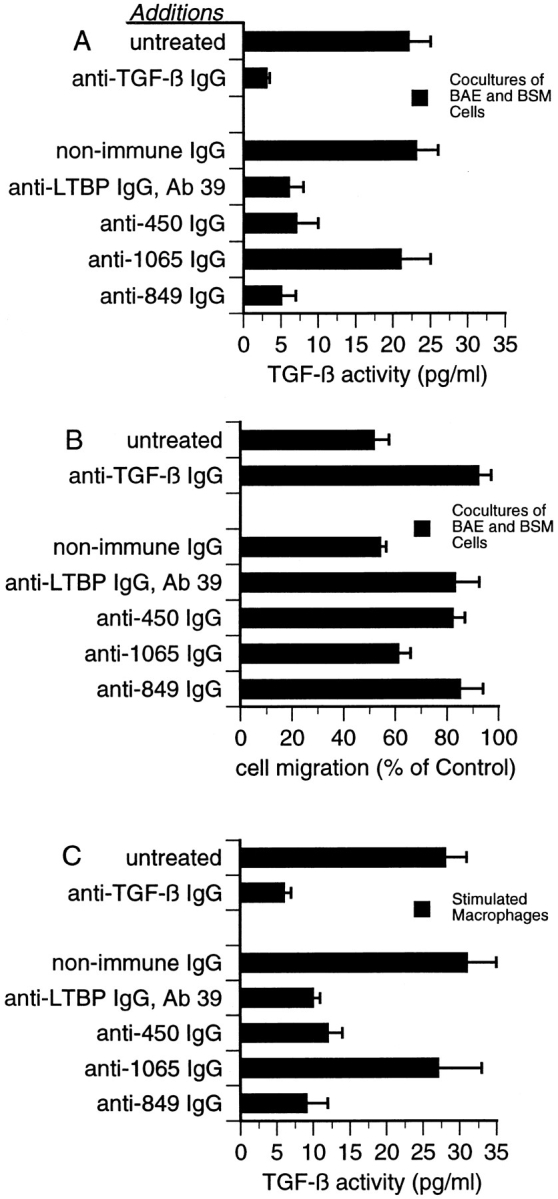
Effect of antibodies to LTBP-1 on latent TGF-β activation by cocultures of BAE and BSM cells, and stimulated macrophages. (A) The MLEC-luciferase assay was used to detect TGF-β activity generated by cocultures of BAE and BSM cells. Serum-free CM harvested from either untreated cultures or cultures treated with 10 μg/ml Ab 39 IgG, 50 μg/ml nonimmune IgG, or 50 μg/ml of affinity-purified anti-450, anti-1065, or anti849 IgG were assayed using the MLEC-luciferase assay. To verify that luciferase production was TGF-β–dependent, 20 μg/ml neutralizing anti–TGF-β IgG was added to CM from untreated cultures. Data presented are from one out of four experiments yielding similar results. (B) The wound migration assay was used to detect TGF-β activity generated by cocultures of BAE and BSM cells. Serum-free CM harvested from untreated cocultures or cocultures treated with the same antibodies as in A were assayed for TGF-β activity using the wound migration assay described in Materials and Methods. Control (100%) is the migration observed in wounded BAE cultures treated with media from homotypic BAE and BSM cells mixed 4:1. These data are presented as the percentage of migration observed in wounded cultures treated with control media. Data are representative of four experiments yielding similar results. (C) The MLEC-luciferase assay was used to detect TGF-β activity generated by stimulated macrophages. Serum-free CM from either untreated or antibodytreated cultures of LPS-stimulated thioglycollate-elicited peritoneal macrophages were harvested and assayed. Antibody treatments were as in A and B. Data presented are from one out of three experiments yielding similar results.
The results obtained using the MLEC-luciferase assay to measure mature TGF-β levels present in coculture CM were confirmed using the wound migration assay, in which the inhibitory effect of TGF-β on endothelial cell migration is quantitated. We observed that CM from cocultures of BAE and BSM cells treated with Ab 39, anti-450, or anti-849 IgG did not inhibit the migration of BAE cells, nor did anti–TGF-β IgG–containing CM from untreated cocultures (Fig. 9 B). CM from cocultures treated with nonimmune or anti-1065 IgG inhibited the migration of endothelial cells by ∼50% (Fig. 9 B). These results are similar to those obtained using the MLEC-luciferase assay, indicating that less TGF-β is present in CM prepared from cocultures treated with Ab 39, anti-450, or anti-849 IgG.
To determine whether the inhibitory effect of anti-450 or anti-849 IgG on latent TGF-β activation was specific to cocultures of BAE and BSM cells, we examined whether these antibodies abrogated TGF-β formation by LPSstimulated thioglycollate-elicited peritoneal macrophages, another cell system previously demonstrated to convert large latent complex to TGF-β (44). Serum-free CM from stimulated macrophages was prepared and assayed for TGF-β activity using the MLEC-luciferase assay. Levels of mature TGF-β present in CM from cultures treated with Ab 39, anti-450, or anti-849 IgG were similar to those present in anti–TGF-β IgG containing CM harvested from untreated cultures (Fig. 9 C). Like cocultures, stimulated macrophages were unaffected by anti-1065 IgG with respect to their ability to produce mature TGF-β (Fig. 9 C). Assays of heat-activated CM from untreated or antibodytreated stimulated macrophages revealed that all cultures generated similar levels of total TGF-β (data not shown). Results obtained using the MLEC-luciferase assay could not be confirmed using the wound migration assay because the wound migration assay was unresponsive to TGF-β present in CM from stimulated macrophages. Addition of either recombinant TGF-β1 or neutralizing anti– TGF-β IgG to CM from stimulated macrophages did not affect the migration of endothelial cells (data not shown). This may be due to the presence of migration stimulatory factors in macrophage CM (2). Nevertheless, anti-450 or anti-849 IgG blocked TGF-β generation by stimulated macrophages as determined by the MLEC-luciferase assay, suggesting that these antibody effects on large latent complex activation were not specific to cocultures of BAE and BSM cells and that the role of LTBP-1 in activation involves interactions with its amino- and carboxyl-terminal regions.
Discussion
The reactants involved in the activation of latent TGF-β by cocultures of endothelial and smooth muscle cells, retinoid-treated BAE cells, and stimulated macrophages include plasmin, cation-independent mannose 6-phosphate/ insulin-like growth factor type II receptor, tissue transglutaminase, and LTBP-1 (27). Interactions between the reactants involved in the activation of latent TGF-β have not been characterized in cell-free systems, with the exception that plasmin can cleave LAP, destabilizing noncovalent interactions between LAP and TGF-β, and that recombinant small latent complex binds to the cell surface cationindependent mannose 6-phosphate/insulin-like growth factor type II receptor via mannose 6-phosphate residues present on LAP (16, 35, 36). In this article, we have identified and characterized interactions between two reactants, LTBP-1 and tissue transglutaminase, that may be involved in latent TGF-β activation by cocultures of BAE and BSM cells as well as LPS-stimulated thioglycollate-elicited macrophages (31, 44, 54). These interactions include an enzyme–substrate relationship between transglutaminase and LTBP-1 and transglutaminase-dependent anchoring of large latent complex to the ECM. In addition, a second functional domain of LTBP-1 involved in latent TGF-β activation was identified.
Matrix incorporation of LTBP-1 or large latent complex (reported by others and reproduced by us) appears to occur through transglutaminase-dependent cross-linking of LTBP-1 and matrix protein(s) (60). We observed that both large latent complex and LTBP-1 are substrates for transglutaminase, the LTBP-1 of large latent complex contains residues involved in cross-linking large latent complex, and the inhibition of transglutaminase severely attenuates the incorporation of LTBP-1 into the matrix. Transglutaminase-reactive sites of LTBP-1S appear to be located within residues 294 and 441 of the amino terminus, as an LTBP-1S mutant truncated at its amino terminus by 441 amino acids was not cross-linked to the matrix, whereas a 293–amino acid truncated LTBP-1S retained its matrix association.
Transglutaminase is more selective for glutamine residues than acyl acceptor sites (25). A consensus sequence for glutamine residues that serve as amine acceptor sites in transglutaminase-catalyzed cross-linking has not been described. However, LTBP-1S shares some structural features present in other transglutaminase substrates. Sequence analysis of reactive glutamine residues reveals that they are exposed in loop structures, as the glutamines tend to be surrounded by positively or negatively charged amino acids (4). In 60% of the substrates, glutamine residues are located at the amino or carboxyl terminus. From sequence alignment analysis of human LTBP-1S and LTBP-2, rat LTBP-1 and murine LTBP-3 (28, 43, 45, 64), glutamine374 and a positively flanking amino acid within the second cysteine-rich repeat of LTBP-1S is conserved among all LTBPs, suggesting that it may be a transglutaminase reactive site. Glutamine-374 is present in ΔN293 LTBP-1S, which was incorporated into ECM but is absent in ΔN441 LTBP-1S, which appeared not to be cross-linked to ECM. Identification of the transglutaminase-reactive glutamine(s) is a current subject of investigation using biochemical and molecular approaches.
The matrix protein(s) to which LTBP-1 is cross-linked has not been identified. However, LTBP-1 has been described to colocalize with fibronectin (Taipale, J., J. Saharinen, K. Hedman, and J. Keski-Oja. 1994. Mol. Biol. Cell. 5[Suppl.]:311a) and with collagen-free fibrillar structures generated by fetal rat calvarial cells (13). LTBP-2 appears to closely associate with elastin-associated microfibrils (22). Thus, fibronectin and proteins of elastic fibers, such as microfibril-associated glycoprotein, are candidate proteins, as they all contain residues susceptible to transglutaminase-catalyzed isopeptide bond formation (10, 37).
In addition to localizing the transglutaminase reactivity of LTBP-1S to amino-terminal residues 294–441, LTBP-1S domains participating in latent TGF-β activation were identified. Both the amino and carboxyl regions of LTBP-1S appear to be involved in protein interactions required for activation as antibodies to these two domains abrogated TGF-β generation by cocultures of BAE and BSM cells as well as by LPS-stimulated thioglycollate-elicited macrophages. Previously, we reported that protein A–purified anti450 IgG (the antibody to the amino terminus of LTBP-1S) did not affect activation of large latent complex by stimulated macrophages (44). This discrepancy results from the fact that in the earlier work total IgG was used, whereas in the experiments reported here affinity-purified IgG was tested. Based upon the concentration of affinity-purified anti-450 IgG required to inhibit activation of large latent complex, the amount of IgG used previously would have been insufficient to observe an effect. Anti-450 IgG also inhibited matrix association of LTBP-1 and large latent complex, suggesting that the role of the amino terminus in latent TGF-β activation may be to mediate the incorporation of large latent complex into the matrix.
We have demonstrated that the carboxyl-terminal region of LTBP-1 is required for large latent complex activation, as antibodies to this region blocked TGF-β generation (44). The carboxyl-terminal sequence of LTBP-1 does not appear to contain sites involved in covalently attaching LTBP-1 to the ECM, as addition of these antibodies to cultures did not affect the matrix content of LTBP-1. This region does contain a putative protease-sensitive site at residue 1257 located after the third cysteine-rich repeat (28, 49, 60). Cleavage at this site might facilitate activation of soluble forms of large latent complex.
Matrix incorporation of large latent complex may create a concentrated pool of large latent complex. Others have proposed that segregating pools of latent TGF-β may be important in regulating the conversion of large latent complex to mature TGF-β at specific sites (23). Alternatively, we speculate that soluble large latent complex may be resistant to plasmin activation. It has been demonstrated that activation of latent TGF-β in fibroblast CM by plasmin requires nonphysiological levels of plasmin (35). We also find that recombinant large latent complex is not readily activated by plasmin in solution (data not shown). Therefore, cross-linking to the matrix may sequester large latent complex that is not readily activated, and subsequent release from the matrix by proteolysis (60) could generate a modified complex susceptible to plasmin activation.
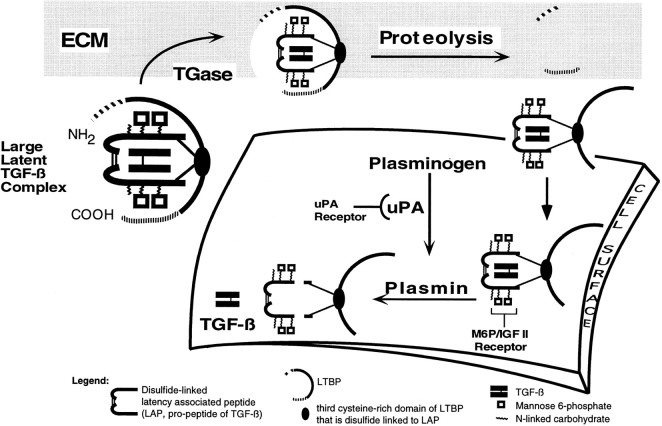
We propose that the activation of large latent complex, to release, TGF-β occurs by the sequence of reactions illustrated in Fig. 10. Cross-linking large latent complex to the ECM is an early step in activation of large latent complex. From our antibody inhibition studies on latent TGF-β activation, both the amino and carboxyl domains of LTBP-1 are involved in protein interactions necessary for activation. The role of the amino terminus of LTBP-1S appears to be in the cross-linking of large latent complex to the matrix, as we observed that the amino terminus contains transglutaminase reactive sites and transglutaminase activity is required for activation (31). We propose that matrix-bound large latent complex is released by cleaving LTBP-1S at a potential tribasic protease site (arginine415) on the carboxyl side of the transglutaminase reactive residue(s). This site has been proposed by Taipale et al. to be plasmin sensitive (60). We hypothesize that the carboxyl domain of LTBP-1S is involved in forming noncovalent interactions, perhaps with matrix, as antibodies to this domain did not interfere with the cross-linking to the matrix but did block TGF-β generation. An additional cleavage of LTBP-1S at the carboxyl terminus (residue 1257) as proposed by Taipale et al. (60) would release a complex with an LTBP-1 molecule containing only its core EGFlike repeats plus cysteine-rich repeats 3–4 bound to small latent complex. This proteolytic processing of large latent complex may be required for subsequent activation steps, such as binding to cell surface mannose 6-phosphate/insulin-like growth factor type II receptors (16, 32). We speculate that cross-linking to the matrix occurs before targeting to the cell surface mannose 6-phosphate/insulin-like factor type II receptor, as the addition of excess mannose 6-phosphate does not interfere with the matrix association of LTBP-1 or large latent complex (data not shown) but abrogates TGF-β generation (16). Once on the cell surface, fragmented latent complex is susceptible to plasmin-dependent activation by proteolytic cleavage of LAP (27, 36). The TGF-β released then binds to its receptor.
Figure 10.
Model of plasmindependent activation of large latent complex. A diagrammatic representation of large latent complex is presented as covalently interacting with the ECM by transglutaminase (TGase)–dependent cross-linking of LTBP-1 and matrix protein(s). For activation of large latent complex to proceed, large latent complex is solubilized by proteolysis of LTBP-1. It is proposed that the release of large latent complex occurs by cleavage occurring at the amino terminus of LTBP-1 downstream of the TGase reactive site(s). In addition, there may be a second cleavage at the carboxyl terminus of LTBP-1 resulting in matrix release and exposure of mannose 6-phosphate (M6P) residues of LAP involved in localizing latent TGF-β to the cell surface. Proteolytically processed LTBP-1 is proposed to target to the cell surface where cell-associated plasmin cleaves LAP to liberate mature TGF-β from the complex.
This model suggests several testable questions. These include the potential significance of the putative plasmin cleavage sites in LTBP-1S, the availability of the mannose 6-phosphate residues of LAP in the complex, and the location of plasmin-sensitive sites of LAP. It is also unknown whether large latent complexes that differ in their LTBP isoforms are activated by a similar mechanism. Answers to these questions can be approached by altering specific residues in the components of large latent complex and monitoring rates of large latent complex activation.
Acknowledgments
The authors thank Ms. Melinda Vassallo for her excellent technical assistance and Dr. John S. Munger for his critical review of the manuscript and stimulating discussions.
Abbreviations used in this paper
- BAE
bovine aortic endothelial
- BSM
bovine smooth muscle
- CM
conditioned medium
- ECM
extracellular matrix
- GST
glutathione S–transferase
- LAP
latency associated peptide
- LPS
lipopolysaccharide
- LTBP-1
latent TGF-β
- binding protein-1; MDC
monodansylcadaverine
- MLEC
mink lung epithelial cells
- PAI-1
plasminogen activator inhibitor-1
- TGF-β
transforming growth factor-β
- WT
wild type
Footnotes
This research was supported by grants CA 2753 (D.B Rifkin), CA 34282 (D.B Rifkin), EY 06537 (I. Nunes), T32GM 07238 (I. Nunes), and CA 09161 (C.N. Metz) from the National Institutes of Health. P.-E. Gleizes was the recipient of a fellowship from the Association pour la Recherche Contre le Cancer.
Address all correspondence to Irene Nunes, Department of Cell Biology, MSB 650, NYU Medical Center, 550 First Ave., New York, NY 10016. Tel.: (212) 263-5327. Fax: (212) 263-8139.
Christine Metz's current address is The Picower Institute for Medical Research, 350 Community Drive, Manhasset, NY 11030.
References
- 1.Abe M, Harpel JG, Metz CN, Nunes I, Loskutoff DJ, Rifkin DB. An assay for transforming growth factor-β using cells transfected with a plasminogen activator inhibitor-1 promoter luciferase construct. Anal Biochem. 1994;216:276–284. doi: 10.1006/abio.1994.1042. [DOI] [PubMed] [Google Scholar]
- 2.Adams D, Hamilton TA. The cell biology of macrophage activation. Annu Rev Immunol. 1984;2:283–318. doi: 10.1146/annurev.iy.02.040184.001435. [DOI] [PubMed] [Google Scholar]
- 3.Aeschlimann D, Paulsson M. Cross-linking of laminin-nidogen complexes by tissue transglutaminase. J Biol Chem. 1991;266:15308–15317. [PubMed] [Google Scholar]
- 4.Aeschlimann D, Paulsson M, Mann K. Identification of gln726 in nidogen as the amine acceptor in transglutaminase-catalyzed crosslinking of laminin-nidogen complexes. J Biol Chem. 1992;267:11316–11321. [PubMed] [Google Scholar]
- 5.Barcellos-Hoff MH, Derynck R, Tsang ML, Weatherbee JA. Transforming growth factor-β activation in irradiated murine mammary gland. J Clin Invest. 1994;93:892–899. doi: 10.1172/JCI117045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bendixen E, Wolfgang B, Harpel PC. Transglutaminases catalyze cross-linking of plasminogen to fibronectin and human endothelial cells. J Biol Chem. 1993;268:21962–21967. [PubMed] [Google Scholar]
- 7.Bishop PD, Teller DC, Smith RA, Lasser GW, Gilbert T, Seale RL. Expression, purification, and characterization of human factor XIII in Saccharomyces cerevisiae. . Biochemistry. 1990;29:1861–1869. doi: 10.1021/bi00459a028. [DOI] [PubMed] [Google Scholar]
- 8.Borth W, Chang VT, Bishop P, Harpel PC. Lipoprotein(a) is a substrate for factor XIIIa and tissue transglutaminase. J Biol Chem. 1991;266:18149–18153. [PubMed] [Google Scholar]
- 9.Brown P, Wakefield L, Levinson A, Sporn M. Physicochemical activation of recombinant latent transforming growth factor-beta's 1,2,3. Growth Factors. 1990;3:35–43. doi: 10.3109/08977199009037500. [DOI] [PubMed] [Google Scholar]
- 10.Brown-Augsburger P, Broekelmann T, Mecham L, Mercer R, Gibson MA, Cleary EG, Abrams WR, Rosenbloom J, Mecham R P. Microfibril-associated glycoprotein (MAGP) binds to the carboxyterminal domain of tropoelastin and is a substrate for transglutaminase. J Biol Chem. 1994;269:28443–28449. [PubMed] [Google Scholar]
- 11.Bungay PJ, Potter JM, Griffin M. The inhibition of glucosestimulated insulin secretion by primary amines. Biochem J. 1984;219:819–827. doi: 10.1042/bj2190819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dallas S, Park-Snyder S, Miyazono K, Twardzik D, Mundy GR, Bonewald LF. Characterization and autoregulation of latent transforming growth factor β (TGF-β) complexes in osteoblast-like cell lines. J Biol Chem. 1994;269:6815–6822. [PubMed] [Google Scholar]
- 13.Dallas SL, Miyazono K, Skerry TM, Mundy GR, Bonewald LF. Dual role for the latent transforming growth factor-β binding protein in storage of latent TGF-β in the extracellular matrix and as a structural matrix protein. J Cell Biol. 1995;131:539–549. doi: 10.1083/jcb.131.2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daopin S, Piez KA, Ogawa Y, Davies DR. Crystal structure of transforming growth factor-β2: an unusual fold for the superfamily. Science (Wash DC) 1992;257:369–373. doi: 10.1126/science.1631557. [DOI] [PubMed] [Google Scholar]
- 15.Dasch JR, Pace DR, Waegell W, Inenaga D, Ellingsworth L. Monoclonal antibodies recognizing transforming growth factor-beta. Bioactivity neutralization and transforming growth factor beta2 affinity purification. J Immunol. 1989;142:1536–1541. [PubMed] [Google Scholar]
- 16.Dennis PA, Rifkin DB. Cellular activation of latent transforming growth factor β requires binding to the cation-independent mannose 6-phosphate/insulin-like growth factor type II receptor. Proc Natl Acad Sci USA. 1991;88:580–584. doi: 10.1073/pnas.88.2.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Derynck R, Jarrett JA, Chen EY, Eaton DH, Bell JR, Assoian RK, Roberts AB, Sporn MB, Goedel DV. Human transforming growth factor-β complementary DNA sequences and expression in normal and transformed cells. Nature (Lond) 1985;316:701–705. doi: 10.1038/316701a0. [DOI] [PubMed] [Google Scholar]
- 18.Flaumenhaft R, Abe M, Mignatti P, Rifkin DB. Basic fibroblast growth factor-induced activation of latent transforming growth factor beta in endothelial cells: regulation of plasminogen activator activity. J Cell Biol. 1992;118:901–909. doi: 10.1083/jcb.118.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flaumenhaft R, Abe M, Sato Y, Miyazono K, Harpel J, Heldin C, Rifkin DB. Role of the latent TGF-β binding protein in the activation of latent TGF-β by co-cultures of endothelial and smooth muscle cells. J Cell Biol. 1993;120:995–1002. doi: 10.1083/jcb.120.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gentry L, Webb NR, Lim GJ, Brunner AM, Ranchalis JE, Twardzick DR, Lioubin MN, Marquardt H, Purchio AF. Type 1 transforming growth factor beta: amplified expression and secretion of mature and precursor polypeptides in Chinese hamster ovary cells. Mol Cell Biol. 1987;7:3418–3427. doi: 10.1128/mcb.7.10.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gentry LE, Lioubin MN, Purchio AF, Marquardt H. Molecular events in the processing of recombinant type I pre-pro-transforming growth factor beta to the mature polypeptide. Mol Cell Biol. 1988;8:4162–4168. doi: 10.1128/mcb.8.10.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson MA, Hatzinikolas G, Davis EC, Baker E, Sutherland GR, Mecham R. Bovine latent transforming growth factor-β1-binding protein 2: molecular cloning, identification of tissue isoforms, and immunolocalization to elastin-associated microfibrils. Mol Cell Biol. 1995;15:6932–6942. doi: 10.1128/mcb.15.12.6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22a.Gleizes P-E, Beauis RC, Mazzieri R, Shen B, Rifkin DB. Identification and characterization of an eight-cysteine repeat of the latent transforming growth factor-β binding protein-1 that mediates bonding to the latent transforming growth factor-β1. J Biol Chem. 1996;271:29891–29896. doi: 10.1074/jbc.271.47.29891. [DOI] [PubMed] [Google Scholar]
- 23.Grainger DJ, Wakefield L, Bethell HW, Farndale RW, Metcalfe JC. Release and activation of platelet latent TGF-β in blood clots during dissolution with plasmin. Nat Med. 1995;1:932–937. doi: 10.1038/nm0995-932. [DOI] [PubMed] [Google Scholar]
- 24.Greenberg CS, Birckbichler PJ, Rice RH. Transglutaminases: multifunctional cross-linking enzymes that stabilize tissues. FASEB (Fed Am Soc Exp Biol) J. 1991;5:3071–3077. doi: 10.1096/fasebj.5.15.1683845. [DOI] [PubMed] [Google Scholar]
- 25.Grootjans JJ, Groenen TA, de Jong WW. Substrate requirements for transglutaminases. J Biol Chem. 1995;270:22855–22858. doi: 10.1074/jbc.270.39.22855. [DOI] [PubMed] [Google Scholar]
- 26.Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, NY. 678–679.
- 27.Harpel JG, Metz CN, Kojima S, Rifkin DB. Control of transforming growth factor-β activity: latency vs. activation. Prog Growth Factor Res. 1993;4:321–335. doi: 10.1016/0955-2235(92)90014-9. [DOI] [PubMed] [Google Scholar]
- 28.Kanzaki T, Olofsson A, Moren A, Wernstedt C, Hellman U, Miyazono K, Claesson-Welsh L, Heldin CH. TGF-β1 binding protein: a component of the large latent complex of TGF-β with multiple repeat sequences. Cell. 1990;61:1051–1061. doi: 10.1016/0092-8674(90)90069-q. [DOI] [PubMed] [Google Scholar]
- 29.Kielty CM, Rantamaki T, Child AH, Shuttleworth CA, Peltonen L. Cysteine-to-arginine point mutation in a “hybrid” eight-cysteine domain of FBN1: consequences for fibrillin aggregation and microfibril assembly. J Cell Sci. 1995;108:1317–1323. doi: 10.1242/jcs.108.3.1317. [DOI] [PubMed] [Google Scholar]
- 30.Kojima S, Rifkin DB. Mechanism of retinoid-induced activation of latent transforming growth factor-β in bovine endothelial cells. J Cell Physiol. 1993;155:323–332. doi: 10.1002/jcp.1041550213. [DOI] [PubMed] [Google Scholar]
- 31.Kojima S, Kiyomitsu N, Rifkin DB. Requirement for transglutaminase in the activation of latent transforming growth factor-β in bovine endothelial cells. J Cell Biol. 1993;121:439–448. doi: 10.1083/jcb.121.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovacina KS, Steele-Perkins G, Purchio AF, Lioubin M, Miyazono K, Heldin C, Roth R. Interactions of recombinant and platelet transforming growth factor-β1 precursor with the insulin-like growth factor II/mannose 6-phosphate receptor. Biochem Biophys Res Commun. 1989;160:393–403. doi: 10.1016/0006-291x(89)91669-0. [DOI] [PubMed] [Google Scholar]
- 33.Lorand L, Conrad SM. Transglutaminases. Mol Cell Biochem. 1984;58:9–35. doi: 10.1007/BF00240602. [DOI] [PubMed] [Google Scholar]
- 34.Lyons RM, Moses HL. Transforming growth factor-β and the regulation of cell proliferation. Eur J Biochem. 1990;187:467–473. doi: 10.1111/j.1432-1033.1990.tb15327.x. [DOI] [PubMed] [Google Scholar]
- 35.Lyons RM, Keski-Oja J, Moses HL. Proteolytic activation of latent transforming growth factor-β from fibroblast-conditioned medium. J Cell Biol. 1988;106:1659–1665. doi: 10.1083/jcb.106.5.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyons RM, Gentry LE, Purchio AF, Moses HL. Mechanism of activation of latent recombinant transforming growth factor β1 by plasmin. J Cell Biol. 1990;110:1361–1367. doi: 10.1083/jcb.110.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez J, Chalupowicz DG, Roush RK, Sheth A, Barsigian C. Transglutaminase-mediated processing of fibronectin by endothelial cell monolayers. Biochemistry. 1994;33:2538–2545. doi: 10.1021/bi00175a024. [DOI] [PubMed] [Google Scholar]
- 38.Massague J. The transforming growth factor-β family. Annu Rev Cell Biol. 1990;6:597–641. doi: 10.1146/annurev.cb.06.110190.003121. [DOI] [PubMed] [Google Scholar]
- 39.McConahey PJ, Dixon FJ. Radioiodination of proteins by the use of the chloramine-T method. Methods Enzymol. 1980;70:210–213. doi: 10.1016/s0076-6879(80)70050-2. [DOI] [PubMed] [Google Scholar]
- 40.Miyazono K, Hellman U, Wernstedt C, Heldin C. Latent high molecular weight complex of transforming growth factor β1. J Biol Chem. 1988;263:6407–6415. [PubMed] [Google Scholar]
- 41.Miyazono K, Olofsson A, Colosetti P, Heldin C. A role of the latent TGF-β1 binding protein in the assembly and secretion of TGF-β1. EMBO (Eur Mol Biol Organ) J. 1991;10:1091–1101. doi: 10.1002/j.1460-2075.1991.tb08049.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miyazono K, Ichijo H, Heldin CH. Transforming growth factor-beta: latent forms, binding proteins and receptors. Growth Factors. 1993;8:11–22. doi: 10.3109/08977199309029130. [DOI] [PubMed] [Google Scholar]
- 43.Moren A, Olofsson A, Stenman G, Sahlin P, Kanzaki T, ClaessonWelsh L, ten Dijke P, Miyazono K, Heldin CH. Identification and characterization of LTBP-2, a novel latent transforming growth factor-β-binding protein. J Biol Chem. 1994;269:32469–32478. [PubMed] [Google Scholar]
- 44.Nunes I, Shapiro RL, Rifkin DR. Characterization of latent TGF-β activation by murine peritoneal macrophages. J Immunol. 1995;155:1450–1459. [PubMed] [Google Scholar]
- 45.Okada F, Yamaguchi K, Ichihara A, Nakamura T. Purification and structural analysis of a latent transforming growth factor-β rat platelets. J Biochem. 1989;106:304–310. doi: 10.1093/oxfordjournals.jbchem.a122849. [DOI] [PubMed] [Google Scholar]
- 46.Olofsson A, Ichijo H, Moren A, ten Dijke P, Miyazono K, Heldin CH. Efficient association of an amino-terminally extended form of human latent transforming growth factor-β binding protein with the extracellular matrix. J Biol Chem. 1995;270:31294–31297. doi: 10.1074/jbc.270.52.31294. [DOI] [PubMed] [Google Scholar]
- 47.Ramirez F. Fibrillin mutations in Marfan syndrome and related phenotypes. Curr Opin Genet Dev. 1996;6:309–315. doi: 10.1016/s0959-437x(96)80007-4. [DOI] [PubMed] [Google Scholar]
- 48.Rosenbloom J, Abrams WR, Mecham R. Extracellular matrix 4: the elastic fiber. FASEB (Fed Am Soc Exp Biol) J. 1993;7:1208–1218. [PubMed] [Google Scholar]
- 49.Saharinen J, Taipale J, Keski-Oja J. Association of the small latent transforming growth factor-β with an eight cysteine repeat of its binding protein LTBP-1. EMBO (Eur Mol Biol Organ) J. 1996;15:245–253. [PMC free article] [PubMed] [Google Scholar]
- 50.Sakai LY, Keene DR, Glanville RW, Bachinger HP. Purification and partial characertization of fibrillin, a cysteine-rich structural component of connective tissue microfibrils. J Biol Chem. 1991;266:14763–14770. [PubMed] [Google Scholar]
- 51.Sane DC, Moser TL, Greenberg CS. Vitronectin in the substratum of endothelial cells is cross-linked and phosphorylated. Biochem Biophys Res Commun. 1991;174:465–469. doi: 10.1016/0006-291x(91)91439-j. [DOI] [PubMed] [Google Scholar]
- 52.Sato Y, Rifkin DB. Autocrine activities of basic fibroblast growth factor: regulation of endothelial cell movement, plasminogen activator synthesis, and DNA synthesis. J Cell Biol. 1988;107:1199–1205. doi: 10.1083/jcb.107.3.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sato Y, Rifkin D. Inhibition of endothelial cell movement by pericytes and smooth muscle cells: activation of a latent transforming growth factor-β1-like molecule by plasmin during co-culture. J Cell Biol. 1989;109:309–315. doi: 10.1083/jcb.109.1.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sato Y, Tsuboi R, Lyons R, Moses H, Rifkin DB. Characterization of the activation of latent TGF-β by co-cultures of endothelial cells and pericytes or smooth muscle cells: a self-regulating system. J Cell Biol. 1990;111:757–763. doi: 10.1083/jcb.111.2.757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sato Y, Okada F, Abe M, Seguchi T, Kuwano M, Sato S, Furuya A, Hanai N, Tamaoki T. The mechanism for the activation of latent TGF-β during co-culture of endothelial cells and smooth muscle cells: cell-type specific targeting of latent TGF-β to smooth muscle cells. J Cell Biol. 1993;123:1249–1254. doi: 10.1083/jcb.123.5.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schultz-Cherry S, Ribeiro S, Gentry L, Murphy-Ullrich JE. Thrombospondin binds and activates the small and large forms of latent transforming growth factor-β in a chemically defined system. J Biol Chem. 1994;269:26775–26782. [PubMed] [Google Scholar]
- 57.Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia colias fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 58.Sporn MB, Robert AB. Transforming growth factor-β: recent progress and new challenges. J Cell Biol. 1992;119:1017–1021. doi: 10.1083/jcb.119.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taipale J, Koli K, Keski-Oja J. Release of transforming growth factor-β1 from the pericellular matrix of cultured fibroblasts and fibrosarcoma cells by plasmin and thrombin. J Biol Chem. 1992;267:25378–25384. [PubMed] [Google Scholar]
- 60.Taipale J, Miyazono K, Heldin CH, Keski-Oja J. Latent transforming growth factor-β1 associates to fibroblast extracellular matrix via latent TGF-β binding protein. J Cell Biol. 1994;124:171–181. doi: 10.1083/jcb.124.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Taipale J, Lohi J, Saarinen J, Kovanen PT, Keski-Oja J. Human mast cell chymase and leukocyte elastase release latent transforming growth factor-β1 from the extracellular matrix of cultured human epithelial and endothelial cells. J Biol Chem. 1995;270:4689–4696. doi: 10.1074/jbc.270.9.4689. [DOI] [PubMed] [Google Scholar]
- 62.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Upchurch HF, Conway E, Patterson MK, Maxwell MD. Localization of cellular transglutaminase on the extracellular matrix after wounding: characteristics of the matrix bound enzyme. J Cell Physiol. 1991;149:375–382. doi: 10.1002/jcp.1041490304. [DOI] [PubMed] [Google Scholar]
- 64.Yin W, Smiley E, Germiller J, Mecham RP, Florer JB, Wenstrup RJ, Bonadio J. Isolation of a novel latent transforming growth factor-β binding protein gene (LTBP-3) J Biol Chem. 1995;270:10147–10160. doi: 10.1074/jbc.270.17.10147. [DOI] [PubMed] [Google Scholar]
- 65.Zhang H, Apfelroth SD, Hu E, Davis EC, Sanguineti C, Bonadio J, Mecham RP, Ramirez F. Structure and expression of fibrillin-2, a novel microfibrillar component preferentially located in elastic matrices. J Cell Biol. 1994;124:855–863. doi: 10.1083/jcb.124.5.855. [DOI] [PMC free article] [PubMed] [Google Scholar]