Abstract
Mutants in the meiosis-specific RED1 gene of S. cerevisiae fail to make any synaptonemal complex (SC) or any obvious precursors to the SC. Using antibodies that specifically recognize the Red1 protein, Red1 has been localized along meiotic pachytene chromosomes. Red1 also localizes to the unsynapsed axial elements present in a zip1 mutant, suggesting that Red1 is a component of the lateral elements of mature SCs. Anti-Red1 staining is confined to the cores of meiotic chromosomes and is not associated with the loops of chromatin that lie outside the SC. Analysis of the spo11 mutant demonstrates that Red1 localization does not depend upon meiotic recombination. The localization of Red1 has been compared with two other meiosisspecific components of chromosomes, Hop1 and Zip1; Zip1 serves as a marker for synapsed chromosomes. Double labeling of wild-type meiotic chromosomes with anti-Zip1 and anti-Red1 antibodies demonstrates that Red1 localizes to chromosomes both before and during pachytene. Double labeling with anti-Hop1and anti-Red1 antibodies reveals that Hop1 protein localizes only in areas that also contain Red1, and studies of Hop1 localization in a red1 null mutant demonstrate that Hop1 localization depends on Red1 function. These observations are consistent with previous genetic studies suggesting that Red1 and Hop1 directly interact. There is little or no Hop1 protein on pachytene chromosomes or in synapsed chromosomal regions.
Meiosis generates haploid germ cells in diploid eukaryotic organisms. During meiosis, chromosomes pair and recombine, and these events ensure the proper segregation of genetic material to the progeny germ cells. Pairing between homologous chromosomes culminates in the formation of the synaptonemal complex (SC)1 (reviewed by von Wettstein et al., 1984). The SC is a meiosis-specific proteinaceous structure consisting of two electron-dense parallel structures called “lateral elements,” which are separated by a less dense “central region.” Each lateral element arises from a pair of sister chromatids and is called an axial element before it becomes part of mature SC. Most of the chromatin is located in loops that lie outside the SC, with each loop attached at its base to a lateral element.
In S. cerevisiae, a number of mutants have been isolated that are defective in the genetic events of meiosis and also fail to make SC (Roeder, 1995). One of the genes thus identified, ZIP1, encodes a component of the central region of the SC (Sym et al., 1993; Sym and Roeder, 1995). zip1 mutants make chromosomes that are homologously paired (Nag et al., 1995), but not intimately synapsed (Sym et al., 1993). Each pair of axial elements is closely associated at a number of discrete sites, termed axial associations, that have been postulated to be sites of synaptic initiation (Rockmill et al., 1995; Sym et al., 1993). Zip1 localizes along the lengths of pachytene chromosomes, but is not associated with unsynapsed axial elements (Sym et al., 1993).
The red1 mutant was isolated in a screen for mutants that make inviable meiotic products (Rockmill and Roeder, 1988). red1 mutants fail to make any SC or any obvious precursors to the SC, raising the possibility that Red1 is a structural component of the SC. The red1 mutant undergoes 12–25% of the wild-type level of reciprocal exchange (Mao-Draayer et al., 1996; Rockmill and Roeder, 1990), but the crossovers that occur do not ensure proper chromosome disjunction (Rockmill and Roeder, 1990). Studies of chromosome pairing indicate that homologue alignment is impaired in red1 strains and/or that the associations between homologous chromosomes are less stable than in wild type (Nag et al., 1995). The Red1 protein does not display significant homology to any proteins in current data bases (Thompson and Roeder, 1989).
Hop1 is a meiosis-specific protein that localizes to chromosomes (Hollingsworth et al., 1990). Genetic evidence suggests that the Red1 and Hop1 proteins physically interact with each other. The red1 and hop1 mutations affect the same subset of meiotic recombination events (Rockmill and Roeder, 1990). Overproduction of Red1 suppresses or enhances certain non-null hop1 alleles (Friedman et al., 1994; Hollingsworth and Johnson, 1993), and Hop1 overproduction suppresses some red1 alleles (Smith, A. V., and G. S. Roeder, unpublished observations).
Studies of the zip1 mutant have provided substantial information about the function of the central region of the SC. zip1 mutants exhibit wild-type levels of gene conversion and a two- to threefold reduction in reciprocal exchange (Sym and Roeder, 1994). The crossovers that occur ensure the proper disjunction of chromosomes at meiosis I, indicating that chiasmata function normally (Sym and Roeder, 1994). The zip1 mutant displays a modest defect in sister chromatid cohesion (Sym and Roeder, 1994). The only absolute defect observed in zip1 strains is a loss of crossover interference (Sym and Roeder, 1994). Thus, a primary function of Zip1, and by implication of the central region of the SC, is the regulation of crossover distribution.
Analysis of the zip1 mutant does not address the functions of axial and lateral elements, which might play critical roles in sister chromatid cohesion and chiasma function. To investigate the functions of axial/lateral elements, genes that encode components of these elements must first be identified. To determine whether Red1 is such a protein, we generated antibodies that specifically recognize the Red1 protein and used these antibodies to localize Red1 within meiotic cells. Our results strongly suggest that Red1 is associated with the axial cores of meiotic chromosomes. Additionally, we demonstrate that Red1 and Hop1 colocalize and that Hop1 localization depends on Red1. We speculate that the Red1 protein plays a role in establishing meiotic sister chromatid cohesion.
Materials and Methods
Strains
All yeast strains are isogenic derivatives of BR2495 (Rockmill and Roeder, 1990), which has the following genotype:
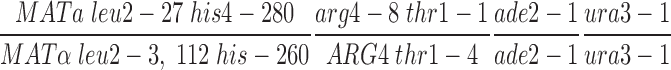
MY129 (Sym and Roeder, 1995), MY231 (Sym and Roeder, 1995), BR2498 (Nag et al., 1995), and S2888 (Rockmill et al., 1995a ) are BR2495 derivatives homozygous for zip1::LEU2, red1::URA3, hop1::TRP1, and spo11::ADE2, respectively. BS354 is BR2495 carrying pB86, which carries RED1, URA3, and the 2 μm-circle origin of DNA replication.
Yeast manipulations were performed and media were prepared using standard procedures (Sherman et al., 1986). E. coli strain XL-1 Blue (Stratagene, La Jolla, CA) was used for plasmid constructions and protein synthesis. An XbaI–EcoRI fragment from pR847 (Thompson and Roeder, 1989) containing the RED1 gene was inserted into YEp352 (Hill et al., 1986) cut with XbaI and EcoRI to construct plasmid B86 (from B. Rockmill).
Antibody Production
To generate fusion proteins for antibody production, RED1 and HOP1 were fused in-frame to glutathione S-transferase. A BglII–HindIII fragment of the RED1 gene encoding amino acids 426 to 827 (Thompson and Roeder, 1989) was cloned into the BamHI-HindIII sites of the expression vector pGEX-KG (Guan and Dixon, 1991) to generate pAV3. A XbaISacI fragment of pNH33-2 encoding amino acids 117 to 461 of Hop1 (Hollingsworth et al., 1990) was cloned into pGEX-KG to generate pAV5. The resulting glutathione S-transferase Red1 and Hop1 fusion proteins were produced in E. coli and purified according to the methods described by Guan and Dixon (1991) and Smith and Johnson (1988). Proteins were sent to the Pocono Rabbit Farm and Laboratory (Canadensis, PA) for injection into rabbits. Additionally, Red1 fusion protein was injected into mice. Antibodies were affinity purified according to Snyder (1989).
Cytology
Chromosome spreads and immunofluorescence were performed as described by Sym et al. (1993). Except as noted, spreads were prepared 15 h after transfer to sporulation medium. Anti-Red1 and anti-Hop1 antibodies were used at a 1:100 dilution. The rabbit anti-Zip1 antibody was diluted 1:100, and the mouse anti-Zip1 antibody (from P. Moens) was diluted 1:1,000. The mouse anti-nucleolus antibody (monoclonal antibody 2.3b; from C. Copeland, H. Friedman, J. Woolford, and M. Snyder) was diluted 1:5. The rabbit anti-Histone2B antibody (from A. Carmen and M. Grunstein) was used at 1:500. Rabbit antibodies were detected with goat anti–rabbit antibodies conjugated to Texas red (Jackson ImmunoResearch Labs, West Grove, PA) diluted 1:200. Mouse antibodies were detected with goat anti–mouse antibodies conjugated to FITC (Jackson ImmunoResearch) diluted 1:200. After antibody staining, spreads were stained with 1 μg/ml 4′-6-diamidino-2-phenylindole (DAPI) to visualize the DNA. A Leitz DMRB microscope equipped with fluorescence and a PL APO 100× objective was used to observe antibody-stained preparations. Images were captured using a Photometrics Imagepoint CCD camera.
Time course analysis of meiotic prophase progression was carried out as follows. Strain BR2495 was grown to saturation overnight at 30°C in rich medium (YPAD; Sherman et al. [1986]). The culture was diluted 1:1 into fresh YPAD and incubated with shaking for 8 h at 30°C. Cells from 10 ml of culture were then washed once with H2O and resuspended in 100 ml of 2% KAc and placed in a one-liter flask and shaken at 250 rpm at 30°C. At 1-h intervals, starting 10 h after transfer to 2% KAc, 10 ml of culture were removed and used to prepare spread chromosomes on each of the four slides. The spreading procedure required 40 min after the removal of cells from KAc. The times given in the text and figure legends indicate when the spreading procedure was initiated. At each time point, two slides were used for double labeling Red1 and Zip1; one was used for double labeling Red1 and Hop1, and one was used for double labeling Hop1 and Zip1.
DNase I digestion of meiotic spreads was carried out as follows. Wildtype spreads from BR2495 were prepared 15 h after transfer to KAc. Spreads were then incubated for 1 h at 37°C in 100 U/ml DNase I (Boehringer Mannheim) in 50 mM Tris (pH 7.5), 10 mM MnCl2, and 50 μg/ml BSA. Control spreads were incubated in buffer lacking DNase I. Subsequent antibody incubations were performed as described above.
Results
Red1 Is a Component of Meiotic Chromosomes
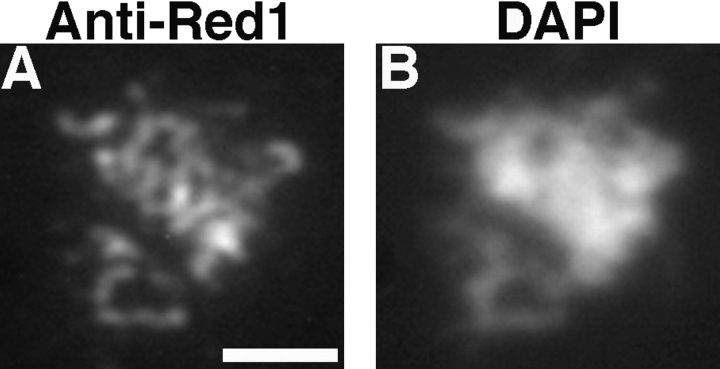
To localize the Red1 protein, anti-Red1 antibodies were generated and used in immunofluorescence experiments. Meiotic chromosomes were prepared from wild-type meiotic cells after 15 h in sporulation medium, when the maximum number of cells are at the pachytene stage. Spread chromosomes were stained with affinity-purified antiRed1 antibodies followed by secondary antibodies conjugated to Texas red. Chromosomal DNA was visualized by staining with DAPI. At pachytene, when chromosomes are maximally condensed and fully synapsed, there is extensive staining of chromosomes by anti-Red1 antibodies (Fig. 1, A and C). These antibodies specifically recognize Red1, since no staining is observed on spread meiotic chromosomes prepared from a red1 null mutant (strain MY231; data not shown).
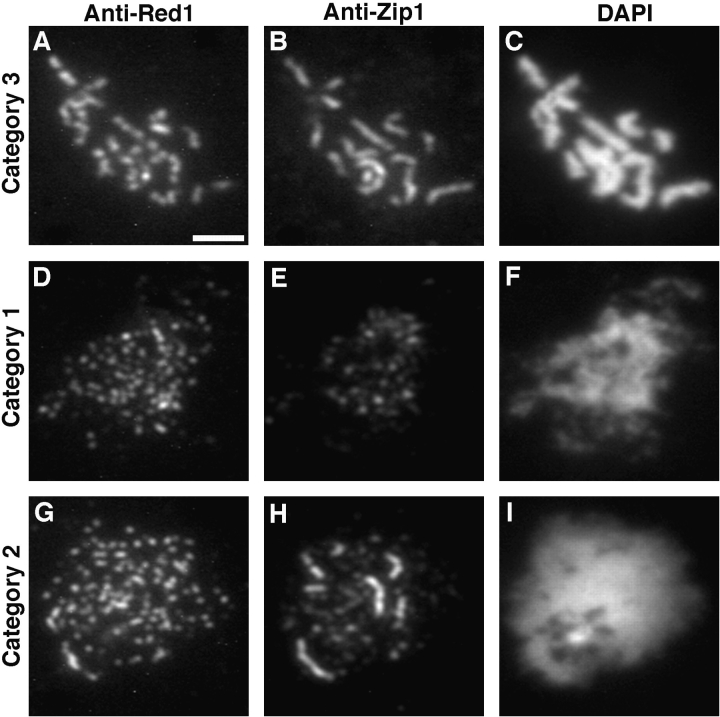
Figure 1.
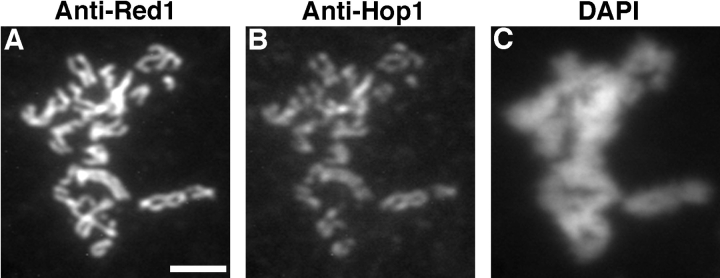
Red1 and Zip1 protein localization on wild-type chromosomes. Spread meiotic chromosomes from wild-type cells (BR2495) stained with mouse anti-Red1 antibodies (A, D, and G), rabbit anti-Zip1 antibodies (B, E, and H) and DAPI (C, F, and I). (A–C) Spread pachytene nucleus from 15-h time point. (D–F) Spread nucleus from 10-h time point. (G–I) Spread nucleus from 13-h time point. Categories 1, 2, and 3 are described in the text. Bar, 2 μm.
In wild-type cells, Red1 staining is generally not continuous along the lengths of pachytene chromosomes. Though continuous linear stretches of Red1 localization are observed, frequent gaps in staining are also seen. This pattern contrasts with the continuous localization of the Zip1 protein along pachytene chromosomes (Sym et al., 1993). To compare directly the anti-Red1 and anti-Zip1 staining patterns, wild-type meiotic chromosomes were double-labeled with mouse anti-Red1 antibodies and rabbit anti-Zip1 antibodies, which were subsequently detected with appropriate secondary antibodies. In pachytene nuclei, Zip1 staining extends along the entire lengths of chromosomes and does not display obvious gaps in staining (Fig. 1 B). At the same stage, Red1 displays many stretches of continuous staining, interrupted by nonstaining regions (Fig. 1 A). Only rarely are nuclei observed in which Red1 staining appears to be continuous along the total lengths of chromosomes.
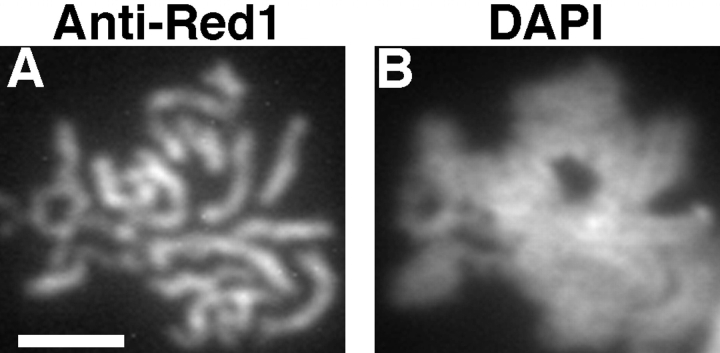
Introduction of a multicopy plasmid carrying the RED1 gene into wild-type cells results in continuous localization of Red1 along the lengths of pachytene chromosomes (Fig. 2). Thus, overproduction of the RED1 gene product alters the pattern of Red1 localization.
Figure 2.
Red1 protein localization in strains overproducing Red1. Spread meiotic chromosomes from a wild-type cell overproducing Red1 (BS354) stained with rabbit anti-Red1 antibodies (A) and DAPI (B). Bar, 2 μm.
Red1 Associates with Chromosome Cores, Not Chromatin Loops
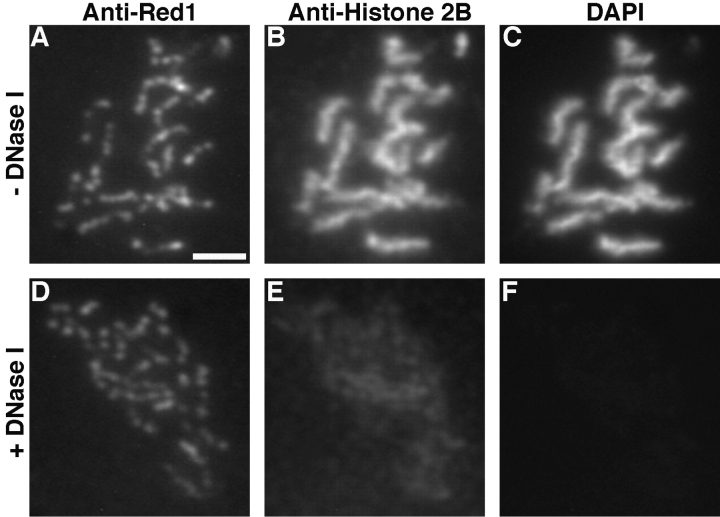
To determine whether Red1 protein is confined to the structural cores of chromosomes or associated with chromatin loops, spreads of wild-type meiotic chromosomes were digested with DNase I before double labeling with anti-Red1 antibodies and antibodies to histone 2B. Digestion with DNase I destroys the chromatin loops, while leaving the proteinaceous chromosomal cores intact (Heyting et al., 1985). Digestion with high amounts of DNase I greatly reduces the level of both histone staining and DAPI staining, while retaining significant levels of Red1 staining (Fig. 3). Therefore, Red1 protein is associated primarily with chromosomal cores.
Figure 3.
Red1 and histone localization following DNase I digestion of meiotic chromosomes. Wild-type spreads (BR2495) incubated for 1 h at 37°C with 0 U/ml (A–C) or 100 U/ml (D–F) DNase I. Spreads were stained with mouse anti-Red1 (A and D) and rabbit antihistone 2B (B and E) antibodies in addition to DAPI staining (C and F). Bar, 2 μm.
Red1 Associates with Chromosomes before and during Pachytene
Meiotic chromosomes prepared from wild-type cells after 15 h in sporulation medium display distinct patterns of anti-Red1 and anti-Zip1 antibody staining that correlate with the extent of chromosomal condensation. The staining patterns observed can be classified into three categories. Spread nuclei with relatively uncondensed chromosomes display numerous non-overlapping foci of Red1 and Zip1 staining (category 1, Fig. 1, D–F). In nuclei with intermediate levels of chromosomal condensation, Zip1 staining reveals some linear stretches as well as smaller dot-like foci (category 2, Fig. 1, G–I). In such nuclei, Red1 stains numerous foci and some more continuous stretches. At pachytene, when chromosomes are fully condensed, Zip1 staining is continuous along the total lengths of chromosomes and Red1 staining extends over the lengths of chromosomes with the discontinuities described above (category 3, Fig. 1, A–C). Distinct patterns of localization that correlate with different DAPI-staining patterns suggest that these patterns represent successive stages during meiotic prophase progression.
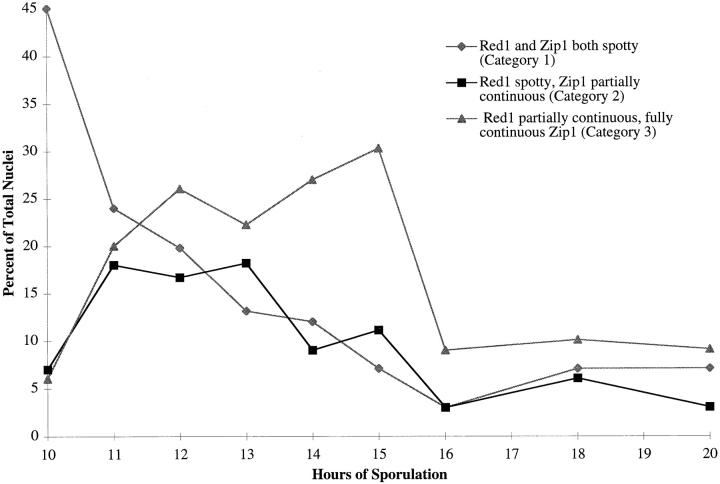
To stage the patterns of localization, wild-type cells were sporulated and nuclei were surface spread at 1-h intervals. Subsequently, chromosomes were double labeled with anti-Red1 and anti-Zip1 antibodies. At each time point, 100 nuclei were photographed and classified into the three categories described above on the basis of antibody staining. At 10 h after transfer to sporulation medium, 45% of all nuclei were in category 1, while both categories 2 and 3 represented a small fraction (∼7%) of the total (Fig. 4); the remaining nuclei failed to stain with either anti-Zip1 or anti-Red1 antibodies. Over time, the number of nuclei in category 1 rapidly decreased, while the numbers in categories 2 and 3 increased. Nuclei in category 2 were at the highest level between 11 and 13 h after transfer to sporulation medium. Category 3 (pachytene) nuclei peaked at 15 h. These results suggest that the order of stages in meiotic prophase is category 1 nuclei followed by category 2 followed by category 3. The data demonstrate that Red1 is present on chromosomes both before and during the pachytene stage.
Figure 4.
Red1 and Zip1 localization patterns during meiotic prophase progression. At each time point, 100 nuclei were examined and scored for Red1 and Zip1 localization patterns. Categories of staining are described in the text. The percentage of nuclei that stain with antibodies is always <100% because not all cells enter meiosis and those cells that undergo sporulation do not proceed through meiosis in synchrony.
Red1 and Hop1 Display Extensive Colocalization
To compare the Red1 and Hop1 localization patterns, rabbit anti-Hop1 antibodies were generated and used in conjunction with mouse anti-Red1 antibodies. The anti-Hop1 antibodies stain spread chromosomes from wild type, but not from a hop1 null mutant (BR2498), indicating that they are specific for the Hop1 protein (data not shown). Anti-Red1 and anti-Hop1 antibodies were used to double label meiotic chromosomes from wild-type cells at different time points after the introduction into sporulation medium.
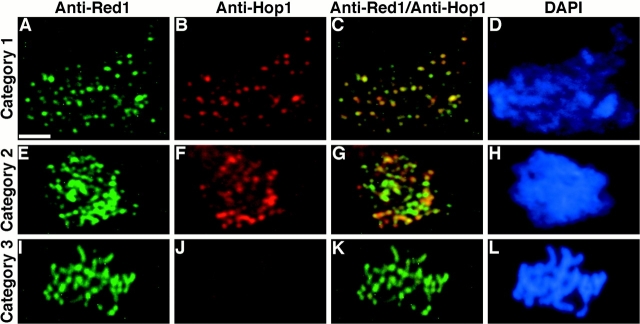
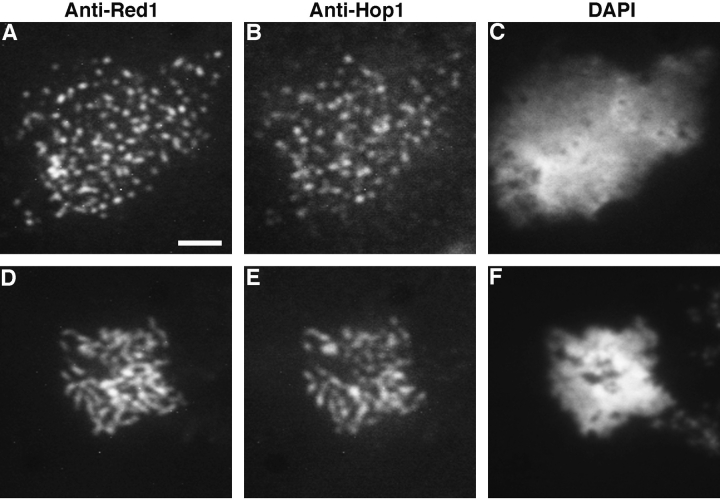
Staining of meiotic chromosomes at early stages of prophase revealed extensive overlap of the Red1- and Hop1staining patterns. In nuclei with relatively uncondensed chromosomes as judged by DAPI staining, both anti-Red1 and anti-Hop1 antibodies stained numerous discrete areas on meiotic chromosomes (Fig. 5, A–D). In such nuclei, all regions of Hop1 staining also displayed Red1 staining, though the total number of Hop1-staining foci was slightly fewer than the number of Red1 spots. In all nuclei examined, there were spots of Red1 staining that failed to show any detectable Hop1 staining.
Figure 5.
Red1 and Hop1 protein localization on wild-type chromosomes. Spread meiotic chromosomes from wild-type cells (BR2495) stained with mouse anti-Red1 antibodies (A, E, and I), rabbit anti-Hop1 antibodies (B, F, and J) and DAPI (D, H, and L). (A–D) Spread nucleus from 10-h time point with category 1 staining pattern. (E–H) Spread nucleus from 13-h time point with category 2 staining pattern. (I–L) Spread pachytene nucleus from 15-h time point with category 3 staining pattern. (C, G, and K) Fusion of anti-Red1 and anti-Hop1 staining patterns; areas of overlap appear as yellow spots. Bar, 2 μm.
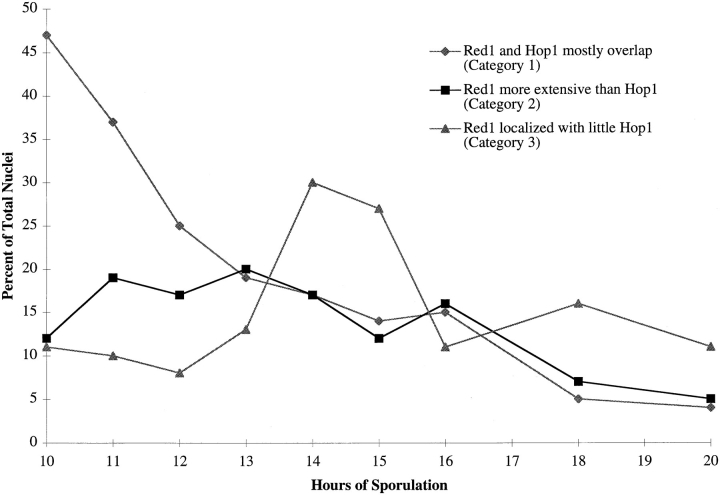
Examination of spread nuclear preparations from the meiotic time course described above revealed progressive changes in the Red1- and Hop1-staining patterns. Antibody-staining nuclei could be classified into three distinct categories, referred to here as categories 1-3, though these are not necessarily the same as the previously described categories of Red1 and Zip1 staining (see Discussion). In category 1, Red1 and Hop1 display extensive and nearly complete overlap (Fig. 5, A–D). Nuclei in category 2 display significant amounts of Hop1 staining, but Hop1 staining is much less extensive than the Red1 staining (Fig. 5, E–H). Nuclei in category 3 exhibit extensive anti-Red1 antibody staining, but very low levels of Hop1 staining (Fig. 5, I–L). At 10 h after transfer to sporulation medium, 47% of all nuclei were in category 1 and categories 2 and 3 were at very low levels (Fig. 6). Over time, both categories 2 and 3 increased, while category 1 decreased. Nuclei in category 2 were at the highest level between 11 and 14 h after transfer to sporulation medium, but the level remained fairly constant throughout the time course. Nuclei in category 3 peaked at 14–15 h. Thus, category 1 precedes category 2, which appears to precede category 3. These results suggest that Hop1 localizes to chromosomes before pachytene, but Hop1 is largely dissociated from chromosomes by the pachytene stage.
Figure 6.
Red1 and Hop1 localization patterns during meiotic prophase progression. At each time point, 100 nuclei were examined and scored for Red1 and Hop1 localization. Categories of staining are described in the text.
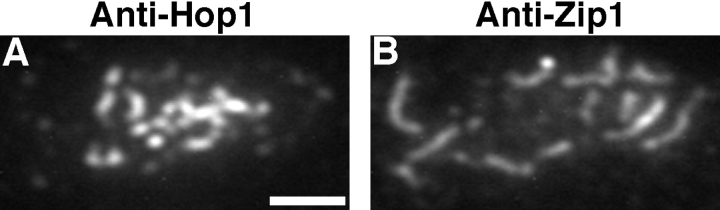
To compare Hop1 staining with Zip1 staining, nuclei were double labeled with mouse anti-Zip1 and rabbit antiHop1 antibodies. At early stages of meiotic prophase, nuclei displayed numerous foci of Hop1 staining and foci of Zip1, but these foci were non-overlapping. At later stages when Zip1 localization was more continuous, little or no Hop1 antibody was seen in regions of extensive Zip1 staining, while strong Hop1 staining was observed in regions that lacked Zip1 (Fig. 7). Thus, there appears to be little or no Hop1 protein in chromosomal regions that are synapsed.
Figure 7.
Hop1 and Zip1 protein localization on wild-type chromosomes. Spread zygotene nucleus from a wild-type cell (BR2495) from 13-h time point stained with rabbit anti-Hop1 antibodies (A) and mouse anti-Zip1 antibodies (B). Bar, 2 μm.
Hop1 Localization Depends on Red1
As described above, Hop1 staining is observed only in areas that are also stained with anti-Red1 antibodies, suggesting that Hop1 localization to meiotic chromosomes depends on prior Red1 localization. To test this hypothesis, Hop1 localization was examined in a red1 null mutant. No Hop1 staining was detected on meiotic chromosomes prepared from red1 cells after 15 h in sporulation medium (data not shown). In contrast, anti-Red1 antibodies stained meiotic chromosomes from hop1 mutant cells (data not shown). These results demonstrate that Hop1 localization to chromosomes depends on Red1 function, but Red1 localization is independent of Hop1.
Red1 and Hop1 Localize to the Unsynapsed Axial Elements in a zip1 Mutant
The localization of Red1 and Hop1 was examined in zip1 mutant spreads in order to examine the relationship between Red1 and Hop1 localization and chromosome synapsis. In a zip1 mutant, chromosomes condense and individual distinct chromosomes are evident by DAPI staining.
Double labeling with anti-Red1 and anti-Hop1 antibodies revealed staining associated with the axial cores of the paired homologous chromosomes in a zip1 mutant (Fig. 8). Thus, Red1 appears to be associated primarily with the lateral elements of the SC, and not with the central region. In spread chromosomes from the zip1 mutant, both Red1 and Hop1 are frequently localized continuously along the lengths of condensed chromosomes (Fig. 8), in contrast to the discontinuous localization observed on wild-type chromosomes. This continuous localization is likely a consequence of the fact that the zip1 mutant arrests in meiotic prophase with condensed chromosomes typical of the pachytene stage (Nag et al., 1995). Assembly of Red1 and Hop1 onto chromosomes may continue at the arrest point. Consistent with this hypothesis, many of the nuclei with condensed chromosomes observed after 15 h of sporulation display discontinuous Red1 and Hop1 localization, while the proportion with continuous localization increases at later time points (data not shown).
Figure 8.
Red1 and Hop1 localization on zip1 chromosomes. Spread meiotic chromosomes from a zip1 cell (MY129) stained with mouse anti-Red1 antibodies (A), rabbit anti-Hop1 antibodies (B), and DAPI (C). Bar, 2 μm.
DAPI staining seldom allows for visualization of the axial associations between homologous chromosomes in zip1 nuclear spreads, since the chromatin is primarily located in loops that lie outside the SC. However, the core structures stained with anti-Red1 and anti-Hop1 antibodies are narrower than the DAPI-staining material, and axial associations are evident in favorable spreads (Fig. 8).
Zip1 Localizes to Chromosomes in a red1 Mutant
red1 mutants fail to make any SC or SC precursors detectable in the electron microscope (Rockmill and Roeder, 1990). However, there is some Zip1 protein associated with the spread meiotic chromosomes from a red1 null mutant (Fig. 9). The staining is often highly discontinuous and consists primarily of numerous small foci. However, in a small number of nuclei, staining is fairly continuous, despite defective chromosome condensation as visualized by DAPI staining. Fig. 9 shows an example of particularly continuous Zip1 localization.
Figure 9.
Zip1 localization on red1 chromosomes. Spread meiotic chromosomes from red1 cells (MY231) stained with rabbit antiZip1 antibodies (A) and DAPI (B). Bar, 2 μm.
Red1 and Hop1 Localization Do Not Depend on Double-strand Break Formation
spo11 mutants do not undergo any of the meiosis-specific, double-strand breaks that initiate meiotic recombination (Cao et al., 1990). To determine whether Red1 and Hop1 localization depends on events in the recombination pathway, meiotic chromosomes from spo11 mutant cells were double-labeled with anti-Red1 and anti-Hop1 antibodies. Both antibodies stain meiotic chromosomes from the spo11 mutant (Fig. 10). In nuclei in which chromosomes are not very condensed, anti-Red1 antibodies stain numerous discrete foci, and anti-Hop1 antibodies stain spots that directly overlap the areas of Red1 localization (Fig. 10, A–C). In some nuclei in which chromosomes are more condensed, both Red1 and Hop1 staining is more continuous (Fig. 10, D–F). Thus, Red1 and Hop1 can localize to meiotic chromosomes independently of the initiation of meiotic recombination.
Figure 10.
Red1 and Hop1 localization on spo11 chromosomes. Spread meiotic chromosomes from spo11 cells (S2888) stained with mouse anti-Red1 antibodies (A and D), rabbit anti-Hop1 antibodies (B and E), and DAPI (C and F). Bar, 2 μm.
Red1, But Not Hop1 or Zip1, Is in the Nucleolus
At pachytene, one chromosomal region is observed that stains with anti-Red1 antibodies, but not with antibodies to Zip1. Within this region, the anti-Red1 staining consists of two closely opposed, but not synapsed axial segments from a single chromosomal region (Fig. 1, A–C). Previous studies suggested that the region that fails to stain with anti-Zip1 antibodies is the nucleolus (Sym et al., 1993). Double labeling of wild-type meiotic chromosomes with antinucleolar and anti-Red1 antibodies revealed that Red1 protein is found in the nucleolus (data not shown). To determine if Hop1 is in the nucleolus, meiotic chromosomes from zip1 mutant cells were double labeled with antiHop1 and anti-nucleolar antibodies. No Hop1 staining was observed in the nucleolus (data not shown).
Discussion
Red1 Is Associated with the Lateral Elements of the Synaptonemal Complex
We have examined localization of the RED1 gene product, which is required for SC formation in S. cerevisiae. Red1 localizes to wild-type pachytene chromosomes and to the unsynapsed chromosomes present in the zip1 mutant. Red1 remains associated with pachytene chromosomes that have been digested with DNase, indicating that Red1 is associated primarily with the core of the SC, not with chromatin loops. When SCs purified from rat spermatocyte nuclei are treated with DNase, lateral elements remain intact as determined by electron microscopy (Heyting et al., 1985). Thus, our results strongly suggest that Red1 is associated with unsynapsed axial elements and with the lateral elements of mature SCs.
Red1 staining is usually discontinuous along the lengths of pachytene chromosomes. This is not the result expected for a building block of lateral elements, which appear to run continuously along the lengths of chromosomes when spread nuclei are stained with silver nitrate and examined in the electron microscope (reviewed by von Wettstein et al., 1984). However, Red1 is required for the formation of axial/lateral elements (Rockmill and Roeder, 1990), and it is associated with these structures. We propose that Red1 serves to nucleate the formation of axial/lateral elements and that other proteins are responsible for the continuous linear densities observed in the electron microscope. The primary building blocks of axial/lateral elements might be proteins present in the mitotic chromosome scaffold. Topoisomerase II is found in the cores of both mitotic and meiotic chromosomes, indicating that these structures are related (Earnshaw and Heck, 1985; Klein et al., 1992; Moens and Earnshaw, 1989). Alternatively, axial/lateral elements might be composed primarily of meiosis-specific proteins whose proper assembly onto chromosomes depends on Red1.
Proteins associated with the cores of sister chromatid pairs have been identified in hamsters and rats via biochemical methods (Dobson et al., 1994; Heyting et al., 1987). Red1 localization displays some similarity to the localization patterns of these previously described proteins, but there are important differences. Like Red1, the Scp3 lateral element protein from rats (Lammers et al., 1994) and the homologous Cor1 protein from hamsters (Dobson et al., 1994) are associated with meiotic chromosome cores both before and during pachytene. However, these proteins appear to localize more continuously than Red1 along the lengths of pachytene chromosomes. In addition, Cor1 associates with meiotic chromosomes longer than Red1; some Cor1 protein remains along chromosomes axes until metaphase I and Cor1 staining persists at kinetochores until anaphase II (Dobson et al., 1994).
Hop1 Localizes to Zygotene Chromosomes in a Red1-dependent Manner
Genetic analysis suggests that Red1 and Hop1 participate in the same pathway or process (Friedman et al., 1994; Hollingsworth and Johnson, 1993; Rockmill and Roeder, 1990). Consistent with this hypothesis, we have found that Red1 and Hop1 localize to the same sites on chromosomes at early stages of meiotic prophase. Localization of Hop1 to chromosomes depends on Red1, suggesting that Red1 acts before Hop1 in SC formation. Although Red1 is necessary for Hop1 localization, it must not be sufficient because Red1, but not Hop1, is found in the nucleolus. The Red1 dependence of Hop1 localization is consistent with the observation that the red1 defect in SC formation is more severe than that of hop1. A red1 null mutant fails to make any detectable axial or lateral elements (Rockmill and Roeder, 1990), while the hop1 mutant does make axial elements (Loidl et al., 1994).
Not all aspects of HOP1 function are strictly dependent on RED1 function. A hop1 mutation reduces meiotic recombination in a red1 strain background, indicating that Hop1 can promote recombination in the absence of Red1 (Rockmill and Roeder, 1990). In red1 cells, Hop1 might be transiently associated with chromosomes and/or it might be present on chromosomes at a level that is too low to be detected by antibody staining. An additional possibility is that Hop1 associates with chromosomes in red1 strains, but the association is unstable and does not survive the spreading procedure. It is also possible that Hop1 does not need to associate with chromosomes to carry out its role in recombination in red1 strains.
Changes in Red1 Localization during Meiotic Prophase Progression
Through comparison of the Red1, Hop1, and Zip1 localization patterns, we have identified three successive stages of meiotic prophase. We have defined three categories of nuclei based on Red1 and Zip1 localization, and three categories based on Red1 and Hop1 localization. We believe that the two sets of staining patterns correspond to each other based on the fact that the nuclei in each category (1, 2, or 3) appear and disappear with similar kinetics. Comparison of Hop1 and Zip1 localization provides additional evidence that the categories are analogous; as the localization of Zip1 becomes more continuous, Hop1 staining becomes fainter in regions that localize Zip1 and is eventually undetectable.
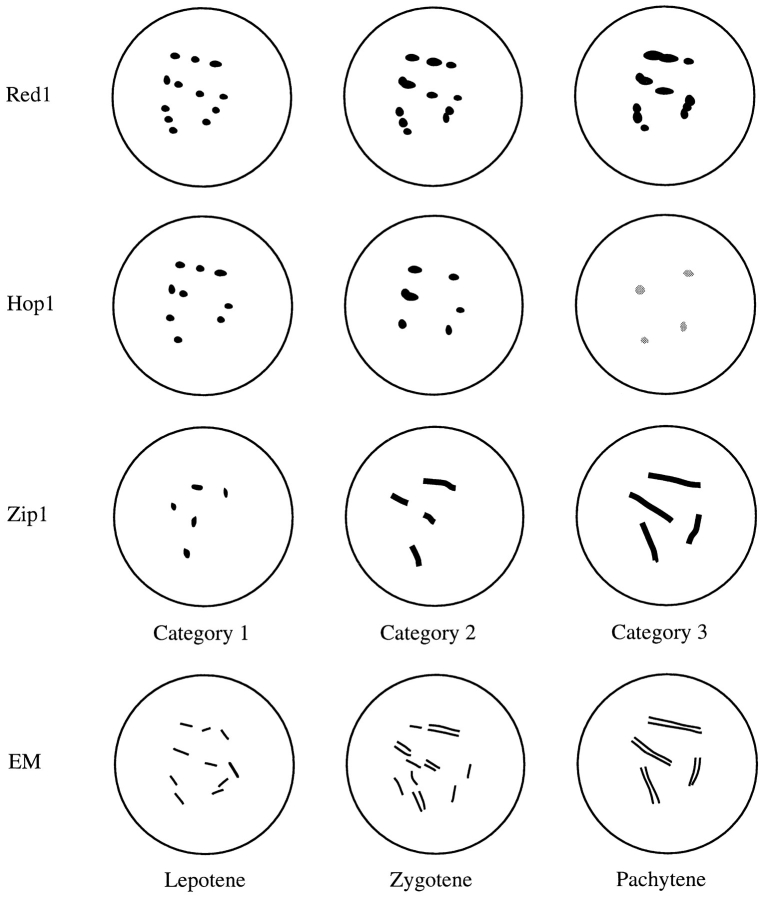
We speculate that the three categories defined by protein localization correspond to the three stages of prophase progression defined previously by electron microscopic analysis of silver-stained spread meiotic chromosomes (Alani et al., 1990; Padmore et al., 1991)(summarized in Fig. 11). In the first stage, Red1 and Hop1 are in numerous overlapping foci, while Zip1 is in different foci. This stage likely corresponds to leptotene nuclei that contain only very short stretches of axial elements (referred to as precursor group A nuclei by Padmore et al. [1991]). As meiosis progresses, Red1 and Zip1 become more continuous in their localization, while Hop1 extends over less of the chromosomes. We postulate that these nuclei are comparable to the zygotene nuclei that contain short stretches of tripartite SC together with unsynapsed axial elements (referred to as precursor group B nuclei by Padmore et al. [1991]). At the time of maximum chromosome condensation, Zip1 is localized continuously along the lengths of chromosomes. At this stage, Red1 is found along much of the lengths of chromosomes, but there is little or no Hop1. This stage corresponds to the pachytene stage with fully formed tripartite SCs. Zip1 and Red1 disappear from chromosomes at similar times, suggesting that little or no Red1 is found on chromosomes after pachytene.
Figure 11.
Summary of immunofluorescence and electron micrographic stages during meiotic prophase.
The Hop1 protein is localized along the lengths of paired meiotic chromosomes from a zip1 mutant, while little Hop1 localizes to synapsed meiotic chromosomes from wild type. These observations suggest that Hop1 disassociates from chromosomes as they synapse.
Red1, But Not Hop1, Is Found in the Nucleolus
The Red1 protein is found in the nucleolus, while Hop1 is not. The nucleolus differs from the remainder of the genome because the chromosomal segments present in the nucleolus fail to undergo meiotic recombination and SC formation (Petes, 1979; Petes and Botstein, 1977; Sym and Roeder, 1994). These observations suggest that Hop1 performs a function that is specific to regions of the genome that synapse and recombine, while Red1 performs a more general role. One function that might be required throughout the genome is cohesion between sister chromatids (see below).
Zip1 Polymerizes in the Absence of Detectable Lateral Elements
The red1 null mutant fails to make any SC or SC precursors detectable by electron microscopic examination of silver-stained spread nuclei, a result that was interpreted to mean that red1 mutants are asynaptic (Rockmill and Roeder, 1990). However, chromosomes from red1 cells display significant levels of Zip1 staining including some linear stretches. This Zip1 localization suggests a limited capacity for chromosome synapsis in the absence of detectable axial elements. Chromosomes in red1 strains may have undergone substantial meiosis-specific modifications that escape detection with electron microscopic methods currently in use.
It has been proposed that Zip1 plays a role in meiotic recombination independent of its role in SC polymerization (Storlazzi et al., 1996). This hypothesis is based on: (a) the observation that a zip1 mutation reduces recombination in a red1 strain background, and (b) the assumption that there is no SC polymerization in red1 strains. The linear stretches of Zip1 staining observed in spreads prepared from red1 strains argue that the latter critical assumption is incorrect.
Red1 Localizes to Chromosomes Independently of Meiotic Recombination
Red1 localizes to chromosomes in a spo11 mutant, which does not sustain any of the double-strand breaks that initiate meiotic recombination but undergoes extensive axial element formation (Cao et al., 1990; Loidl et al., 1994). In contrast, the localization to chromosomes of the RecA homologues, Dmc1 and Rad51 (Bishop, 1994), depends on early events in the recombination pathway. The Spo11 independence of Red1 localization is consistent with the hypothesis that Red1 serves as a structural component of meiotic chromosomes and is not directly involved in recombination. These results are also compatible with temporal analyses indicating that axial elements begin to develop before double-strand breaks occur (Padmore et al., 1991).
A Role for Red1 in Meiotic Sister Chromatid Cohesion
The Red1 protein localizes to the cores of meiotic chromosomes before and independent of both synapsis and recombination. These observations and others lead us to propose that Red1 plays a role in meiotic sister chromatid cohesion. The establishment of cohesion between sister chromatids is presumably a necessary step in the establishment of a proteinaceous core that is shared by sisters (i.e., an axial/lateral element).
During meiosis in wild type, recombination between nonsister chromatids is about ten times as frequent as recombination between sisters (Game et al., 1989; Haber et al., 1984). A defect in sister chromatid cohesion may result in a failure to discriminate sister from nonsister chromatids, and thus abolish the normal constraints against sister chromatid exchange. This might lead to an increase in the frequency of recombination between sisters relative to recombination between nonsister chromatids. This is the phenotype observed for the red1 mutant. red1 reduces the rate of interhomolog recombination (Rockmill and Roeder, 1990), but increases sister chromatid exchange (Thompson, D., personal communication) as assessed genetically.
Measurements of meiotic crossing over in different intervals indicate that a red1 null mutation reduces interhomologue exchange at least fourfold and as much as 13fold, with an average reduction of eightfold (Mao-Draayer et al., 1996; Rockmill and Roeder, 1990; Novak, J., and G. S. Roeder, unpublished observations). The magnitude of this reduction is greater than that expected if red1 sustains a wild-type number of meiotic recombination events, and these are distributed at random with respect to sister-sister vs nonsister-nonsister exchanges. Thus, the data suggest that sister chromatid exchange events are favored in the red1 mutant. It may also be the case that the absolute number of meiotic exchange events is decreased in red1 strains, as suggested by Mao-Draayer et al. (1996). A loss of sister chromatid cohesion may perturb meiotic chromosome structure such that chromosomes serve as less effective substrates for the enzymes that initiate meiotic recombination.
Only ∼1% of the spores produced by a red1 mutant are viable, and even chromosomes that have recombined missegregate (Rockmill and Roeder, 1988, 1990). Thus, red1 spore inviability cannot be accounted for simply by a failure of some chromosome pairs to cross over and a consequent failure to form the chiasmata necessary for reductional chromosome segregation. Since sister chromatid cohesion is thought to be important for chiasma function (Maguire, 1990), a defect in cohesion could result in an inability of crossovers to ensure proper homologue separation at the first division of meiosis. Thus, a defect in meiotic sister chromatid cohesion can account for multiple aspects of the red1 mutant phenotype, including the increase in sister chromatid exchange, a decrease in interhomolog recombination, the decrease in spore viability, and the missegregation of recombinant chromosomes.
The cytological studies reported here strongly suggest that the Red1 protein is associated with axial and lateral elements. Thus, further studies of the red1 mutant phenotype should shed light on the role of these structures in meiotic chromosome behavior.
Acknowledgments
We are grateful to Andrew Carmen, Mike Snyder, and Peter Moens for generous gifts of antibodies. We thank Penelope Chua, Lise Hoffmann, Janet Novak, and Beth Rockmill for critical comments on the manuscript.
This work was supported by American Cancer Society grant VM-7G to G.S. Roeder and by a Postdoctoral Fellowship (DRG-1239) from the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation to A.V. Smith.
Footnotes
Please address all correspondence to G.S Roeder, Department of Biology, Yale University, P.O. Box 208103, New Haven, CT 06520-8103. Tel.: (203) 432-3501. Fax: (203) 432-3263. E-Mail: shirleen.roeder@yale.edu
1. Abbreviation used in this paper: SC, synaptonemal complex.
References
- Alani E, Padmore R, Kleckner N. Analysis of wild-type and rad50mutants of yeast suggests an intimate relationship between meiotic chromosome synapsis and recombination. Cell. 1990;61:419–436. doi: 10.1016/0092-8674(90)90524-i. [DOI] [PubMed] [Google Scholar]
- Bishop DK. RecA homologues Dmc1 and Rad51 interact to form discrete nuclear complexes prior to meiotic chromosome synapsis. Cell. 1994;79:1081–1092. doi: 10.1016/0092-8674(94)90038-8. [DOI] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. . Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Dobson MJ, Pearlman RE, Karaiskakis A, Spyropoulos B, Moens PB. Synaptonemal complex proteins: occurrence, epitope mapping and chromosome disjunction. J Cell Sci. 1994;107:2749–2760. doi: 10.1242/jcs.107.10.2749. [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Heck MM. Localization of topoisomerase II in mitotic chromosomes. J Cell Biol. 1985;100:1716–1725. doi: 10.1083/jcb.100.5.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman DB, Hollingsworth NM, Byers B. Insertional mutations in the yeast HOP1gene: evidence for multimeric assembly in meiosis. Genetics. 1994;136:449–464. doi: 10.1093/genetics/136.2.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Game JC, Sitney KC, Cook VE, Mortimer RK. Use of a ring chromosome and pulsed-field gels to study interhomolog recombination, double-strand DNA breaks and sister-chromatid exchange in yeast. Genetics. 1989;123:695–713. doi: 10.1093/genetics/123.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan K, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Haber JE, Thorburn PC, Rogers D. Meiotic and mitotic behavior of dicentric chromosomes in Saccharomyces cerevisiae. . Genetics. 1984;106:185–205. doi: 10.1093/genetics/106.2.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyting C, Dietrich AJ, Redeker EJ, Vink AC. Structure and composition of synaptonemal complexes, isolated from rat spermatocytes. Eur J Cell Biol. 1985;36:307–314. [PubMed] [Google Scholar]
- Heyting C, Moens PB, van Raamsdonk W, Dietrich AJ, Vink AC, Redeker EJ. Identification of two major components of the lateral elements of synaptonemal complexes of the rat. Eur J Cell Biol. 1987;43:148–154. [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. colishuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Johnson AD. A conditional allele of the Saccharomyces cerevisiae HOP1 gene is suppressed by overexpression of two other meiosis-specific genes: RED1 and REC104. . Genetics. 1993;133:785–797. doi: 10.1093/genetics/133.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth NM, Goetsch L, Byers B. The HOP1gene encodes a meiosis-specific component of yeast chromosomes. Cell. 1990;61:73–84. doi: 10.1016/0092-8674(90)90216-2. [DOI] [PubMed] [Google Scholar]
- Klein F, Laroche T, Cardenas ME, Hofmann JF-X, Schweizer D, Gasser SM. Localization of RAP1 and topoisomerase II in nuclei and meiotic chromosomes of yeast. J Cell Biol. 1992;117:935–948. doi: 10.1083/jcb.117.5.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers JH, Offenberg HH, van Marle A, Vink AC, Dietrich AJ, Heyting C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol Cell Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J, Klein F, Scherthan H. Homologous pairing is reduced but not abolished in asynaptic mutants of yeast. J Cell Biol. 1994;125:1191–1200. doi: 10.1083/jcb.125.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguire MP. Sister chromatid cohesiveness: vital function, obscure mechanism. Biochem Cell Biol. 1990;68:1231–1242. doi: 10.1139/o90-183. [DOI] [PubMed] [Google Scholar]
- Mao-Draayer Y, Galbraith AM, Pittman DL, Cool M, Malone RE. Analysis of meiotic recombination pathways in the yeast Saccharomyces cerevisiae. . Genetics. 1996;144:71–86. doi: 10.1093/genetics/144.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Earnshaw WC. Anti-topoisomerase II recognizes meiotic chromosome cores. Chromosoma. 1989;98:317–322. doi: 10.1007/BF00292383. [DOI] [PubMed] [Google Scholar]
- Nag DK, Scherthan H, Rockmill B, Bhargava J, Roeder GS. Heteroduplex DNA formation and homolog pairing in yeast meiotic mutants. Genetics. 1995;141:75–86. doi: 10.1093/genetics/141.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. . Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Petes TD. Meiotic mapping of yeast ribosomal deoxyribonucleic acid on chromosome XII. J Bacteriol. 1979;138:185–192. doi: 10.1128/jb.138.1.185-192.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petes TD, Botstein D. Simple Mendelian inheritance of the reiterated ribosomal DNA of yeast. Proc Natl Acad Sci USA. 1977;74:5091–5095. doi: 10.1073/pnas.74.11.5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. RED1: a yeast gene required for the segregation of chromosomes during the reductional division of meiosis. Proc Natl Acad Sci USA. 1988;85:6057–6061. doi: 10.1073/pnas.85.16.6057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Roeder GS. Meiosis in asynaptic yeast. Genetics. 1990;126:563–574. doi: 10.1093/genetics/126.3.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Engebrecht J, Scherthan H, Loidl J, Roeder GS. The yeast MER2gene is required for meiotic recombination and chromosome synapsis. Genetics. 1995a;141:49–59. doi: 10.1093/genetics/141.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Sym M, Scherthan H, Roeder GS. Roles for two RecA homologs in promoting meiotic chromosome synapsis. Genes Dev. 1995b;9:2684–2695. doi: 10.1101/gad.9.21.2684. [DOI] [PubMed] [Google Scholar]
- Roeder GS. Sex and the single cell: meiosis in yeast. Proc Natl Acad Sci USA. 1995;92:10450–10456. doi: 10.1073/pnas.92.23.10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman, F., G.R. Fink, and J.B. Hicks. 1986. Methods in Yeast Genetics: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.
- Smith DB, Johnson KS. Single-step purification of polypeptides expressed in Escherichia colias fusions with glutathione S-transferase. Gene (Amst) 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- Snyder M. The SPA2protein of yeast localizes to sites of cell growth. J Cell Biol. 1989;108:1419–1429. doi: 10.1083/jcb.108.4.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storlazzi A, Xu L, Schwacha A, Kleckner N. Synaptonemal complex (SC) component Zip1 plays a role in meiotic recombination independent of SC polymerization along the chromosomes. Proc Natl Acad Sci USA. 1996;93:9043–9048. doi: 10.1073/pnas.93.17.9043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell. 1994;79:283–292. doi: 10.1016/0092-8674(94)90197-x. [DOI] [PubMed] [Google Scholar]
- Sym M, Roeder GS. Zip1-induced changes in synaptonemal complex structure and polycomplex assembly. J Cell Biol. 1995;128:455–466. doi: 10.1083/jcb.128.4.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Engebrecht J, Roeder GS. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell. 1993;72:365–378. doi: 10.1016/0092-8674(93)90114-6. [DOI] [PubMed] [Google Scholar]
- Thompson EA, Roeder GS. Expression and DNA sequence of RED1, a gene required for meiosis I chromosome segregation in yeast. Mol Gen Genet. 1989;218:293–301. doi: 10.1007/BF00331281. [DOI] [PubMed] [Google Scholar]
- von Wettstein D, Rasmussen SW, Holm PB. The synaptonemal complex in genetic segregation. Annu Rev Genet. 1984;18:331–413. doi: 10.1146/annurev.ge.18.120184.001555. [DOI] [PubMed] [Google Scholar]