Abstract
The kinesin superfamily is a large group of proteins (kinesin-like proteins [KLPs]) that share sequence similarity with the microtubule (MT) motor kinesin. Several members of this superfamily have been implicated in various stages of mitosis and meiosis. Here we report our studies on KLP67A of Drosophila. DNA sequence analysis of KLP67A predicts an MT motor protein with an amino-terminal motor domain. To prove this directly, KLP67A expressed in Escherichia coli was shown in an in vitro motility assay to move MTs in the plus direction. We also report expression analyses at both the mRNA and protein level, which implicate KLP67A in the localization of mitochondria in undifferentiated cell types. In situ hybridization studies of the KLP67A mRNA during embryogenesis and larval central nervous system development indicate a proliferation-specific expression pattern. Furthermore, when affinity-purified anti-KLP67A antisera are used to stain blastoderm embryos, mitochondria in the region of the spindle asters are labeled. These data suggest that KLP67A is a mitotic motor of Drosophila that may have the unique role of positioning mitochondria near the spindle.
In many cell types, mitochondria are localized in accordance with substrate availability or energy requiring processes. In higher eukaryotes, this localization is known to depend upon the microtubule (MT)1 cytoskeleton. The first convincing cytological demonstration of MT dependence was the coimmunolocalization of mitochondria to MTs in cultured cells (Heggeness et al., 1978). Direct observation of the translocation of mitochondria along MTs in extruded squid axoplasm has also been reported (Brady et al., 1982). At the ultrastructural level, mitochondria are also found surrounded by MTs in brain cells (Raine et al., 1971) and in the astral MTs of mouse oocytes (Van Blerkom, 1991). Finally, direct evidence for the requirement of MTs in mitochondrial localization has come from genetic analyses in Schizosaccharomyces pombe (Yaffe et al., 1996).
Recent work indicates that mitochondrial translocation may involve at least one member of the kinesin superfamily. Kinesin-like proteins (KLPs) all share a protein domain that has sequence homology to the kinesin heavy chain (KHC) globular head or motor domain. The remainder of each KLP molecule is in most cases divergent and suggested to be responsible for the attachment of specific cargoes within the cell (Vale and Goldstein, 1990). In the case of mitochondrial movement, the mouse kinesin superfamily protein 1B (KIF1B) protein has recently been found associated with mitochondria in neurons (Nangaku et al., 1994). Since KIF1B expression is enriched in neurons it may be, in part, specialized for the anterograde axonal transport system.
In this paper, we describe the characterization of KLP67A and show that it is a plus end–directed motor. This KLP was initially discovered in a PCR-based screen for new members of the kinesin superfamily in Drosophila (Stewart et al., 1991). Expression and immunolocalization studies suggest that KLP67A may be a new type of mitochondrial motor that is specialized for mitochondrial movement in proliferative cells. Together with previous data on KIF1B, the results reported here suggest that eukaryotes may use different mitochondrial motors adapted for specific MT systems such as the axonal MT network, the interphase cytoskeleton, and the mitotic spindle.
Materials and Methods
DNA Sequence Analysis
KLP67A was originally identified as a fragment in a PCR-based screen for new members of the kinesin superfamily in Drosophila (Stewart et al., 1991). We used the original PCR fragment as a probe to screen a Drosophila 0–4-h embryonic cDNA library (Brown and Kafatos, 1988). Several isolates from this library were found to each contain inserts of ∼3 kb in size. A 2.9-kb HindIII fragment was subcloned from one of these clones into the Bluescript vector (Stratagene, La Jolla, CA) and a deletion series was constructed using the Exo/mung kit (Stratagene) according to the manufacturer's instructions. Deletion clones were sequenced on both strands by the dideoxy chain termination method using a sequencing kit (United States Biochem Corp., Cleveland, OH). The noncoding strand was sequenced using a T7 primer. The coding strand was sequenced using a T3 primer as well as newly synthesized oligonucleotide primers. The resulting DNA sequence predicts a 2,442-bp open reading frame flanked by a 138-bp 5′ untranslated region and a 383-bp 3′ untranslated region. The original KLP67A cDNA clone within pNB40 was used to determine the last 70 bp of sequence through the poly A tail.
Expression and Purification of KLP67A in E. coli
The vector pGEX-2T (Pharmacia LKB Biotechnology, Piscataway, NJ) was used to express KLP67A as part of a glutathione S-transferase (GST) fusion protein (Guan and Dixon, 1991). KLP67A was subcloned into this vector in two steps. In the first step, a NcoI/HindIII fragment derived from the KLP67A cDNA was cloned into NcoI/HindIII-digested pGEX-2T. This subclone did not contain the first 350 bp of the KLP67A coding sequence. In a second step, the 5′ most 350-bp segment was ligated in the form of an NcoI-digested PCR fragment. An NcoI site was created at the 5′ end of this PCR product through the use of a primer that encoded the initiator methionine as part of the NcoI recognition sequence. The modified E. coli KLP67A therefore begins with MET ALA rather than MET PRO. This prediction was verified by DNA sequence analysis of pGEX-KLP67A. The final construct was transformed into E. coli 71/18. Induction with isopropylthiogalactoside (IPTG) resulted in the production of a 122-kD GST fusion protein.
E. coli 71/18 cells containing pGEX-KLP67A were grown at 22°C for 4–5 h, IPTG was added to a final concentration of 1 mM, and the culture was incubated for an additional 1–2 h at 22°C. Cells were harvested by centrifugation at 10,000 rpm, resuspended in sonication buffer (0.1 M sodium phosphate buffer, pH 7, 5 mM EDTA, 1 mM PMSF), recentrifuged, and resuspended in 10 ml sonication buffer per 500 ml starting culture. Cells were lysed by sonication followed by centrifugation to pellet insoluble material. The supernatant was incubated with glutathione-coupled agarose beads (Sigma Chemical Co., St. Louis, MO) for 30 min on ice. The agarose beads were washed three times with PEM 80 (80 mM K2Pipes/1 mM EGTA/1 mM MgCl2, pH 7.2), 1 mM DTT, 0.1M NaCl and centrifuged briefly in a microfuge. After the final wash, the agarose beads were resuspended in 0.1 M glutathione in PEM 80, pH 7.2, and incubated on ice for 20 min. The beads were pelleted in a microfuge and the supernatant was used immediately for motility assays.
Motility Assays
Soluble GST–KLP67A fusion protein was tested for motility activity with MTs prepared from bovine brain tubulin as described previously (Stewart et al., 1993). A rabbit anti-GST antiserum was necessary to bind the fusion protein to glass coverslips. The order in which the components of the assay were combined was found to be critical. The GST–KLP67A fusion protein (final concentration ∼10 μg/ml) was added to a glass coverslip, followed by the anti-GST antiserum (1:100 dilution), the MTs (final concentration 1–5 μg/ml), and ATP to a final concentration of 10 mM. MT movement was observed by video-enhanced differential interference contrast microscopy. The direction of movement was determined using rhodamine-labeled MTs as described previously (Stewart et al., 1993).
In Situ Hybridization to Embryos
In situ hybridization was carried out according to the method of Tautz and Pfeifle (1989). After dechorionation by washing for 3 min in hypochlorite, embryos were fixed in 4% paraformaldehyde in 120 mM NaCl, 10 mM NaPO4 (PBS) with an equal volume of heptane. They were next devitalized by vigorous shaking in 1:1 heptane/methanol (Mitchison and Sedat, 1983) and then pretreated for hybridization as described by Tautz and Pfeifle (1989). After digestion with SalI, a digoxigenin-labeled anti-sense RNA probe was transcribed from a KLP67A cDNA clone in the vector pNB40 (Brown and Kafatos, 1988). Before hybridization, probe RNA was reduced in size by hydrolysis in 40 mM NaHCO3, 60 mM Na2CO3, pH 10, for 2 h at 60°C. The probe was precipitated and resuspended in 50 μl water; 5 μl was used for hybridization, which was carried out for 16 h at 47°C. Hybridization solution and subsequent treatments were as described by Tautz and Pfeifle (1989). Hybridization was detected by an alkaline phosphatase conjugated antidigoxigenin antibody (Boehringer Mannheim Corp., Indianapolis, IN) used at a dilution of 1:1,000. Embryos were washed in PBS and then incubated for 15 min with 10 μg/ml 4,6-diamidino-2-phenylindole (DAPI; Sigma Chemical Co.) for visualization of DNA. Embryos were washed 3× 5 min in PBS and then mounted in 85% glycerol and viewed using a microscope (model Axiophot; Carl Zeiss, Inc., Thornwood, NY). Embryos used to compare RNA levels between different stages were drawn from the same collection and fixation batch.
In Situ Hybridization to Larval Tissue
Brains and imaginal discs were dissected from third instar larvae in 0.7% NaCl, transferred to 4% paraformaldehyde in PBS, and fixed for a further 20 min. Pretreatment and hybridization was carried out according to the method of Tautz and Pfeifle (1989) except that the tissue was digested for 8 min with 50 μg/ml Proteinase K before hybridization. A probe was prepared by incorporation of digoxigenin-dUTP by random oligo-labeling (Feinberg and Vogelstein, 1984) using a digoxigenin DNA labeling kit (Boehringer Mannheim Corp.). Tissue was dehydrated through an ethanol series (30, 50, 70, 90, and 100%, 2 min each), mounted in Canada Balsam, and viewed with a microscope (model Axiophot; Carl Zeiss, Inc.) using differential interference contrast optics.
Antibody Production
Four rat antisera were raised against GST–KLP67A. GST–KLP67A was purified by two different methods for immunization. Soluble GST–KLP67A was prepared as for the motility assays described above. Approximately 10 μg of the fusion protein bound to 500 μl of glutathione agarose was mixed with RIBI adjuvant (Immunochemical Research Inc., Hamilton, MT) and injected subcutaneously into rat 1. The other three animals were injected with ∼10 μg each of gel-purified GST–KLP67A suspended in RIBI adjuvant. Rats were boosted on day 14 with an identical preparation. Test bleeds 10 d later showed that three of the four rats (excluding rat 4) had identical immunoreactivity patterns to Drosophila embryos as described in the results section. 4 mo later, all four rats received another boost of the same immunogen to maintain the anti-KLP67A antibody titer. After this second boost, rat 4 antiserum exhibited an immunostaining pattern identical to those from the other three rats.
Gel-purified GST–KLP67A-70 (see below) was also used to immunize two rabbits. After affinity purification, one of these rabbit antisera displayed a staining pattern identical to that of the four rat antisera described below. The second antisera weakly stains spindle fibers. At this time we do not know if this second staining pattern is due to a cross-reacting KLP.
Affinity Purification of Antisera
Each of the rat antisera were affinity purified using two different halves of the protein. To make the construct encoding the carboxy half of KLP67A, full-length GST–KLP67A (described above) was digested with BamHI and NotI and repaired with the Klenow fragment of DNA polymerase and ligated. E. coli transformed with this subclone expresses an IPTG-inducible 70-kD protein (GST–KLP67A-70). To affinity purify antisera specific to this part of the protein, ∼100 μg of GST–KLP67A-70 were run on a 7.5% polyacrylamide gel and transferred to Immobilon (Millipore Corp., Bedford, MA). The filter was stained with Ponceau S (Sigma Chemical Co.) and the band containing GST–KLP67A-70 was excised and used for the affinity purification (Olmsted, 1981). The amino half of the molecule was used in the same way for affinity purification. To make the construct encoding this half of the protein, the pGEX construct containing GST–KLP67A was digested with NotI and HindIII, repaired as described above, and ligated. Antisera specific to the amino part of the protein were affinity purified as described above. Rat 1 antisera resulted in the most intense staining pattern. Antisera from the other rats exhibited an identical albeit weaker staining pattern after affinity purification using either half of the protein.
Immunolocalization
KLP67A was immunolocalized within methanol-fixed Drosophila embryos as described (Warn and Warn, 1986), with the following modifications. After fixation, the embryos were stored in 100% methanol at −20°C from overnight to up to 1 mo with no change in tubulin staining quality, although mitochondria would gradually enlarge and detach from the MTs and appear on the surface of the embryo. Washes after primary and secondary antibody incubations were overnight at 4°C with PBS, 0.1% Triton X-100 (PBST). Primary antibody incubations were done using a 1:200 dilution of affinity-purified antisera together with one of the following: a 1: 10 dilution of mouse anti–tubulin monoclonal 3A5 (Piperno and Fuller, 1985) , a 1:200 dilution of a rabbit anti–mitochondrial ATP synthase antisera (Pena and Garesse, 1993), or a 1:200 dilution of an anticytochrome oxidase antibody (Molecular Probes, Eugene, OR). Dilutions were made in PBST. The secondary antibodies used were fluorescein-conjugated goat anti–mouse or anti–rabbit IgG and Cy3-conjugated goat anti-rat IgG (Cappel, Malvern, PA). Secondary antibodies were used at a dilution of 1:2,000 in PBST. Embryos were also fixed with formaldehyde as described (Theurkauf, 1992).
To test for colocalization of KLP67A with mitochondria, embryos were fixed with methanol minus EGTA or were used after 2 mo at −20°C in methanol. This resulted in a release of the mitochondria from the cytoskeleton and onto the surface of the embryo, which was necessary to visualize the mitochondria with an anti–cytochrome oxidase antibody (Molecular Probes, Eugene, OR). In embryos fixed as described above for MT preservation, the anti-cytochrome oxidase antibody staining pattern was very difficult to visualize because of the high cytoplasmic background seen with this antibody. It was still possible, however, to verify the colocalization of KLP67A and cytochrome oxidase to mitochondria in the area surrounding the spindle poles.
Immunolocalization experiments using CHO cells were done using cells grown on glass coverslips. To label mitochondria, MitoTracker Green (Molecular Probes) was used according to the manufacturer's directions. After labeling, the cells were washed with PBS and fixed in ice-cold methanol. Antibody labeling was as described (Doxsey et al., 1994). CHO cells were observed and photographed using a microscope (model Axiophot; Carl Zeiss, Inc.).
Western Blot Analysis
Drosophila lysate was prepared as described (Saxton et al., 1988) and loaded onto a 10% SDS–polyacrylamide gel. The protein was transferred to Immobilon (Millipore Corp.), and the blot was then processed as described (Pereira et al., 1992) except that a HRP-conjugated secondary antibody (Kirkegaard and Perry Laboratories, Gaithersburg, MD) was used to detect binding of the primary antibody. For the primary antibody incubation, rat 4 antisera affinity purified against GST–KLP67A-70 was used at a 1:10 dilution. The HRP-conjugated anti–rat secondary antibody was used at a dilution of 1:5,000. The HRP substrate was LumiGLO and was used according to the manufacturer's instructions (Kirkegaard and Perry Laboratories).
Results
The complete amino acid sequence of KLP67A (Fig. 1 A) was deduced from the DNA sequence of a KLP67A cDNA isolated from a Drosophila embryonic cDNA library (Brown and Kafatos, 1988). A fragment of the amino acid sequence of the KLP67A motor domain was previously published and was designated KLP3; it has now been renamed according to its cytological location, 67A (Stewart et al., 1991). The KLP67A protein sequence includes 814 amino acid residues. The putative initiator methionine is located at nucleotide 138. Further analysis of the non–motor domain sequence using standard computer programs (Garnier et al., 1978; Devereux et al., 1984) reveals that the non– motor domain, particularly between amino acid residues 340–660, has the potential to form an α-helix (Fig. 1 B). Within this same stretch of amino acids, there is also a high probability of α-helical coiled coil formation as predicted by the method of Lupas et al. (1991) as can be seen in Fig. 1 C. The secondary structure of the 200–amino acid carboxy-terminal region is unknown. Therefore, unlike the KHC, KLP67A cannot be easily predicted to have a threedomain structure. This is also the case for several other members of this superfamily such as nod (Zhang et al., 1990). Determination of the KLP67A secondary structure therefore awaits biochemical analyses of the native protein.
Figure 1.
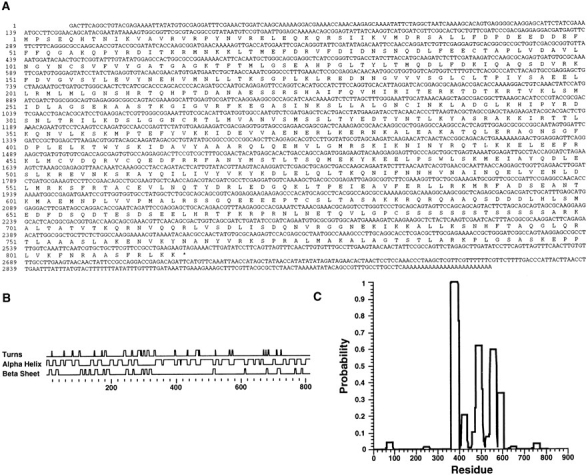
KLP67A cDNA sequence. (A) The sequence of a KLP67A cDNA clone and its deduced amino acid sequence is shown. These sequence data are available from EMBL/Genbank/DDBJ under accession number U89264. The amino acid sequence of a portion of the presumptive motor domain was previously compared to 27 known members of the kinesin superfamily (Goldstein, 1993). (B) Structural predictions from the UWGCG Peptide Structure program. (C) α-Helical coiled coil probability predicted from the algorithm of Lupas et al. (1991).
Many members of the kinesin superfamily can be grouped into families on the basis of sequence similarity within their motor domains (Goldstein, 1993). For example, pairwise sequence comparisons of the predicted motor domains among all members of the superfamily reveal several families that are more similar among themselves than with other members of the superfamily (e.g., the bimC family, the KHC family, and the KAR3 family). KLP67A does not fall into any of these families, nor does it show significantly more sequence similarity with the motor or tail domain of any other kinesin superfamily member than it does with the KHC. Direct comparisons to KIF1B (Nangaku et al., 1994) also revealed no sequence similarity outside of the conserved motor domain.
KLP67A Is a Plus End–directed MT Motor
The amino acid sequence of KLP67A predicts that this protein is an MT motor. To verify this prediction, a GST– KLP67A fusion protein was produced in E. coli, partially purified by affinity chromatography, and used in an MT motility assay. MT motility was observed in vitro after addition of GST–KLP67A fusion protein, MTs, and ATP to the assay. The average speed of movement from three assays was .05 (±.02) μm/s, about 10 times slower than the speed measured for Drosophila kinesin (Kuznetsov et al., 1989) and KIF1B (Nangaku et al., 1994) but similar to the speed of Eg5, another kinesin-related protein (Sawin et al., 1992). Although a “no-insert” bacterial lysate control was not done specifically during this series of motility experiments, previous work has demonstrated that E. coli lacks an endogenous MT motility activity (data not shown).
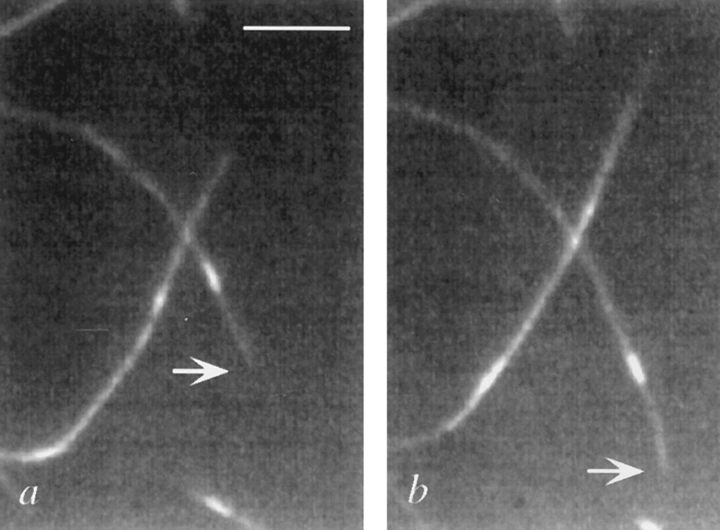
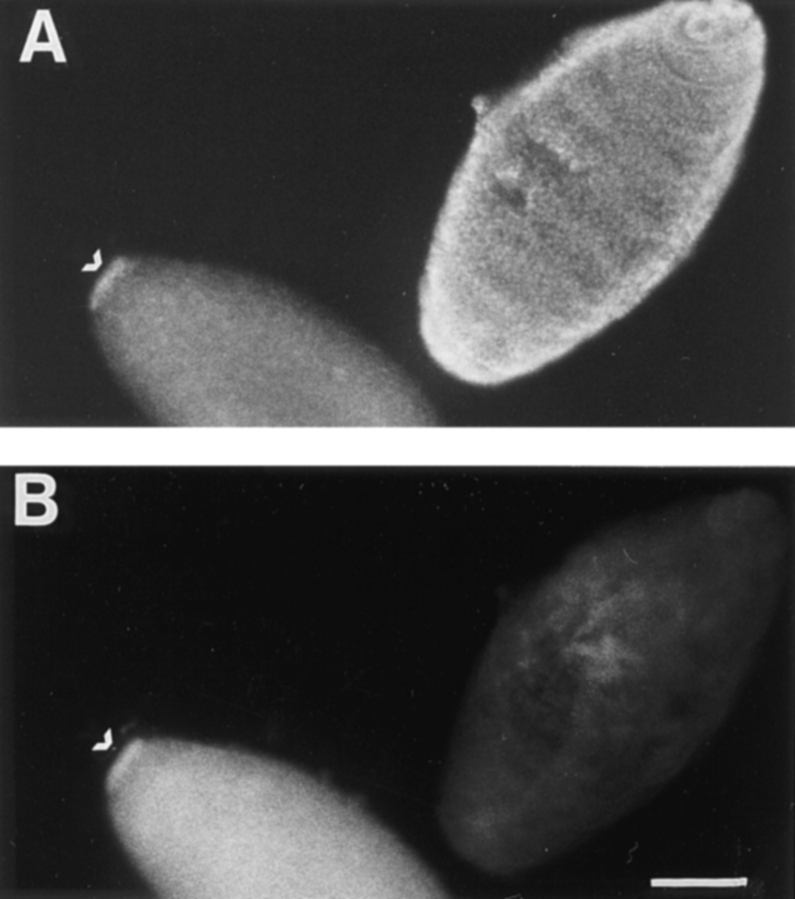
Kinesin and related proteins exhibit unidirectional movement along the MT. MTs have an intrinsic polarity that is generally defined by the relative rates of addition and loss of tubulin subunits to their ends. This polarity is also related to MT orientation within the cell. The more stable minus end of the MT is embedded in a centrosome or nucleation site, and the more rapidly growing plus end is in the spindle midzone or at the cell periphery. A specific motor protein therefore will reflect this polarity by causing movement toward or away from the cell periphery or spindle pole. Since the directionality of a motor protein is critical for predictions of its cellular function, we determined the direction of movement of KLP67A. To do this, a motility assay similar to that described above was carried out. In this case, the minus ends of the MTs were labeled with rhodamine-conjugated tubulin (Hyman, 1991; Stewart et al., 1993). Plus end–directed movement by KLP67A would cause MT movement with the rhodamine seed leading, while minus end–directed movement would cause the rhodamine seed to trail. In 12 assays, the addition of GST–KLP67A clearly resulted in plus end–directed movement. For example, in Fig. 2 a the minus end of a rhodamine-seeded MT is indicated by an arrow. Several minutes later the minus end has moved several microns beyond the point of reference, as shown in Fig. 2 b.
Figure 2.
Movement of polarity-marked microtubules in the plus direction. The microtubule minus end is nearest the rhodamine seed and is indicated by the arrow. (a) Microtubule position at t = 0. (b) Microtubule position at t = 6 min. The velocities are not as fast as those in the non–fluorescence-based assays, presumably owing to photodamage. Bar, 5 μm.
Expression Pattern of KLP67A mRNA
The expression pattern of a gene often provides information about functions of its encoded protein. We therefore examined the pattern of KLP67A mRNA expression during embryogenesis by in situ hybridization (see Materials and Methods). During the synchronous divisions of the blastoderm embryo, KLP67A transcripts are uniformly distributed (Fig. 3 a). Their presence at high levels before the onset of zygotic gene expression indicates that they are maternally provided. Many maternal RNAs appear to be degraded throughout the embryo upon cellularization, and at this time KLP67A RNA is depleted (Fig. 3 b). After the completion of cellularization, mitotic synchrony is lost, but groups of cells referred to as mitotic domains continue to divide in unison. These divisions take place in a precise temporal and spatial pattern (Foe, 1989). We observed that KLP67A transcripts assume a distribution that corresponds with this pattern.
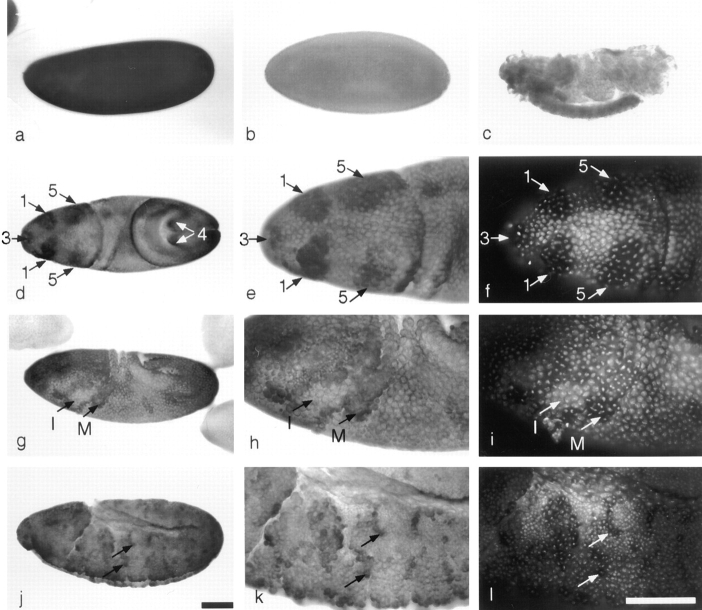
Figure 3.
The distribution of KLP67A transcripts in embryogenesis shown by in situ hybridization. (a) Syncytial embryo showing high levels of uniformly distributed KLP67A RNA. In the cellularizing embryo, the RNA is depleted (b). RNA is present in the embryonic central nervous system after mitosis has ceased in other tissues (c). In the cycle 14 embryo, KLP67A transcripts accumulate in mitotic domains, indicated by arrows in d, e, and f, numbers refer to domains described by Foe (1989). Cells undergoing mitosis are distinguished by staining of DNA using DAPI, in f, i, and l. In later cycle 14 the distribution of KLP67A RNA continues to reflect the pattern of mitotic domains (g). Comparison of h (KLP67A) and i (DNA staining) indicates that cells undergoing mitosis contain elevated levels of KLP67A transcripts. In g, h, and i, adjacent regions of cells in interphase (I), and mitosis (M) are shown. In the cycle 15 embryo, KLP67A RNA accumulation corresponds with the pattern of mitosis (j). Two groups of cells that contain high levels of KLP67A RNA are indicated in j and k. DNA is visualized and the same groups of cells are indicated in l. All embryos are oriented anterior to left, dorsal up, except b, d, e, and f, where dorsal views are shown. Bars: (a–d, g, and j; shown in j) 100 μm; (e, f, h, i, k, and l; shown in l) 100 μm.
To determine accurately the relationship between the accumulation of KLP67A transcripts and the pattern of mitosis, we performed in situ hybridization and stained embryos with DAPI to visualize DNA and thus allow cells that are undergoing mitosis to be distinguished from those in interphase. The early mitotic domains 1 and 5 are located on the dorsal side of the embryo, anterior to the cephalic furrow. Both are divided bilaterally, and, when they are undergoing mitosis (Fig. 3 d, arrows), KLP67A transcripts are present at levels higher than in the surrounding interphase cells (Fig. 3, d and e). Cells in domains 3 and 4, which lie at the anterior and posterior ends of the embryo, respectively, also accumulate high levels of KLP67A RNA at this time (Fig. 3, e and f, arrows). Similarly, in other cycle 14 domains, the accumulation of KLP67A transcripts continues to follow closely the pattern of mitosis (Fig. 3, g, h, and i). In cycle 15, the mitotic domains now consist of smaller groups of contiguous cells that still accumulate KLP67A transcripts to high levels as they divide (Fig. 3, j, k, and l, arrows). After the completion of the sixteenth mitotic cycle, divisions are confined to the cells of the developing central nervous system (CNS) where KLP67A transcripts are still present, whereas in other nonproliferative regions of the embryo, the transcripts can no longer be detected (Fig. 3 c).
During larval development, mitosis is largely restricted to the proliferative regions of the brain and imaginal tissue, which have distinctive patterns of cell division (White and Kankel, 1978). We therefore performed in situ hybridization (see Materials and Methods) to determine if KLP67A transcripts were distributed correspondingly. In the eyeantenna disc, cells become incorporated into the ommatidia preclusters as a wave of differentiation, referred to as the morphogenetic furrow, moves across the disc from posterior to anterior. Anterior to the furrow, asynchronous mitoses provide cells for incorporation into the preclusters. Then, posterior to the furrow, specific cells of each newly formed precluster undergo a final round of synchronous mitosis (Ready et al., 1976; Wolff and Ready, 1991).
We find that the distribution of KLP67A transcripts reflects this pattern of cell division. Anterior to the furrow are regions of cells, containing KLP67A transcripts, that are likely to correspond to those undergoing asynchronous mitoses. Posterior to the furrow, transcripts are restricted to a narrow stripe of cells, including those undergoing synchronous mitoses (Fig. 4 a, arrows). Transcripts cannot be detected in cells more posterior to the furrow, where mitosis has ceased. Similarly, in the optic lobes of the larval CNS, KLP67A transcripts can only be detected in cells whose distribution is characteristic of those in the proliferative regions (White and Kankel, 1978; Truman and Bate, 1988) (Fig. 4 b, arrows). Hence in embryogenesis and in the tissues of the third instar larva that we examined, the distribution of KLP67A transcripts closely follows the patterns of mitosis.
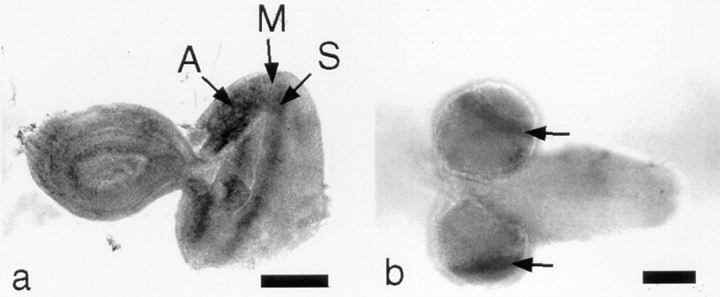
Figure 4.
Distribution of KLP67A transcripts in tissues of the 3rd instar larva, shown by in situ hybridization. (a) Transcripts are present in the anterior region of the eye disc where asynchronous mitoses are taking place (A). Transcripts are concentrated in a stripe corresponding to cells undergoing synchronous mitoses (S), immediately posterior to the morphogenetic furrow (M). (b) In the larval brain, transcripts are concentrated in the proliferative regions of the optic lobes (arrows). In a and b, anterior is to the left. Bars: (a) 100 μm; (b) 200 μm.
Immunolocalization of KLP67A
The observation that KLP67A mRNA is expressed in a pattern indicative of a mitotic function prompted an examination of the distribution of KLP67A within the rapidly dividing nuclei of the precellular blastoderm. For these analyses, four rat antisera were raised against GST–KLP67A as described in Materials and Methods. All four antisera showed identical staining patterns on embryos before as well as after affinity purification against two different parts of the protein (see Materials and Methods). Analysis of the specificity of these antisera by Western blotting indicates that they give reactivity with a band of the appropriate size in embryo lysates. Rat 4 antisera, however, resulted in the strongest signal in Western blot experiments. For example, rat 4 antisera affinity purified against the KLP67A non–motor domain recognizes a predominant protein species of ∼90-kD from Drosophila embryo lysates (Fig. 5, lane 2). This size is very close to the molecular weight of 92 kD predicted from the KLP67A amino acid sequence. This species is not recognized by preimmune sera from any of the four rats (data not shown). A slightly smaller species is recognized in lysates prepared from CHO cells (Fig. 5, lane 1).
Figure 5.
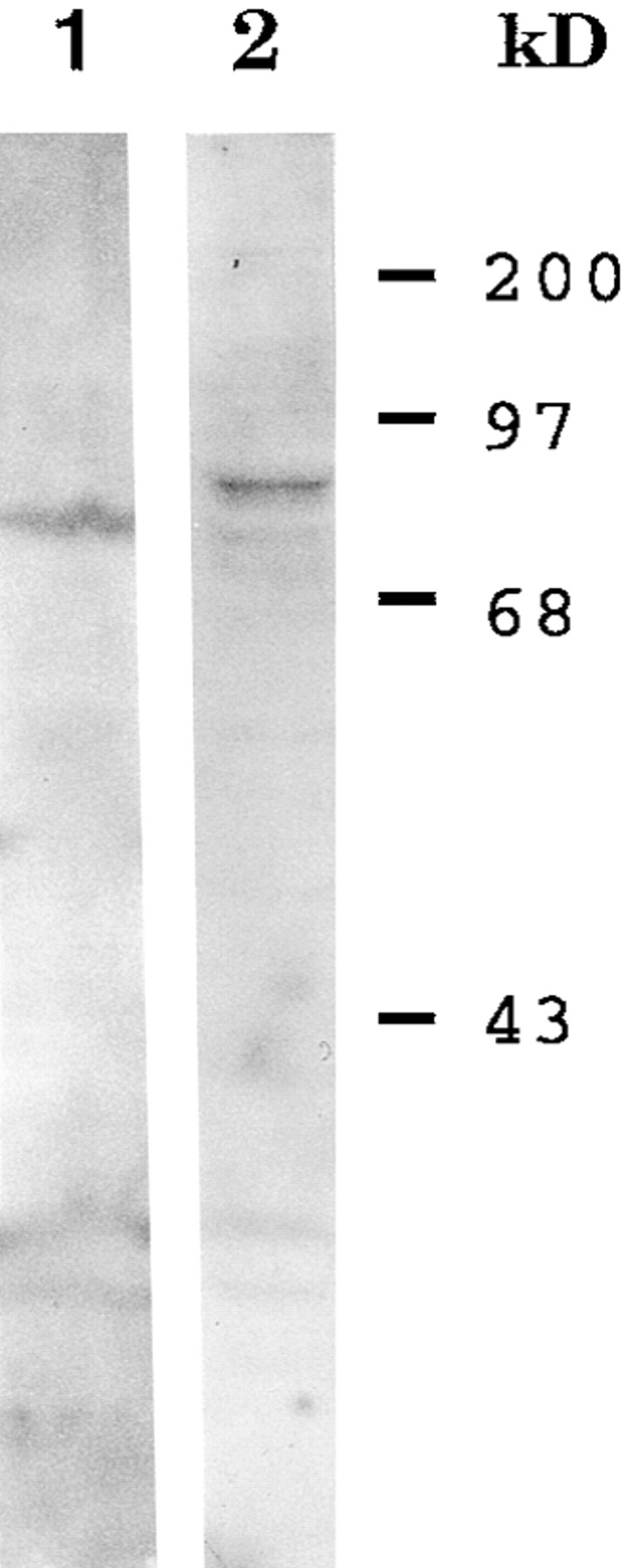
Western blot of Drosophila embryo (lane 2) and CHO cell lysates (lane 1) probed with rat 4 anti-KLP67A rat antisera affinity purified against GST–KLP67A-70.
Initially, immunofluorescence experiments with these anti-KLP67A antisera were conducted with D. melanogaster infected with the endosymbiotic bacteria Wolbachia. These bacteria are found in many laboratory stocks of Drosophila grown in the absence of tetracycline (O'Neill and Karr, 1990). This allowed the fortuitous discovery that KLP67A is associated with bacteria found on the plus ends of astral MT fibers (Pereira, A., and T. Karr, unpublished observation; the localization of KLP67A to Wolbachia will be presented elsewhere). In embryos that are uninfected by Wolbachia, KLP67A staining is still observed on particles in the region of the astral fibers, but these particles are at least 10-fold smaller than Wolbachia. An example of this localization within a blastoderm embryo double labeled with anti-KLP67A and anti–tubulin antisera is shown in Fig. 6. The antigen distribution relative to the spindle poles is most evident when a stepwise series of optical sections of the spindles is viewed simultaneously. Thus, four 0.8-μm optical sections through a sample of late anaphase stage spindles are shown. Late anaphase B is shown specifically because at this stage the poles are more widely separated and the concentration of particles at the poles is more evident than at other stages of mitosis. The small particles stained by anti-KLP67A are predominantly seen in regions containing astral fibers radiating from each spindle pole. To examine the possibility that these small particles are mitochondria, embryos were also double labeled with affinity-purified anti-KLP67A antiserum and an antiserum specific for the Drosophila mitochondrial ATP synthase β subunit (Pena and Garesse, 1993). A higher magnification view (Fig. 7) shows that KLP67A staining is primarily associated with particles that are also stained with the ATP synthase antisera. However, KLP67A appears to associate with a subset of mitochondria since not all of the mitochondria that are stained by the ATP synthase antisera are also stained by anti-KLP67A. Similarly, localization of these particles to the region of the astral fibers is also observed when embryos are double labeled with the ATP synthase and tubulin antisera (data not shown).
Figure 6.
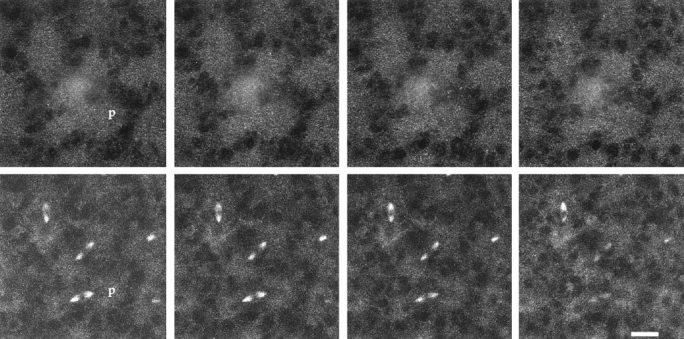
A Z series of four consecutive optical sections through a Drosophila precellular blastoderm embryo double labeled with affinity-purified rat 4 KLP67A (top four panels) and antitubulin antibodies (bottom four panels). The leftmost section corresponds to the approximate middle of the spindle and the next three sections are progressing downward away from the surface of the embryo. Note that anti-KLP67A stains circular areas of small particles concentrated at the spindle poles. One such area is indicated by the “p” in the first pair of panels. Bar, 15 μm.
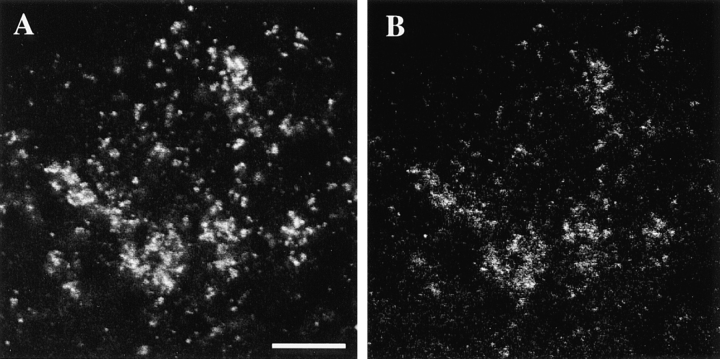
Figure 7.
Double labeling of a blastoderm embryo similar to that shown in Fig. 6 with a Drosophila mitochondrial ATP synthase (A) and affinity-purified rat 4 anti KLP67A (B) antiserum. Bar, 5 μm.
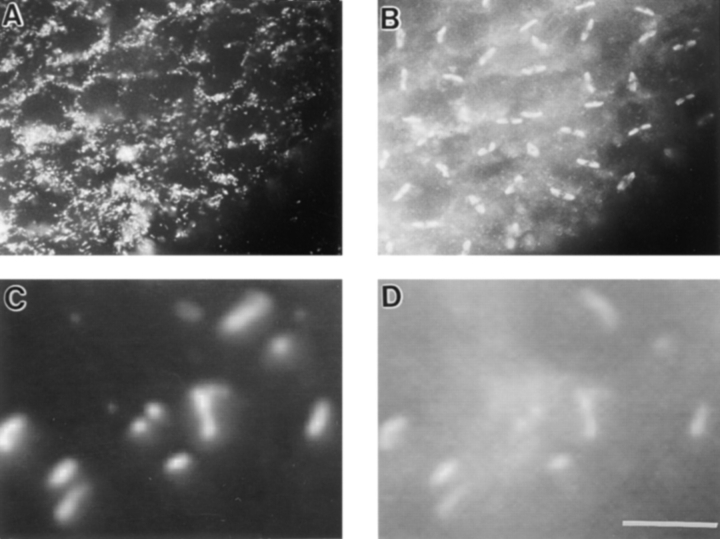
An alternative embryo preparation allowed these particles to be released from the cytoskeleton, thereby becoming more visible. It was found that in embryos fixed in methanol without EGTA, in embryos that have been stored for a prolonged time at −20°C, or in embryos with poor astral MT preservation, anti-KLP67A stains mitochondrialike particles that are released from the cytoskeleton and appear in aggregates on the surface of the egg (Fig. 8). Since this aberration allows the particles to be visualized more clearly, such a preparation was used to show that the small particles detected by anti-KLP67A are indeed mitochondria. Thus, Fig. 8 also shows the surface of one such embryo double labeled with anti-KLP67A (Fig. 8 C) and the mitochondrial marker anti–cytochrome oxidase (Fig. 8 D). Although the cytochrome oxidase staining is not as intense and clear as that of the KLP67A staining, it can be seen that the two colocalize completely to mitochondria on the surface of this embryo.
Figure 8.
Immunolocalization of KLP67A in the absence of astral fiber fixation. (A) Surface of a blastoderm embryo immunostained for KLP67A. (B) The corresponding DAPI-stained image of anaphase chromosomes. Double labeling of the embryo preparation shown in Fig. 7 with anti-KLP67A (C) and anti–cytochrome oxidase (D). Bar, 5 μm.
It was also apparent that KLP67A staining is enriched at the posterior pole of the blastoderm embryo (Fig. 9 B). This observation is consistent with ultrastructural analyses of the Drosophila oocyte and early embryo, which have shown that a large number of mitochondria are concentrated at the posterior pole of the oocyte, where they are attached or in close apposition to the polar granules until about 30 min after fertilization (Mahowald, 1971). Indeed, the high concentration of mitochondria in this region causes intense DAPI staining in this area as well (Fig. 9 A); KLP67A colocalizes with small DAPI-stained particles at the posterior pole of the embryo (data not shown). This association suggests that KLP67A may be used for the transport of mitochondria and possibly other polar granule components to the posterior pole during oogenesis.
Figure 9.
Early and late Drosophila embryos double labeled with DAPI (A) and affinity-purified rat 1 antisera (B) directed against KLP67A. The arrowhead indicates the posterior polar end of the stage 1 embryo. The second embryo is approximately at stage 15. Bar, 100 μm.
Some KLP67A staining is detectable in gastrula stage embryos where the mRNA expression pattern is enriched in mitotic domains. Surprisingly, however, at this stage KLP67A staining is of apparently equivalent intensity in interphase as in mitosis (data not shown). Whether this apparent inconsistency is a result of the level of the KLP67A protein not following the mRNA amount, or whether it is because immunofluorescence localization is not a reliable indicator of actual protein amount in a cell is unclear at present. Finally, staining for KLP67A late in embryogenesis is difficult to detect (e.g., the stage 15 embryo in Fig. 9, A and B, upper right) relative to the high level of KLP67A visible in early embryos (the stage 1 embryo in Fig. 9, A and B, lower left can be used for comparison). Whether this is due to an absence of KLP67A protein at later stages of embryogenesis or due to a detection problem is not clear.
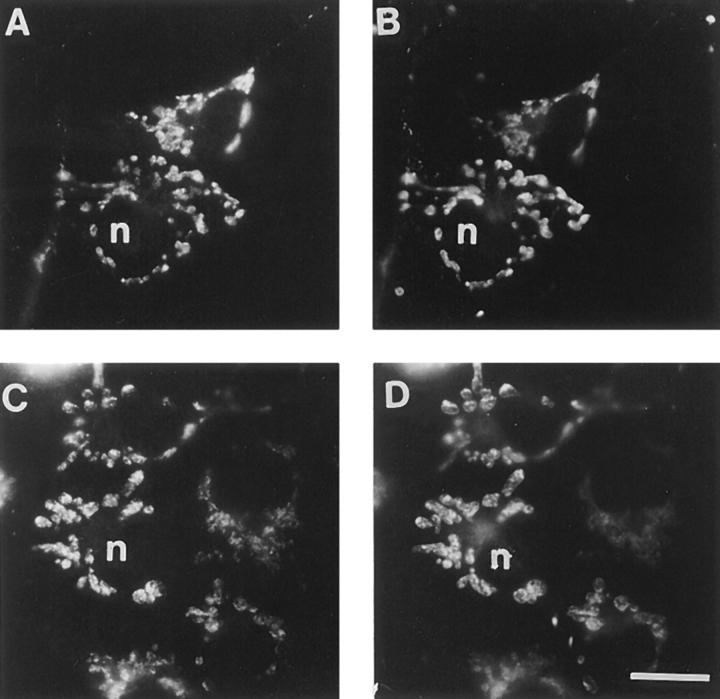
Finally, anti-KLP67A was also used to stain mammalian cells. CHO cells labeled with anti-KLP67A and a vital dye that is specifically taken up by mitochondria, Mitotracker, show colocalization to mitochondria as shown in Fig. 10. This staining pattern was observed with two different antisera, one affinity purified against the carboxyl half of KLP67A and the other affinity purified against the amino half. This latter result is a strong verification that KLP67A is evolutionarily conserved and is associated with mitochondria in both invertebrates and vertebrates.
Figure 10.
Immunolocalization of KLP67A within CHO cells double labeled with Mitotracker (B and D). (A) Cells stained with anti-KLP67A affinity purified against the motor domain portion of KLP67A. (C) Cells stained with anti-KLP67A affinity purified against the non–motor domain portion of KLP67A. The nucleus is indicated by an “n” for registration purposes. Bar, 5 μm.
Discussion
MTs are used for translocating a number of different organelles within the cell. Several members of the kinesin superfamily are now known to be responsible for these movements. Here we show that KLP67A is a plus end– directed MT motor that is associated with mitochondria in both Drosophila and mammalian cells. Although KLP67A may persist after embryonic cells have ceased to divide (see below), our data suggest that it may be found primarily in proliferating tissue such as cultured cells and in early Drosophila embryos. A survey of some third instar larval tissue has shown that KLP67A is found in the imaginal discs and proliferation centers of the CNS but not in some differentiated tissue such as neuropil and muscle (Pereira, A., unpublished observations). It is therefore possible that a mitochondrial motor, similar to KIF1B of mouse, is expressed in nonproliferating differentiated cell types of Drosophila.
KLP67A is notable in that it does not readily fall into any of the known KLP families that have been defined strictly by sequence homology (Goldstein, 1993). Often, a functional homology among family members also exists. For example, the bimC family consists of several different KLPs that all separate spindle poles during spindle assembly (discussed in Heck et al., 1993). Other families of KLPs also appear to share similar cellular functions as well (Pereira and Goldstein, 1994). It is therefore curious that the Drosophila KLP67A associates with mitochondria in vivo, yet it is not in the same family as the mouse mitochondrial motor, KIF1B (Nangaku et al., 1994). The sequence divergence of KLP67A and KIF1B may be due to several different factors. At the ultrastructural level, mitochondria exhibit striking morphological differences depending on the cell type examined. It is therefore possible that the receptors for motor binding may be different on differentiated versus embryonic mitochondria. The divergence may also have resulted from the requirement for KIF1B to move rapidly down the relatively long distance of the axon or myofibril compared to the short distance of the astral MTs traveled by KLP67A. Indeed, although velocity in vitro may not be a reliable indicator of velocity in vivo, the measured velocity of KLP67A driven movement in vitro (∼0.05 μm/s) is 10-fold slower than that reported for KIF1B and for mitochondrial movement within the axon (Nangaku et al., 1994).
The expression analysis of KLP67A indicates that while KLP67A protein and mRNA levels appear to be equivalent throughout the blastoderm embryo, the KLP67A mRNA exhibits extraordinarily tight regulation later in embryogenesis. Specifically, KLP67A transcripts are present at high levels in the syncytial embryo, and they appear to be degraded at the time of cellularization before the onset of zygotic transcription. After cellularization of the early Drosophila blastoderm embryo, the differential regulation of the start of the 14th mitosis gives rise to what are called mitotic domains (Foe, 1989). KLP67A mRNA is exclusively expressed in these mitotic domains and may therefore be regulated during the cell cycle. This pattern of transcript distribution is reminiscent of RNAs encoding proteins known to be required for progression through mitosis in Drosophila, such as cyclin A, cyclin B (Lehner and O'Farrell, 1990; Dalby and Glover, 1993), cdc2 (Jimenez et al., 1990; Lehner and O'Farrell, 1990), and polo (Llamazares et al., 1991). While the levels of these other RNA species are elevated in regions of active mitosis, they do not appear to be as tightly coupled as KLP67A to the pattern of mitotic domains in either the 14th or later cycles. In fact, the pattern of KLP67A transcript distribution in these cycles is more similar to that of another kinesin-like protein, KLP61F, which is required for correct separation of centrosomes in mitosis (Heck et al., 1993). However, the levels of KLP61F RNA also appear less tightly coupled to the pattern of mitotic domains than those of KLP67A. Of the RNAs encoding proteins known to be required for correct progression through mitosis in Drosophila, only the string RNA (encoding a tyrosine phosphatase) (Edgar and O'Farrell, 1989) appears to show coupling to the spatial and temporal pattern of mitosis that is as close as that of KLP67A transcripts. Thus, the KLP67A expression pattern may be a strong indication of function during mitosis. The patterns of transcript accumulation observed could be determined by both cell cycle–regulated transcription and message stability. There is, however, an apparent contradiction between our failure to observe enrichment of KLP67A protein in these mitotic domains of mRNA expression. It is possible that our immunofluorescent methods are simply not reliable enough to preserve an actual difference in the amount of KLP67A between interphase and mitotic cells, i.e., there may be differences in the amounts of KLP67A extracted from these two cell types during fixation, or the antibody staining may not reflect the amounts of protein depending on organization of the protein in the cells. Alternatively, there may be near equivalent amounts of KLP67A in mitotic and interphase cells at this stage of development; this apparent equivalence could result from persistence of the KLP67A protein from earlier stages when the mRNA is more abundant. In this case, the mitotic cycling of KLP67A mRNA may be used to ensure that there are sufficient amounts of KLP67A for the dividing cell. In this regard, at least two other genes have been reported to have a pattern of cycling mRNA during the cell cycle but constant protein expression similar to that of KLP67A. The yeast SPC110/NUF1 transcript is cell cycle–regulated, yet the corresponding protein levels are not (Kilmartin et al., 1993). Also in yeast, the CDC9 (DNA ligase) transcript is tightly regulated, while the stability of the protein allows it to persist through the cell cycle (Johnson et al., 1986). In each of these cases, cell cycle control may ensure that the gene product will be present in sufficient quantities during mitosis, whereas protein stability may result in its persistence through interphase.
A requirement for KLP67A in mitotic cells is suggested by the immunolocalization results. KLP67A is found associated with mitochondria near to or on the astral fibers throughout mitosis in the early Drosophila embryo. A similar localization of mitochondria to the astral fibers in mouse oocytes has been demonstrated, but the reason for this localization is unknown (Van Blerkom, 1991). One possibility is that the mitochondria are required for spindle function. The propinquity of the mitochondria to the spindle poles may allow a locally high concentration of ATP or calcium for anaphase or other movements. It is also conceivable that the mitochondria have a structural role as a support for the astral pulling force during spindle assembly and anaphase B (Waters et al., 1993). Alternatively, the spindle may be required to segregate mitochondria during division. In the case of the large acellular Drosophila embryo, a spindle attachment could ensure that the mitochondria will be localized to the dividing cells rather than to the yolk region or areas that are less metabolically active. The genetic analysis of KLP67A should allow us to distinguish between these various possibilities.
Acknowledgments
This work was supported by National Institutes of Health grant GM35252 to L.S.B. Goldstein. B. Dalby was supported by a European Molecular Biology Organization fellowship. S.J. Doxsey is supported through the American Heart Association. L.S.B. Goldstein is an investigator of the Howard Hughes Medical Institute.
Abbreviations used in this paper
- CNS
central nervous system
- DAPI
4,6-diamidino-2-phenylindole
- GST
glutathione S-transferase
- KIF1B
kinesin superfamily protein 1B
- KHC
kinesin heavy chain
- KLP
kinesinlike protein
- IPTG
isopropylthiogalactoside
- MT
microtubule
Footnotes
A.J. Pereira would like to express her gratitude to William S. Gelbart of Harvard University (Cambridge, MA) in whose lab she did a large part of the work reported. She would like to thank Nicholas Lim (Harvard University) for technical assistance, Aruna Purohit (University of Massachusetts, Worcester), Jason Dictenberg (University of Massachusetts, Worcester), Tim Karr (University of Chicago), and Paul Dobner (University of Massachusetts, Worcester) for scientific discussion and advice, and also Kristin White (Brandeis University, Boston, MA) for the use of her confocal microscope and Fuji printer. A.J. Pereira would also like to thank Rafael Garesse (Universidad Autonoma de Madrid, Spain) for an aliquot of anti-Drosophila mitochondrial ATP synthase antisera.
Address all correspondence to L.S.B. Goldstein, HHMI/CMMW RM. 334, University of California at San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0683. Tel.: (619) 534-9702. Fax: (619) 534-9701.
References
- Brady ST, Lasek RJ, Allen RD. Fast axonal transport in extruded axoplasm from squid giant axon. Science (Wash DC) 1982;218:1129–1131. doi: 10.1126/science.6183745. [DOI] [PubMed] [Google Scholar]
- Brown NH, Kafatos F. Functional cDNA libraries from Drosophilaembryos. J Mol Biol. 1988;203:425–437. doi: 10.1016/0022-2836(88)90010-1. [DOI] [PubMed] [Google Scholar]
- Dalby B, Glover DM. Discrete sequence elements control posterior pole accumulation and translational repression of maternal cyclin B RNA in Drosophila. . EMBO (Eur Mol Biol Organ) J. 1993;12:1219–1227. doi: 10.1002/j.1460-2075.1993.tb05763.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey SJ, Stein P, Evans L, Calarco PD, Kirschner M. Pericentrin, a highly conserved centrosome protein involved in microtubule organization. Cell. 1994;76:639–650. doi: 10.1016/0092-8674(94)90504-5. [DOI] [PubMed] [Google Scholar]
- Edgar BA, O'Farrell PH. Genetic control of cell division patterns in the Drosophilaembryo. Cell. 1989;57:177–187. doi: 10.1016/0092-8674(89)90183-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1984;137:266–267. doi: 10.1016/0003-2697(84)90381-6. [DOI] [PubMed] [Google Scholar]
- Foe VA. Mitotic domains reveal early commitment of cells in Drosophilaembryos. Development (Camb) 1989;107:1–22. [PubMed] [Google Scholar]
- Garnier J, Osguthorpe DJ, Robson B. Analysis of the accuracy and implications of simple methods for predicting the secondary structure of globular proteins. J Mol Biol. 1978;120:97–120. doi: 10.1016/0022-2836(78)90297-8. [DOI] [PubMed] [Google Scholar]
- Goldstein LSB. With apologies to Scheherazade: tails of 1001 kinesin motors. Annu Rev Genet. 1993;27:319–351. doi: 10.1146/annurev.ge.27.120193.001535. [DOI] [PubMed] [Google Scholar]
- Guan KL, Dixon JE. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- Heck MMS, Pereira A, Pesavento P, Yannoni Y, Spradling AC, Goldstein LSB. The kinesin-like protein KLP61F is essential for mitosis in Drosophila. . J Cell Biol. 1993;123:665–679. doi: 10.1083/jcb.123.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heggeness MH, Simon M, Singer SJ. Association of mitochondria with microtubules in cultured cells. Proc Natl Acad Sci USA. 1978;75:3863–3866. doi: 10.1073/pnas.75.8.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman, A.A. 1991. Preparation of marked microtubules for the assay of the polarity of microtubule-based motors by fluorescence. J. Cell Sci. 14: (Suppl.)125–127. [DOI] [PubMed]
- Jimenez J, Alphey L, Nurse P, Glover DM. Complementation of fission yeasts cdc2ts and cdc25ts mutants identifies two cell cycle genes from Drosophila: a cdc2 homologue and string. EMBO (Eur Mol Biol Organ) J. 1990;9:3565–3571. doi: 10.1002/j.1460-2075.1990.tb07567.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AL, Barker DG, Johnston LH. Induction of yeast DNA ligase genes in exponential and stationary phase cultures in response to DNA damaging agents. Curr Genet. 1986;11:107–112. doi: 10.1007/BF00378201. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT. A spacer protein in the Saccharomyces cerevisiaespindle pole body whose transcript is cell cycle-regulated. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov SA, Vaisberg YA, Rothwell SW, Murphy DB, Gelfand VI. Isolation of a 45-KDA fragment from the kinesin heavy chain with enhanced ATPase and microtubule-binding activities. J Biol Chem. 1989;264:589–595. [PubMed] [Google Scholar]
- Lehner C, O'Farrell PH. The role of Drosophilacyclins A and B in mitotic control. Cell. 1990;61:535–547. doi: 10.1016/0092-8674(90)90535-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamazares S, Moreira MA, Tavares A, Girdham C, Gonzales C, Karess RE, Glover DM, Sunkel SE. polo encodes a protein kinase homologue required for mitosis in Drosophila. . Genes Dev. 1991;5:2153–2164. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- Lupas A, Dyke MV, Stock J. Predicting coiled coils from protein sequences. Science (Wash DC) 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mahowald AP. Polar granules of Drosophila. . J Exp Zool. 1971;151:329–343. doi: 10.1002/jez.1401760308. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Sedat JT. Localization of antigenic determinants in whole Drosophilaembryos. Dev Biol. 1983;99:261–264. doi: 10.1016/0012-1606(83)90275-0. [DOI] [PubMed] [Google Scholar]
- Nangaku M, Sato-Yoshitake R, Okada Y, Noda Y, Takemura R, Yamazaki H, Hirokawa N. KIF1B, a novel microtubule plus end– directed monomeric motor protein for transport of mitochondria. Cell. 1994;79:1209–1220. doi: 10.1016/0092-8674(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Olmsted JB. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J Biol Chem. 1981;256:11955–11957. [PubMed] [Google Scholar]
- O'Neill SL, Karr TL. Bi-directional incompatibility between conspecific populations of Drosophila simulans. . Nature (Lond) 1990;348:178–180. doi: 10.1038/348178a0. [DOI] [PubMed] [Google Scholar]
- Pena P, Garesse R. The β subunit of the ATP synthase: cDNA cloning, amino acid analysis and identification of the protein in adult flies. Biochem Biophys Res Commun. 1993;195:785–791. doi: 10.1006/bbrc.1993.2114. [DOI] [PubMed] [Google Scholar]
- Pereira, A.J., and L.S.B. Goldstein. 1994. The kinesin superfamily. In Microtubules. J.S. Hyams and C.W. Lloyd, editors. Wiley-Liss, Inc., New York. 269–284.
- Pereira AJ, Doshen J, Tanaka E, Goldstein LSB. Genetic analysis of a Drosophilamicrotubule-associated protein. J Cell Biol. 1992;116:377–383. doi: 10.1083/jcb.116.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G, Fuller MT. Monoclonal antibodies specific for an acetylated form of α-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J Cell Biol. 1985;101:2085–2094. doi: 10.1083/jcb.101.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raine CS, Ghetti B, Shelanski ML. On the association between microtubules and mitochondria within axons. Brain Res. 1971;34:389–393. doi: 10.1016/0006-8993(71)90293-9. [DOI] [PubMed] [Google Scholar]
- Ready DF, Hanson TE, Benzer S. Development of the Drosophilaretina, a neurocrystalline lattice. Dev Biol. 1976;53:217–240. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Mitotic spindle organization by a plus-end-directed microtubule motor. Nature (Lond) 1992;359:540–543. doi: 10.1038/359540a0. [DOI] [PubMed] [Google Scholar]
- Saxton WM, Porter ME, Cohn SA, Scholey JM, Raff EC, Macintosh JR. Drosophilakinesin: characterization of microtubule motility and ATPase. Proc Natl Acad Sci USA. 1988;85:1109–1113. doi: 10.1073/pnas.85.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RJ, Pesavento PA, Woerpel DN, Goldstein LSB. Identification and partial characterization of six members of the kinesin superfamily in Drosophila. . Proc Natl Acad Sci USA. 1991;88:8470–8474. doi: 10.1073/pnas.88.19.8470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart RJ, Thaler JP, Goldstein LSB. Direction of microtubule movement is an intrinsic property of the motor domains of kinesin heavy chain and Drosophilancd protein. Proc Natl Acad Sci USA. 1993;90:5209–5213. doi: 10.1073/pnas.90.11.5209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific mRNAs in the Drosophilaembryo reveals translational control of the segmentation gene hunchback. Chromosoma (Berl) 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE. Behavior of structurally divergent α-tubulin isotypes during Drosophilaembryogenesis: evidence for post-translational regulation of isotype abundance. Dev Biol. 1992;154:205–217. doi: 10.1016/0012-1606(92)90060-t. [DOI] [PubMed] [Google Scholar]
- Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. . Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- Vale RD, Goldstein LSB. One motor, many tails, an expanding repertoire of force-generating enzymes. Cell. 1990;60:883–885. doi: 10.1016/0092-8674(90)90334-b. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J. Microtubule mediation of cytoplasmic and nuclear maturation during the early stages of resumed meiosis in cultured mouse oocytes. Proc Natl Acad Sci USA. 1991;88:5031–5035. doi: 10.1073/pnas.88.11.5031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warn RM, Warn A. Microtubule arrays present during the syncytial and cellular blastoderm stages of the early Drosophilaembryo. Exp Cell Res. 1986;163:201–210. doi: 10.1016/0014-4827(86)90573-2. [DOI] [PubMed] [Google Scholar]
- Waters JC, Cole RW, Rieder CL. The force-producing mechanism for centrosome separation during spindle formation in vertebrates is intrinsic to each aster. J Cell Biol. 1993;122:361–372. doi: 10.1083/jcb.122.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Kankel DR. Patterns of cell division and cell movement in the formation of the imaginal nervous system in Drosophila melanogaster. . Dev Biol. 1978;65:296–321. doi: 10.1016/0012-1606(78)90029-5. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophilacompound eye: the morphogenetic furrow and the second mitotic wave. Development (Camb) 1991;113:841–850. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Yaffe MP, Harata D, Verde F, Edison M, Toda T, Nurse P. Microtubules mediate mitochondrial distribution in fission yeast. Proc Natl Acad Sci USA. 1996;93:11664–11668. doi: 10.1073/pnas.93.21.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Knowles BA, Goldstein LSB, Hawley RS. A kinesinlike protein required for distributive chromosome segregation in Drosophila. . Cell. 1990;62:1053–1062. doi: 10.1016/0092-8674(90)90383-p. [DOI] [PubMed] [Google Scholar]