Abstract
The structure of the Shaker potassium channel has been modeled as passing through the cellular membrane eight times with both the NH2 and COOH termini on the cytoplasmic side (Durrell, S.R., and H.R. Guy. 1992. Biophys. J. 62:238–250). To test the validity of this model, we have inserted an epitope consisting of eight hydrophilic amino acids (DYKDDDDK) in predicted extracellular and intracellular loops throughout the channel. The channels containing the synthetic epitope were expressed in Xenopus oocytes, and function was examined by two-electrode voltage clamping. All of the mutants containing insertions in putative extracellular regions and the NH2 and COOH termini expressed functional channels, and most of their electrophysiological properties were similar to those of the wild-type channel. Immunofluorescent staining with a monoclonal antibody against the epitope was used to determine the membrane localization of the insert in the channels. The data confirm and constrain the model for the transmembrane topology of the voltage-gated potassium channel.
Voltage-gated cation channels, consisting of potassium, sodium, and calcium channels, play a critical role in the conduction properties of electrically excitable cells. All of these channels share a number of functional properties, including voltage-dependent gating and selectivity for specific cations, and they are all predicted to have a similar structural topology (2). The most accepted model for the structure of these channels is based primarily on interpretations of hydropathy data derived from the amino acid sequence of the Shaker potassium channel (4), which is a 74-kD membrane protein that assembles as a homotetramer to form a functional channel (13). The model predicts a transmembrane protein that consists of six membrane-spanning regions termed S1 through S6, with both the NH2 and COOH termini on the intracellular face of the membrane (4). The majority of these regions are thought to form hydrophobic α-helical segments that cross the width of the membrane. The region between the S5 and S6 segments forms a portion of the channel pore (7, 17, 27) and so must cross at least a portion of the membrane. The fourth membrane-spanning region (S4) has been modeled to consist of two short segments that form amphipathic helices whose combined length crosses the membrane. The S6 segment has been modeled as a hydrophobic α-helix with a break or bend in the middle due to a pair of proline residues.
There is very little structural data concerning the topology of the Shaker potassium channel, but results from functional studies are consistent with the model. The NH2 terminus must be intracellular, since the first 20 amino acids function as an inactivation particle to occlude the inner mouth of the channel pore (8). Deletion of the first 20 amino acids eliminates fast inactivation, and application of a synthetic peptide with the same sequence to the cytoplasmic side of the cell membrane restores inactivation (8, 28). The loop between S4 and S5 is thought to reside on the inside membrane of the cell, because mutations in this region affect inactivation, conductivity, and toxin blocking of the inner mouth of the pore (10, 26). The region between S5 and S6 has been demonstrated to comprise the pore of the channel, because mutations in this region affect both selectivity and conductance of the channel (7, 18, 27). In addition, binding sites for external charybdotoxin and tetraethylammonium ion have been identified at amino acid positions D431 and T449, with a binding site for internal tetraethylammonium ion at position T441 in between the external sites (11, 17, 18). These data demonstrate that the region between the S5 and S6 segments dips in and out of the membrane to form part of the pore of the channel. The COOH terminus is likely to be intracellular, because mutation of a consensus protein kinase A site in that region prevents the effects of intracellular phosphatase on channel inactivation (3).
The only biochemical evidence concerning the topology of the Shaker channel is based on the characterization of glycosylation in Sf9 cells transiently expressing recombinant Shaker channels (23). A triplet of bands on a Western blot was reduced to a single band after treatment of the cells with tunicamycin or the lysate with N-glycanase, indicating that the higher molecular weight species represented N-linked glycosylated forms of the protein. Mutation of two asparagine residues in the S1–S2 loop eliminated all but the lowest molecular weight band that corresponds to the immature protein, indicating that the S1–S2 region is glycosylated in Sf9 cells and is therefore extracellular.
The studies described have focused on small regions of the potassium channel and are consistent with the predicted topology. However, variations of the model have been proposed (6, 12, 24), and definitive localization studies have not been reported. It is essential to have a structural model that is supported by definitive localization studies to correlate functional properties with specific amino acid sequences, which is a major goal of ion channel research. To obtain more definitive data concerning the topology of the potassium channel, we have inserted at positions throughout the channel an epitope that can be localized with an antibody. The functional properties of channels containing the epitope insertions were shown to be similar to those of the wild-type channel. The epitopes were then localized to either the internal or external side of the membrane, thereby establishing the transmembrane topology of the channel in these regions.
Materials and Methods
Construction of the Epitope Insertion Mutations
The Shaker H4 potassium channel–coding region from pH4U5 was transferred to pBSTA (5), a plasmid containing the 5′ and 3′ nontranslated sequences from the Xenopus β-globin transcript, to increase the level of expression to enable detection by immunofluorescent staining. An insert containing the Shaker coding region was cut from pH4U5 in a 2.2-kb EagI/EcoRI fragment (bp 670–2,889). To prepare the fragment for insertion into pBSTA, the ends of the insert were made blunt using T4 DNA polymerase (0.2 U T4 DNA polymerase, 50 mM Tris-HCl, pH 8.0, 5.0 mM MgCl2, 5.0 mM DTT, 100 μM dNTPs) at 11°C for 20 min and 75°C for 10 min. BglII linkers (5′GAAGATCTTC 3′), which were previously phosphorylated with T4 polynucleotide kinase and annealed, were attached to the fragment using T4 DNA ligase. The pBSTA vector was linearized with BglII, treated with calf intestinal phosphatase, and ligated to the 2.2-kb Shaker H4 insert. The new pBSTA vector containing the Shaker H4 coding region was named pKH4T.
Phagemid DNA was prepared from pKH4T grown in CJ236 cells, a dut − ung − strain (Invitrogen Corp., San Diego, CA). A single, transformed colony was used to inoculate 5 ml of 2× YT containing 30 μg/ml chloramphenicol and 50 μg/ml ampicillin. The culture was grown at 37°C for 6 h, and then 300 μl was used to inoculate 30 ml 2× YT containing 250 ng/ml uridine. The culture was grown to early log phase (A600 = 0.05, ∼1 × 109 cells) within 2 h at 37°C, and then infected with 80 μl VCS-M13 helper phage (Stratagene, La Jolla, CA) with a multiplicity of infection of 10:1 and grown at 37°C overnight. The cells were removed by centrifugation at 4K for 20 min, and the supernatant was transferred to a new tube. Phagemids were precipitated by adding 0.25 vol 20% PEG (polyethylene glycol; 8,000) in 3.5 M NH4OAc (pH 7.5), incubating on ice for 30 min, and spinning at 17K for 15 min at 4°C. The pellet was resuspended in 0.6 ml TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and extracted six times with phenol/ chloroform/isoamyl alcohol; and the single-stranded, uracil-containing pKH4T DNA template was ethanol precipitated and resuspended in 50 μl TE.
All of the oligonucleotides for mutagenesis contained the same 24 nucleotides encoding Flag (GACTATAAAGACGATGACGACAAA) flanked by 15–20 nucleotides of potassium channel sequence corresponding to sequences extending 5′ and 3′ from the insertion site within the potassium channel coding region. The oligonucleotides were phosphorylated with T4 polynucleotide kinase, mixed with 1 μg pKH4T single-stranded DNA template in a 10:1 molar ratio in SSC (150 mM NaCl, 15 mM NaCitrate, pH 6.8), heated to 70°C for 5 min, and cooled from 70° to 30°C over 30 min. The complementary strand to the template was synthesized in vitro using T7 DNA polymerase and T4 DNA ligase (2.5 U T7 DNA polymerase, 4.0 U T4 DNA ligase, 40 mM Tris-HCl, pH 7.5, 10 mM MgCl2·6 H2O, 50 mM NaCl, 50 μg/ml BSA, 0.6 mM dNTPs, 1.0 mM ATP, 5.0 mM DTT) for 5 min on ice, 5 min at room temperature, and 2 h at 37°C. The heteroduplex DNA was precipitated with ethanol, resuspended in 20 μl H2O, and transformed into DH5α cells to select against the uracil-containing template strand. Individual colonies were picked and grown in small 5 ml cultures from which DNA was purified using the Wizard Miniprep DNA Purification System (Promega Biotech, Madison, WI). Isolates containing the FLAG epitope were identified by DNA sequencing.
Expression in Xenopus Oocytes and Electrophysiological Recording
Plasmid DNA was linearized with SstII, which cuts at the end of the poly A tail in the 3′ nontranslated sequence from the β-globin gene, and RNA was transcribed in vitro using the T7 mMESSAGE mMACHINE kit (Ambion Inc., Austin, TX). The kit consistently produced 18–25 μg of RNA, which was resuspended in 20 μl of 10 mM Tris-HCl, pH 6.5. Stage V oocytes were removed from adult female Xenopus laevis frogs, prepared as previously described (5), and incubated in ND-96 media (96 mM NaCl, 2.0 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, and 5.0 mM Hepes, pH 7.5) supplemented with 0.1 mg/ml gentamicin, 0.55 mg/ml pyruvate, and 0.5 mM theophylline. 50 nl of undiluted RNA (∼50 ng) was injected into each oocyte for immunofluorescent staining, and ∼50 pg was injected for electrophysiological analysis. After 24–48 h of incubation at 20°C in ND96, potassium currents were recorded at room temperature using a two-electrode voltage clamp (22). The recording solution was ND96 without supplements, and the holding potential was −80 mV. All currents were recorded using P/4 subtraction to eliminate capacitive and linear leak currents (1). The level of sensitivity below which functional expression was considered negative was 50 nA, during a depolarization to +30 mV after injection of 50 ng RNA. This quantity of wild-type potassium channel RNA resulted in currents >100 μA at −50 mV.
Currents were converted to conductance values using the equation: G = I/(V−VR), where G = conductance, I = current, V = voltage, and VR = reversal potential, which was assumed to be −90 mV in all cases. Conductance values were normalized to the maximum conductance and fit with a two state Boltzmann equation: G = 1/[1 + exp (− 0.03937*z*(V − V1/2))], where z = slope factor and V1/2 = voltage for half-maximal depolarization. Time constants for inactivation (τ) were determined by fitting the current traces with a single exponential function: (A = exp[− (t − k)/τ] + C), where A = current, t = time, k = time shift, and C = steady-state asymptote. Recovery from inactivation was determined from a holding potential of −80 mV by a 50 mV inactivating pulse for 50 ms, a variable recovery interval from 10 to 250 ms in 10 ms steps, and a test pulse to +50 mV for 40 ms to assess recovery. The maximum current during each test pulse was normalized to the maximum current during the inactivating pulse and plotted against the recovery interval. Recovery data were fit with a double exponential equation: A = 1 − [a*exp(− t/τ 1) + b*exp(− t/τ2)], where A = current, a and b represent the proportion of recovery with time constants τ1 and τ2, respectively, and t is the recovery interval.
Immunofluorescent Staining
Oocytes expressing >10 μA of potassium current at −20 mV after 24 h were used for immunofluorescent staining. It was important to remove all follicle cells from the oocytes before staining to reduce background. For intracellular staining, oocytes were injected with 50 nl of 200 μg/ml M2 anti-Flag monoclonal antibody (Eastman Kodak Co., Rochester, NY) and incubated in ND96 at 20°C overnight. For extracellular staining, oocytes were incubated overnight at 20°C in 1 ml M2 anti-Flag monoclonal antibody diluted 1:500 in ND96. After incubation with the antibody, both sets of oocytes were rinsed in ND96, fixed in 3.7% formaldehyde in ND96 for 15 min to prevent unbound antibody from crossing the membrane and binding to extracellular epitopes, fixed in ice-cold 100% methanol for 15 min to permeabilize the membrane, and rinsed several times with ND96.
For fluorescent labeling, the oocytes were first incubated overnight at room temperature in biotinylated sheep anti–mouse antibody diluted 1: 1,000 in labeling solution (ND96, 10% sheep serum, 0.2% sodium azide). The oocytes were then stained with three cycles of rhodamine–avidin DCS (1:1,000 in labeling solution, Vector Laboratories Inc., Burlingame, CA) overnight followed by biotinylated anti-avidin D (1:1,000 in labeling solution, Vector Laboratories Inc.) overnight with several rinses in ND96 after each step. The labeled oocytes were protected from light to prevent photobleaching. The stained oocytes were embedded in Tissue-Tek OCT compound (Baxter Scientific Products, Salt Lake City, UT) and frozen in blocks at −20°C. A Reichert Jung Cryocut 1800 cryostat was used to cut 14 μm sections from the OCT blocks, which were mounted on slides. The slides were air dried overnight at room temperature, fixed with a drop of acetone, and then rehydrated for 10 min in PBS. Glass coverslips were mounted with a drop of Vectashield (Vector Laboratories Inc.) and sealed around the edges with nail polish. The slides were stored protected from light at 4°C. The samples were viewed and photographed on a Nikon OPTIPHOT microscope with epifluorescent illumination at 160× magnification using 30 s exposures and Kodak 35 mm Tmax 400 film. Phase contrast images were exposed for 10 s.
Results
Insertion of the Flag Epitope in Many Regions Does Not Disrupt Channel Function
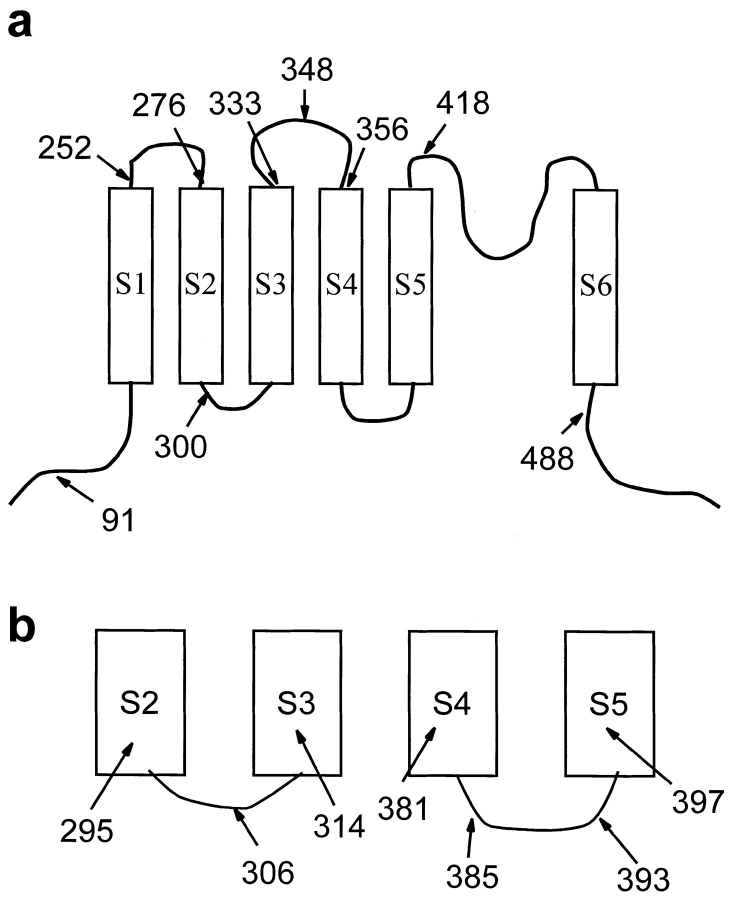
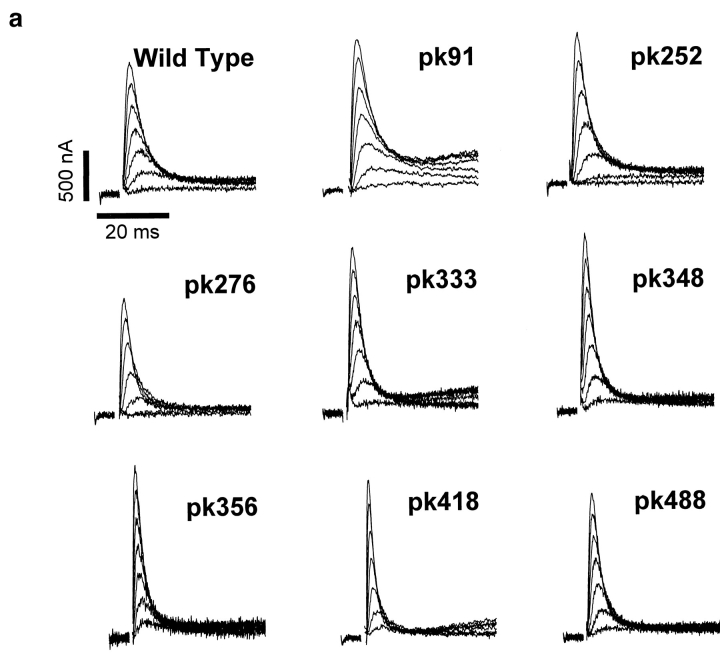
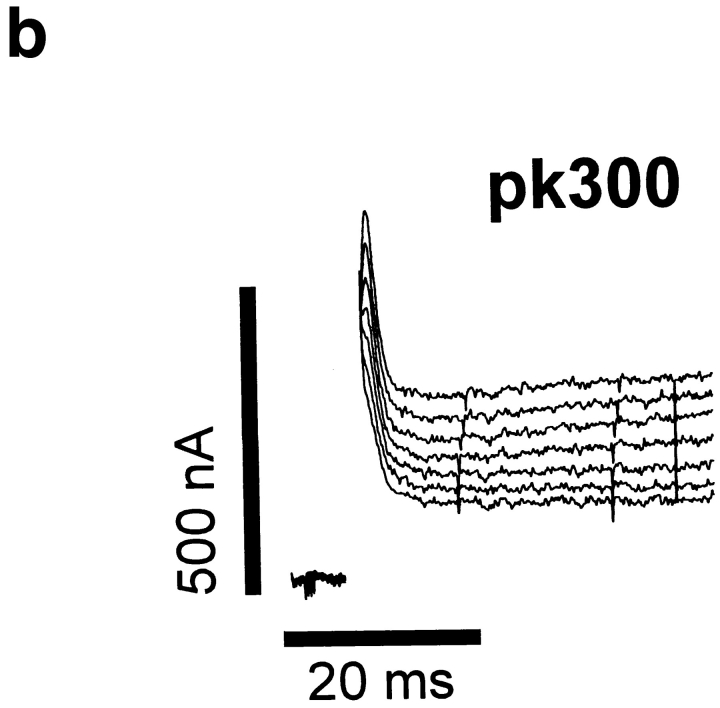
To experimentally test the proposed transmembrane topology of the Shaker potassium channel, the hydrophilic epitope termed Flag (DYKDDDDK) was inserted into putative extracellular and intracellular regions throughout the channel (Fig. 1). The specific sites for insertion were generally chosen to be near clusters of charged amino acids in an attempt to avoid transmembrane regions. This strategy was based on the assumption that insertion of the epitope's charged amino acids into a transmembrane-spanning region would severely alter the structure of the protein, resulting in either a nonfunctional channel or profound changes in the channel's electrophysiological properties. In contrast, insertions that resulted in functional channels would not have significantly altered the normal topology of the channel. To determine the effects of each insertion on channel function, each channel was expressed in Xenopus oocytes, and the electrophysiological properties were characterized by two-electrode voltage clamping. Insertions of Flag into all of the putative extracellular loops resulted in functional channels, as did insertions in either the NH2- or COOH-terminal regions (Figs. 1 a and 2). However, the two putative intracellular loops were very sensitive to insertions, and all but one of the insertions within these two regions resulted in the complete absence of functional potassium channels in oocytes (Fig. 1 b). One insertion in the S2–S3 loop (pk300) did result in a low level of potassium channel activity when expressed in oocytes (Fig. 2 b). However, the level of expression was only ∼200 nA at +30 mV so that we were unable to characterize this mutant in any detail.
Figure 1.
Schematic diagram of the Shaker potassium channel showing the sites at which Flag epitopes were inserted to result in functional (a) or nonfunctional (b) channels. The epitopes were placed after the amino acids indicated. The position for inserts close to the surface of the membrane was choosen to be next to clusters of charged residues to avoid disrupting transmembrane segments. (a) Functional insertions were determined by heterologous expression in Xenopus oocytes (Fig. 2). An insertion in the NH2-terminus (pk91) confirms the intracellular location of this end. The S1–S2 loop is defined by two functional insertions at 252 and 276. Some evidence for the S2–S3 loop is provided by a partially functional insertion at 300. The minimum length of the S3– S4 loop is established by three insertions at 333, 348, and 356. The NH2-terminal boundary of the p-region is determined by one insertion at 418. Finally, the COOH terminus must begin at least by position 488. (b) Numerous insertions into the S2–S3 and S4–S5 loops resulted in nonfunctional channels. No currents >50 nA were detected in oocytes injected with 50 ng of RNA for these mutants, whereas oocytes injected with 50 ng of wild-type RNA produced currents greater than 100 μA at −50 mV.
Figure 2.
Current recordings from oocytes expressing the Flag insertion mutants. (a) 200 pg of RNA encoding the wild-type Shaker and insertion mutants were injected into oocytes, which were incubated at 20°C for 24–48 h before recording currents using a two-electrode voltage clamp, as described in Materials and Methods. The currents were elicited by depolarizations from a holding potential of −80 mV to between −30 mV and +30 mV in 10 mV increments. The slowly activating outward currents for pk91, pk333, and pk418 result from calcium activated chloride channels that are endogenously present in oocytes from some frogs. (b) 50 ng of RNA encoding insertion mutant pk300 was injected into oocytes. Incubation and recording conditions were as described in a.
Functional Analysis of Potassium Channels Containing the Flag Epitope
It is likely that the eight insertion mutations that resulted in functional channels had not severely altered the transmembrane topology of the channel protein. To test this hypothesis, the electrophysiological properties of the channels were examined in detail. The properties that were examined include voltage dependence of conductance, kinetics of inactivation, and recovery from inactivation. Selectivity did not appear to be altered for any of the mutant channels, based on the observation that the reversal potential was consistent for all of the functional channels (data not shown).
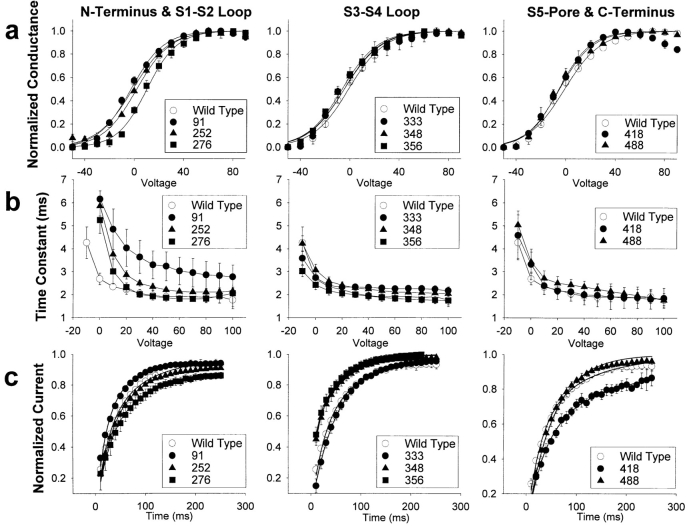
The voltage dependence of conductance, which provides an indication of the integrity of the voltage sensing mechanism of the channel, was characterized by two parameters. The slope of the curve (z) provides a measure of the voltage sensitivity of the channel, and the voltage for halfmaximal conductance (v1/2) provides a measure of the relative stability of the open and closed states of the channel. All of the mutants demonstrated conductance voltage relationships that were comparable to that of the wild-type channel (Fig. 3 a and Table I). Mutant pk276, with an insertion in the putative S1–S2 loop, is the most distinct with a positive shift in v1/2 of ∼10.5 mV, but this degree of shift is quite small compared to mutations in the voltage sensor of the channel (14, 15, 21). Therefore, none of the Flag insertions appeared to significantly disrupt the structure of the voltage sensor and other structures associated with activation of the channel.
Figure 3.
Electrophysiological properties of the Flag insertion mutants. (a) Conductance voltage relationships were determined by depolarizations from a holding potential of −80 mV to between −80 mV and +150 mV in 10 mV increments. Currents were converted to conductance, as described in Materials and Methods. Conductance values were normalized to the maximum conductance, and the average value is plotted against the depolarizing potential. The curve represents the best fit of a two state Boltzmann equation, as described in Materials and Methods. Symbols represent the means, and error bars indicate the standard deviations for data from four to eight oocytes. (b) Time constants for inactivation were determined by fitting the current traces obtained as in a with a single exponential function, as described in Materials and Methods. The average time constants are plotted against the depolarizing potential. Symbols represent the means, and error bars indicate the standard deviations for data from four to six oocytes. (c) Recovery from inactivation was determined by a 50 mV inactivating pulse for 50 ms, a variable recovery interval from 10 to 250 ms, and a test pulse to +50 mV for 40 ms to assess recovery, as described in Materials and Methods. The maximum current during each test pulse was normalized to the maximum current during the inactivating pulse, and the average values are plotted against the recovery interval. The smooth curve represents a fit of the average data to a double exponential equation, as described in Materials and Methods. Symbols represent the means, and error bars indicate the standard deviations for data from four to eight oocytes.
Table I.
Voltage Dependence of Conductance
| Channel | Slope (z) | V1/2 | ||
|---|---|---|---|---|
| mV | ||||
| Wild type | 1.7 ± 0.1 | −1.1 ± 1.1 | ||
| pk91 | 1.9 ± 0.1 | −2.5 ± 1.8 | ||
| pk252 | 2.0 ± 0.1 | 6.2 ± 1.1 | ||
| pk276 | 2.1 ± 0.2 | 10.5 ± 2.6 | ||
| pk333 | 2.2 ± 0.1 | −6.9 ± 1.9 | ||
| pk348 | 1.7 ± 0.1 | −4.6 ± 2.5 | ||
| pk356 | 1.8 ± 0.1 | −6.4 ± 0.1 | ||
| pk418 | 1.8 ± 0.1 | 1.7 ± 1.7 | ||
| pk488 | 1.9 ± 0.1 | −5.2 ± 1.6 |
Normalized conductance data for individual recordings were fit with a two state Boltzmann equation as described in Materials and Methods. The values for the slope (z) and the half-maximal conductance (V1/2) were averaged, and standard deviations were calculated for data from four to eight oocytes for each channel.
Fast inactivation in the potassium channel is thought to involve a ball-and-chain type mechanism, in which the NH2 terminus functions to occlude the pore of the channel (8, 28). All of the mutant channels except pk91 appear to inactivate with kinetics comparable to those of the wildtype channel (Fig. 3 b). It is not surprising that pk91 would demonstrate slower inactivation, because insertions into the NH2-terminal region between amino acid 20 and the beginning of S1 have previously been shown to slow inactivation, presumably by lengthening the chain by which the inactivating ball is tethered to the channel. Therefore, insertion of 8 amino acids after amino acid 91 would be expected to slow inactivation. None of the other insertions markedly affected the kinetics of channel inactivation.
The kinetics of recovery from fast inactivation provides an indication of the rate at which the inactivating ball is released from occluding the channel pore. A number of the mutants demonstrated differences from the wild-type channel with respect to recovery from inactivation (Fig. 3 C). Two of the insertions in the S3–S4 loop accelerated recovery (pk348 and pk356), and the insertion in the S5–S6 region (pk418) slowed recovery. There is no precedent for the effects of the S3–S4 insertions, but the S5–S6 insertion is in a region that has previously been shown to be involved in slow inactivation of the potassium channel (9). It is apparent that Flag insertions in many regions of the channel can affect recovery from inactivation, but all of the changes were subtle, suggesting that there was not a dramatic change in the transmembrane topology of the channel.
Localization of the Flag Epitope by Immunofluorescent Staining
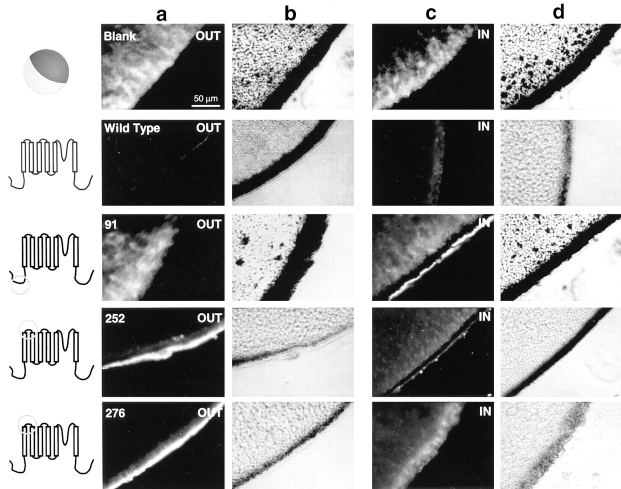
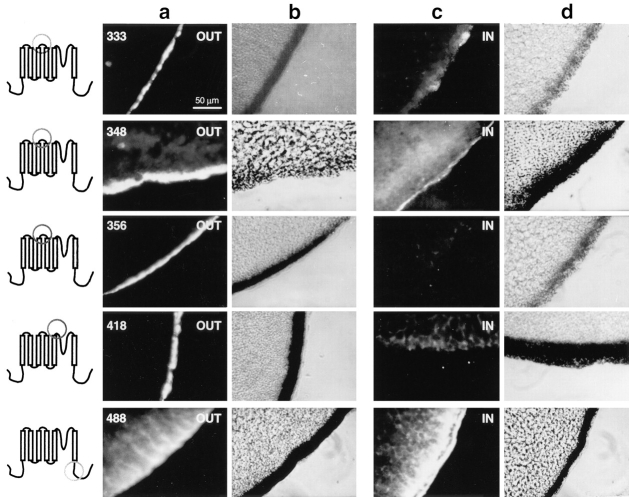
Since insertion of the Flag epitope into eight regions of the channel did not significantly alter channel function, it is likely that the channels containing those insertions had comparable topology to the wild-type channel. Therefore, localization of the Flag epitope should localize the region of the channel in which the epitope was inserted. To determine the transmembrane location of each epitope, separate pools of oocytes were examined for extracellular or intracellular immunofluorescent staining. Extracellular epitopes were targeted by incubating the intact oocytes in the M2 monoclonal anti-Flag antibody. Intracellular epitopes were targeted by injecting the M2 antibody into the intact oocytes. The results are shown in Figs. 4 and 5, with extracellular staining shown in a (fluorescent photomicrograph) and b (phase contrast photomicrograph) and intracellular staining shown in c (fluorescent photomicrograph) and d (phase contrast photomicrograph).
Figure 4.
Immunofluorescent staining of control, wild-type, pk91, pk252, and pk276. Schematic diagrams to the left of each row of microscopy images indicate the region in which each Flag insertion is located. Photomicrographs of oocytes labeled on the extracellular side of the membrane are shown in columns a (fluorescence) and b (phase contrast). Photomicrographs of oocytes labeled on the intracellular side of the membrane are shown in columns c (fluorescence) and d (phase contrast).
Figure 5.
Immunofluorescent staining of pk333, pk348, pk356, pk418, and pk488. Schematic diagrams to the left of each row of microscopy images indicate the region in which each Flag insertion is located. Photomicrographs of oocytes labeled on the extracellular side of the membrane are shown in columns a (fluorescence) and b (phase contrast). Photomicrographs of oocytes labeled on the intracellular side of the membrane are shown in columns c (fluorescence) and d (phase contrast).
The first Flag insertion that was examined was pk91, which contains a Flag epitope in the NH2-terminal region. The intracellular stain of pk91 showed a concentrated band of fluorescence at the membrane, indicating that the M2 antibody localized specifically to the membrane when the epitope was present on the intracellular face of the membrane (Fig. 4). This staining was specific, in that only diffuse background staining was observed when pk91 was incubated with extracellular M2 antibody. The background level of staining was comparable to that observed in uninjected oocytes (Blank) and oocytes injected with RNA encoding the wild-type potassium channel that does not contain a Flag epitope (Wild Type). The level of background staining varied among oocytes from different frogs, as can be seen in the difference in extracellular staining for the Blank versus Wild Type. To control for this variability, each experiment included negative controls consisting of blank and wild-type injected oocytes and positive controls consisting of pk91 (internal) and pk348 (external) injected oocytes. The internal staining of oocytes injected with pk91 demonstrates that the NH2 terminus is intracellular, which is consistent with previous studies indicating that it functions as an inactivating particle.
The two Flag insertions within the S1–S2 loop (pk252 and pk276) both show positive extracellular staining and negative intracellular staining, demonstrating that the S1– S2 loop is on the outer surface of the membrane and consists of at least 24 amino acids (Fig. 4). The Flag insertions within the S3–S4 loop (pk333, pk348, and pk356) all show positive extracellular staining and negative intracellular staining (Fig. 5), indicating that this 23-amino acid stretch is on the outer surface of the membrane. The Flag insertion between S5 and the pore region (pk418) similarly shows positive extracellular staining and negative intracellular staining (Fig. 5), indicating an extracellular location for this region of the channel. Finally, the Flag insertion in the COOH-terminus (pk488) shows negative extracellular staining and positive intracellular staining (Fig. 5), indicating that the COOH-terminus resides on the intracellular face of the membrane. The intensity of specific fluorescence varied with the different insertion mutations, and it did not correlate with either the level of functional expression or the localization (external or internal) of the epitope. For example, pk91 (internal) and pk348 (external) produced the strongest signals, even though some of the other insertion mutants expressed comparable current amplitudes. One possible explanation for the variation in signal intensity is that the epitope in both of these mutants is located farthest away from the surface of the membrane compared to the other insertions, which might indicate that accessibility of the epitope to the antibody decreases with proximity to the membrane. In summary, these results demonstrate that the S1–S2, S3–S4, and S5–pore regions are all located on the extracellular side, and that the NH2- and COOH-termini are both located on the inside of the cell.
Discussion
Functional Studies of the Epitope Insertion Mutants
By inserting a synthetic epitope throughout the Shaker potassium channel and determining the transmembrane localization of that epitope, we have confirmed and constrained the structural model of the channel. The epitope that we inserted (Flag) is extremely hydrophilic, which should have ensured that any insertion within the membrane would result in a nonfunctional channel. An insertion into a transmembrane segment would presumably force that segment to the surface, significantly altering the functional properties of the channel. It was therefore essential to demonstrate that the channels containing the epitope insertions had relatively normal function, to conclude that the results reflected the normal topology of the channel.
Some Flag insertions did alter the electrophysiological properties of the potassium channel. Insertion into the NH2-terminal region (pk91) resulted in slower inactivation (Fig. 3 B), but this result is expected based on the ball-andchain model for potassium channel inactivation (8). One insertion into the S1–S2 loop resulted in a positive shift in the voltage dependence of activation (Fig. 3 A and Table I), indicating that this mutation stabilizes the channel in the closed state relative to the open state. Negative charges in the S2 region have an important role in activation by interacting with positive charges in S4 (21) and contribute to the gating charge of the channel (25) so that a small distortion of the normal topology of that region could interfere with activation. However, it is unlikely that this insertion resulted in a major rearrangement of channel topology, because the mutant channel still functions in a manner very similar to that of the wild-type channel. In addition, another insertion into the S1–S2 loop (pk252) did not alter any of the functional properties that were examined.
Three Flag insertions, pk348 and pk356 in the S3–S4 loop and pk418 in the S5–pore region, altered the kinetics of recovery from inactivation. The results with pk418 are consistent with previous data, in that mutations in the S5– S6 region have been shown to affect the process of C-type potassium channel inactivation (16). Variations in the S3– S4 segment of L-type calcium channels have been shown to affect the kinetics of activation (19), but an effect on recovery from inactivation has not previously been observed. The observation that pk348 and pk356 accelerate recovery but that pk333 (which is closer to the S3 end of the region) does not, suggests that the effect may be mediated through the S4 region. The two insertions may make it more likely for the S4 helix to return to the closed state, displacing the inactivation particle. However, it is unlikely that these two insertions caused a major rearrangement of channel topology, since all other functional properties are similar to those of the wild-type channel.
Insertion of Flag epitopes into two regions of the channel (S2–S3 and S4–S5) eliminated channel activity in oocytes. There are a number of possible explanations for these results. First, these two loops may normally be located within the membrane, so that the hydrophilic insertions would have disrupted the normal topology. If this were the case, it is likely that one or more of the putative membrane-spanning segments (S2, S3, S4, and S5) would have to be located either inside or outside of the membrane. However, Flag insertions in all of those segments also resulted in nonfunctional channels (Fig. 1 B). A second possibility is that the S2–S3 and S4–S5 loops need to move into the membrane during channel activation, and a hydrophilic epitope prevents that movement. Finally, Flag insertions into these two small loops (modeled to contain 9 and 12 amino acids) might disrupt the structure of the channel, preventing either assembly or transport to the membrane in a functional conformation.
Topological Model of the Potassium Channel
The NH2-terminus of the potassium channel has been modeled to extend through A225 before the S1 segment crosses the membrane (4). The intracellular localization of this region was previously demonstrated by the fact that the first 20 amino acids function as an inactivating particle to block the channel from the inside of the cell (8, 28). Our data with pk91 are in complete agreement with the intracellular position of the NH2 terminus. Similarly, the COOHterminus is predicted to be located on the intracellular face of the membrane, starting at H486, and our data with pk488 are consistent with that prediction.
The S1–S2 loop has been modeled as an extracellular loop stretching 28 amino acids from L249 to D277 (4). The two asparagine residues (N259 and N263) within this loop have been shown to be glycosylated (23), consistent with its extracellular position. Our data substantiate that localization and define the extracellular region as extending from at least F252 to T276. The S3–S4 loop is modeled as a 30 amino acid region from A328 to L358. Our results demonstrate that this region is extracellular and define the loop as extending from at least E333 to M356. The loop may extend farther, since insertion of a Flag epitope after residue V363 does not disrupt channel function (data not shown).
The topology of the region between the S5 and S6 segments (S424 to K450) has been defined by binding sites for pore blocking toxins. The residues D431 and T449 have been shown to be binding sites for externally blocking charybdotoxin and tetraethylammonium ion (17, 18). Between these two sites, the residue T441 was shown to be the binding site for internally blocking tetraethylammonium ion (11). These results indicated that the S5–S6 stretch dipped in and out of the membrane forming the pore of the channel. Consistent with this model, mutations in this region affect ion conductance and selectivity, properties inherent in the channel pore (7, 18, 27). Our data verify the extracellular localization of the region between S5 and the pore, limiting the boundary of S5 to E418 rather than S424, as predicted in the model (4). We were unable to establish the extent of the external loop between the pore and S6, because an insert near the S6 segment after V454 was nonfunctional. The external staining of pk418 demonstrates that the exposed extracellular surface extends from at least E418 to the toxin-binding site at D431.
These results are consistent with the model proposed by Durell and Guy (4) for the structure of the Shaker potassium channel. The only regions for which we have not been able to obtain data are the putative intracellular S2– S3 and S4–S5 loops. As insertions in these two regions eliminate channel activity, alternative approaches will have to be used to demonstrate their positions. Both the NH2- and COOH-termini were shown to be intracellular with the expected boundaries. The S1–S2 and S5–pore loops were shown to be extracellular and consistent with the limits of the model. The S3–S4 loop was also extracellular and consistent with the model, although the extent of this loop was six amino acids more than predicted. In summary, these data provide an experimental basis for refining the model of the Shaker potassium channel.
Acknowledgments
We thank Dr. Rozanne Sandri-Goldin and Dr. Sandra Loughlin for the use of equipment, Dr. Roderick MacKinnon for the procedure to isolate phagemid DNA, and Drs. Raymond Smith, Michael Pugsley, Kris Kontis, Linda Hall, and Marianne Smith for helpful discussions during the course of this work.
This work was supported by grants from the National Institutes of Health (NS26729) and the National Science Foundation (IBN9221984). A.L. Goldin is an Established Investigator of the American Heart Association, and T.M. Shih was supported by a University of California Biotechnology Research and Education Program Training Grant.
Footnotes
Please address all correspondence to Alan Goldin, Department of Microbiology and Molecular Genetics, University of California, Irvine, CA 926974025. Tel.: (714) 824-5334; Fax: (714) 824-8598; E-mail: AGoldin@uci.edu
References
- 1.Bezanilla F, Armstrong CM. Inactivation of the sodium channel–sodium current experiments. J Gen Physiol. 1977;70:549–566. doi: 10.1085/jgp.70.5.549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Catterall WA. Structure and function of voltage-gated ion channels. Trends Neurosci. 1993;16:500–506. doi: 10.1016/0166-2236(93)90193-p. [DOI] [PubMed] [Google Scholar]
- 3.Drain P, Dubin AE, Aldrich RW. Regulation of Shaker K+channel inactivation gating by the cAMP-dependent protein kinase. Neuron. 1994;12:1097–1109. doi: 10.1016/0896-6273(94)90317-4. [DOI] [PubMed] [Google Scholar]
- 4.Durell SR, Guy HR. Atomic scale structure and functional models of voltage-gated potassium channels. Biophys J. 1992;62:238–250. doi: 10.1016/S0006-3495(92)81809-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldin AL. Expression of ion channels by injection of mRNA into Xenopusoocytes. Methods Cell Biol. 1991;36:487–509. doi: 10.1016/s0091-679x(08)60293-9. [DOI] [PubMed] [Google Scholar]
- 6.Greenblatt RE, Blatt Y, Montal M. The structure of the voltage-sensitive sodium channel inferences derived from computer-aided analysis of the Electrophorus electricuschannel primary structure. FEBS (Fed Eur Biochem Soc) Lett. 1985;193:125–134. doi: 10.1016/0014-5793(85)80136-8. [DOI] [PubMed] [Google Scholar]
- 7.Hartmann HA, Kirsch GE, Drewe JA, Taglialatela M, Joho RH, Brown AM. Exchange of conduction pathways between two related K+channels. Science (Wash DC) 1991;251:942–944. doi: 10.1126/science.2000495. [DOI] [PubMed] [Google Scholar]
- 8.Hoshi T, Zagotta WN, Aldrich RW. Biophysical and molecular mechanisms of Shakerpotassium channel inactivation. Science (Wash DC) 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 9.Hoshi T, Zagotta WN, Aldrich RW. Two types of inactivation in Shaker K+channels: effects of alterations in the carboxy-terminal region. Neuron. 1991;7:547–556. doi: 10.1016/0896-6273(91)90367-9. [DOI] [PubMed] [Google Scholar]
- 10.Isacoff EY, Jan YN, Jan LY. Putative receptor for the cytoplasmic inactivation gate in the Shaker K+channel. Nature (Lond) 1991;353:86–90. doi: 10.1038/353086a0. [DOI] [PubMed] [Google Scholar]
- 11.Kirsch GE, Taglialatela M, Brown AM. Internal and external TEA block in single cloned K+channels. Am J Physiol. 1991;261:C583–C590. doi: 10.1152/ajpcell.1991.261.4.C583. [DOI] [PubMed] [Google Scholar]
- 12.Kosower EM. A structural and dynamic molecular model for the sodium channel of Electrophorus electricus. . FEBS (Fed Eur Biochem Soc) Lett. 1985;182:234–242. doi: 10.1016/0014-5793(85)80306-9. [DOI] [PubMed] [Google Scholar]
- 13.Li M, Jan YN, Jan LY. Specification of subunit assembly by the hydrophilic amino-terminal domain of the Shaker potassium channel. Science (Wash DC) 1992;257:1225–1230. doi: 10.1126/science.1519059. [DOI] [PubMed] [Google Scholar]
- 14.Liman ER, Hess P, Weaver F, Koren G. Voltage-sensing residues in the S4 region of a mammalian K+channel. Nature (Lond) 1991;353:752–756. doi: 10.1038/353752a0. [DOI] [PubMed] [Google Scholar]
- 15.Lopez GA, Jan YN, Jan LY. Hydrophobic substitution mutations in the S4 sequence alter voltage-dependent gating in Shaker K+channels. Neuron. 1991;7:327–336. doi: 10.1016/0896-6273(91)90271-z. [DOI] [PubMed] [Google Scholar]
- 16.Lopez-Barneo J, Hoshi T, Heinemann SH, Aldrich RW. Effects of external cations and mutations in the pore region on C-type inactivation of Shakerpotassium channels. Receptors and Channels. 1993;1:61–71. [PubMed] [Google Scholar]
- 17.MacKinnon R, Yellen G. Mutations affecting TEA blockade and ion permeation in voltage-activated K+channels. Science (Wash DC) 1990;250:276–279. doi: 10.1126/science.2218530. [DOI] [PubMed] [Google Scholar]
- 18.MacKinnon R, Heginbotham L, Abramson T. Mapping the receptor site for a pore-blocking potassium channel inhibitor. Neuron. 1990;5:767–771. doi: 10.1016/0896-6273(90)90335-d. [DOI] [PubMed] [Google Scholar]
- 19.Nakai J, Adams BA, Imoto K, Beam KG. Critical roles of the S3 segment and S3-S4 linker of repeat I in activation of L-type calcium channels. Proc Natl Acad Sci USA. 1994;91:1014–1018. doi: 10.1073/pnas.91.3.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Papazian DM, Timpe LC, Jan YN, Jan LY. Alteration of voltage-dependence of Shakerpotassium channel by mutations in the S4 sequence. Nature (Lond) 1991;349:305–310. doi: 10.1038/349305a0. [DOI] [PubMed] [Google Scholar]
- 21.Papazian DM, Shao XM, Seoh S-A, Mock AF, Huang Y, Wainstock DH. Electrostatic interactions of S4 voltage sensor in Shaker K+channel. Neuron. 1995;14:1293–1301. doi: 10.1016/0896-6273(95)90276-7. [DOI] [PubMed] [Google Scholar]
- 22.Patton DE, West JW, Catterall WA, Goldin AL. A peptide segment critical for sodium channel inactivation functions as an inactivation gate in a potassium channel. Neuron. 1993;11:967–974. doi: 10.1016/0896-6273(93)90125-b. [DOI] [PubMed] [Google Scholar]
- 23.Santacruz-Toloza L, Huang Y, John SA, Papazian DM. Glycosylation of shaker potassium channel protein in insect cell culture and in Xenopusoocytes. Biochemistry. 1994;33:5607–5613. doi: 10.1021/bi00184a033. [DOI] [PubMed] [Google Scholar]
- 24.Sawaryn A, Drouin H. Reevaluation of hydropathy profiles of voltage-gated ionic channels. Experientia (Basel) 1991;47:962–964. doi: 10.1007/BF01929892. [DOI] [PubMed] [Google Scholar]
- 25.Seoh S-A, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+channel. Neuron. 1996;16:1159–1167. doi: 10.1016/s0896-6273(00)80142-7. [DOI] [PubMed] [Google Scholar]
- 26.Slesinger PA, Jan YN, Jan LY. The S4-S5 loop contributes to the ion-selective pore of potassium channels. Neuron. 1993;11:739–749. doi: 10.1016/0896-6273(93)90083-4. [DOI] [PubMed] [Google Scholar]
- 27.Yool AJ, Schwarz TL. Alteration of ionic selectivity of a K+channel by mutation of the H5 region. Nature (Lond) 1991;349:700–704. doi: 10.1038/349700a0. [DOI] [PubMed] [Google Scholar]
- 28.Zagotta WN, Hoshi T, Aldrich RW. Restoration of inactivation in mutants of Shakerpotassium channels by a peptide derived from ShB. Science (Wash DC) 1990;250:568–571. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]