Abstract
Protein translocation in the mammalian endoplasmic reticulum (ER) occurs cotranslationally and requires the binding of translationally active ribosomes to components of the ER membrane. Three candidate ribosome receptors, p180, p34, and Sec61p, have been identified in binding studies with inactive ribosomes, suggesting that ribosome binding is mediated through a receptor-ligand interaction. To determine if the binding of nascent chain-bearing ribosomes is regulated in a manner similar to inactive ribosomes, we have investigated the ribosome/nascent chain binding event that accompanies targeting. In agreement with previous reports, indicating that Sec61p displays the majority of the ER ribosome binding activity, we observed that Sec61p is shielded from proteolytic digestion by native, bound ribosomes. The binding of active, nascent chain bearing ribosomes to the ER membrane is, however, insensitive to the ribosome occupancy state of Sec61p. To determine if additional, Sec61p independent, stages of the ribosome binding reaction could be identified, ribosome/nascent chain binding was assayed as a function of RM concentration. At limiting RM concentrations, a protease resistant ribosome-membrane junction was formed, yet the nascent chain was salt extractable and cross-linked to Sec61p with low efficiency. At nonlimiting RM concentrations, bound nascent chains were protease and salt resistant and cross-linked to Sec61p with higher efficiency. On the basis of these and other data, we propose that ribosome binding to the ER membrane is a multi-stage process comprised of an initial, Sec61p independent binding event, which precedes association of the ribosome/nascent chain complex with Sec61p.
In mammalian cells, the translocation of nascent chains across the endoplasmic reticulum (ER) membrane is obligatorily cotranslational, and is thought to take place through an aqueous channel composed primarily of the resident ER membrane protein Sec61p and, in some cases, TRAM (Görlich and and Rapoport, 1993; Mothes et al., 1994; Do et al., 1996; Rapoport et al., 1996; Hanein et al., 1996). Furthermore, it is thought that during translocation, the ribosome forms a tight, continuous seal with Sec61p and thereby provides a direct, physically protected path for the nascent chain as it passes from the exit site in the ribosome to the protein conducting channel (Görlich et al., 1992; Crowley et al., 1993).
SEC61 was discovered in a genetic screen designed to identify components of the yeast protein translocation pathway, and encodes a polytopic 54-kD ER membrane protein (Deshaies and Schekman, 1987; Stirling et al., 1992). When purified from mammalian sources, Sec61p is recovered as a complex containing two low molecular weight subunits, β and γ (Görlich and Rapoport, 1993). Various temperature sensitive alleles of SEC61 display, at the nonpermissive temperature, profound defects in the translocation of a broad spectrum of secretory and membrane protein precursors (Rothblatt et al., 1989; Stirling et al., 1992). Both in sequence and topology, Sec61p bears limited homology to SecY, a bacterial protein which, in concert with SecA, SecE, and SecG, directs protein translocation across the inner membrane of E. coli (Brundage et al., 1990; Görlich et al., 1992; Stirling et al., 1992). Sec61p has been shown by both chemical and photocross-linking approaches to be in close physical proximity to translocating secretory and integral membrane precursors, data consistent with the proposal that Sec61p is the protein conducting channel (Thrift et al., 1991; Görlich et al., 1992; High et al., 1993a ,b; Mothes et al., 1994; Nicchitta et al., 1995; Do et al., 1996). Related cross-linking approaches have also demonstrated that phospholipids are physically proximal to the hydrophobic core of the signal sequence, suggesting that the lipid bilayer can be directly accessed from the translocation site (Martoglio et al., 1995).
That there exists within the rough ER a specific machinery dedicated to ribosome binding is embodied in current models of translocation (Görlich et al., 1992; Crowley et al., 1993; Walter and Johnson, 1994; Rapoport et al., 1996; Hanein et al., 1996). Indeed, it is generally assumed that there exist protein components resident to the rough ER which impart an affinity for ribosomes, and thereby yield the morphological distinction between rough and smooth ER (Blobel and Dobberstein, 1975; Kreibich et al., 1978). Historically, experiments designed to identify candidate ribosome receptors in the ER membrane have employed purified, inactive ribsomes and ER membranes stripped of bound ribosomes (Borgese et al., 1974). In these studies, ribosomes were reported to bind to microsomes in a high affinity, saturable manner (Borgese et al., 1974). In rat liver microsomes, high affinity, saturable ribosome binding is markedly salt-sensitive, and is negligible at physiological salt concentrations (Borgese et al., 1974).
Extensive characterization of the ribosome-membrane junction has established that binding is mediated in part by the nascent chain and by protease-sensitive, electrostatic interactions between the ribosome and components of the ER membrane (Adelman et al., 1973). From these data, it appears that ribosome-nascent chain complexes bind to discrete sites, defined by specific receptor proteins, and that both the nascent chain and the ribosome contribute to the binding event. The combination of these two binding components yields a ribosome/nascent chain complex which is resistant to salt extraction and digestion by exogenous protease (Sabatini and Blobel, 1970; Adelman et al., 1973; Connolly and Gilmore, 1986). There also appear to be components of the ribosome which function in the binding event. Recent reports on the regulation of ribosome binding to the ER membrane describe a role for the nascent chain-associated protein complex (NAC)1 in regulating the membrane binding activity of active ribosomes (Lauring et al., 1995a ). In the absence of the signal recognition particle (SRP), NAC has been demonstrated to function as a global inhibitor of ribosome binding (Lauring et al., 1995b ).
Three candidate ribosome receptors, p180, p34, and the Sec61p complex, have been identified by the ribosome binding protocol of Borgese et al. (1974) (Savitz and Meyer, 1990; Tazawa et al., 1991; Ichimura et al., 1992; Savitz and Meyer, 1993; Kalies et al., 1994; Wanker et al., 1995). p180 was identified through analysis of the inhibition of ribosome binding by protein fragments derived from proteolyzed ER membranes (Savitz and Meyer, 1990, 1993). In reconstitution assays, p180 imparted ribosome binding activity to proteoliposomes (Savitz and Meyer, 1990, 1993). Furthermore, proteoliposomes reconstituted from detergent extracts of RM depleted of p180 by immuno-affinity chromatography, exhibited markedly reduced ribosome binding, as well as defects in translocation (Savitz and Meyer, 1993). In addition, expression of p180 in yeast induces ER proliferation and an apparent increase in the number of membrane-associated ribosomes (Wanker et al., 1995). Other groups have reported, however, that ribosome binding activity could be ascribed to a p180 deficient protein fraction and thus the functional contribution of p180 to ribosome binding is considered controversial (Collins and Gilmore, 1991; Nunnari et al., 1991). p34 was identified as an abundant protein component of liver ER membranes which, upon reconstitution into liposomes, exhibited ribosome binding activity (Tazawa et al., 1991; Ichimura et al., 1992). Antibodies directed against p34 have been shown to block ribosome binding and to impair translocation (Tazawa et al., 1991; Ichimura et al., 1992). In a recent study, however, it was reported that p34 was not protected from proteolytic digestion by membrane-bound ribosomes and thus was unlikely to mediate membrane binding of ribosomes (Kalies et al., 1994).
There is substantial experimental evidence in support of a ribosome receptor function for the Sec61p complex. Upon detergent solubilization and centrifugation of rough microsomes (RM), Sec61p was found in the ribosomeenriched pellet fraction, along with a subset of other ribosome-associated membrane proteins, or RAMPS (Görlich et al., 1992). Release of Sec61p from the RAMP fraction was achieved following treatment with high salt concentrations (0.75–1 M KOAc) and puromycin, conditions similar to those employed to release ribosomes from intact RM (Adelman et al., 1973; Görlich et al., 1992). Furthermore, velocity sedimentation studies of solubilized RM indicated that Sec61p solubilized at moderate (0.5 M) salt concentrations remains in association with the 80 S ribosome (Görlich et al., 1992). In more recent studies, Kalies et al. (1994) have provided direct evidence that at physiological salt concentrations, Sec61p is the predominant ribosome binding site in ER membranes (Kalies et al., 1994). Using native membranes as well as proteoliposomes containing the purified Sec61p complex, Kalies et al. (1994) identified high affinity ribosome binding to the Sec61p complex at physiological salt concentrations (Kalies et al., 1994). As would be predicted from the binding data, in native RM the majority of Sec61p was found to be protected from proteolytic degradation by native, bound ribosomes (Kalies et al., 1994). It appears, therefore, that Sec61 is the primary ribosome receptor, although other components, such as p180 and p34 may contribute to the total ribosome binding activity.
Using established criteria for differentiating membrane bound vs free ribosome/nascent chain complexes, we report that the binding of translationally active ribosome/ nascent chain complexes to the ER membrane is insensitive to the ribosome occupancy state of the Sec61p complex, and is not blocked by addition of a large molar excess of free 80 S ribosomes. However, and consistent with previous reports, we observed that of the identified ribosome receptors, only Sec61p was protected from proteolytic digestion by native, bound ribosomes. To reconcile this apparent paradox, we evaluated the hypothesis that the binding of active, nascent chain bearing ribosomes is comprised of Sec61p independent and Sec61p dependent stages. When binding reactions were performed with limiting concentrations of RM, and thus limiting levels of ribosome-unoccupied Sec61p, bound nascent chains, although protease resistant, were sensitive to extraction with high salt, thereby identifying a novel state of nascent chain association with the ER membrane. With nonlimiting concentrations of RM, bound nascent chains were protease and salt resistant. In related experiments, it was observed that the efficiency of nascent chain cross-linking to Sec61p was a function of RM concentration. Thus, at limiting RM concentrations the yield of nascent chain/Sec61p crosslinks was markedly reduced relative to that observed at nonlimiting RM concentrations. On the basis of these data, we propose that ribosome/nascent chain association with the ER membrane is a multi-stage process comprised of an initial Sec61p independent and a subsequent Sec61p dependent stage.
Materials and Methods
Reagents
Hemin, creatine phosphate, and creatine phosphokinase were obtained from Calbiochem (San Diego, CA). Staphylococcal nuclease, calf liver tRNA, puromycin, and proteinase K were obtained from Boehringer Mannheim Biochemicals (Indianapolis, IN). Phenylhydrazine hydrate and trypsin was from Sigma Chem. Co. (St. Louis, MO). Chymotrypsin was from Worthington Scientific Corporation (Freehall, NJ). Restriction enzymes were obtained from either New England Biolabs (Beverly, MA) or Promega (Madison, WI). [35S] Pro-Mix ([35S] methionine and cysteine) was obtained from Amersham (Arlington Heights, IL). Nucleotides were obtained from Pharmacia (Piscataway, NJ).
Membrane Protein Protease Accessibility
Protease accessibility studies in canine and porcine RM was performed as follows: four equivalents (eq.) of RM were diluted in a buffer containing 25 mM K-Hepes, pH 7.2, 25 mM KOAc, and 2.5 mM Mg(OAc)2 to a final volume of 100 μl. Chymotrypsin was added to the indicated concentrations from a 1-mg/ml stock solution. Protease digestions were performed for 30 min at 4°C. After digestion, samples were precipitated by addition of TCA to a final concentration of 10%, and processed for SDS-PAGE. Transfer to nitrocellulose membranes for immunoblot analysis was performed by semi-dry transfer in a 50 mM CAPS, pH 11.0, 20% methanol, 0.075% SDS buffer. Immunoblots were visualized by ECL detection (Amersham Corp.). Immunoblot films were scanned on a Hewlett-Packard Scanjet Plus, and size and contrast adjusted in Photoshop version 3.0 (Adobe Systems, Inc., Mountain View, CA). Quantitation of imaged immunoblots was by means of NIH Image software.
Generation of Anti-Ribosomal Antibodies
Ribosomes were prepared from deoxycholate treated canine RM by the method of Florini and Breuer (1966). 60 S and 40 S subunits were resolved on 10–30% sucrose gradients, following puromycin/0.5 M KOAc treatment, and separated subunit fractions resolved on SDS-PAGE. Strips of SDS-PAGE gels containing either homogenous proteins L3/L4 or protein S9 were excised, minced, mixed in Freunds complete adjuvant, and used for antibody production in chickens. Animal services were performed by contract agreement with Cocalico Biologicals (Reamstown, PA).
Cell-Free Transcription and Translation
The plasmid pGEMBP1 (Connolly and Gilmore, 1986) containing a cDNA insert encoding for bovine preprolactin, was linearized within the coding region with PvuII. Transcription reactions were performed by the procedure of Weitzmann et al. (1990) in a buffer containing 40 mM Tris/ HCl (pH 8.0), 8 mM Mg(OAc)2, 25 mM NaCl, 2 mM spermidine, 10 mM dithiothreitol, 2.5 mM ATP, CTP, UTP, and GTP, 2 U/ml yeast inorganic pyrophosphatase and 1 U/ml T7 RNA polymerase. Cell-free translations were performed in a rabbit reticulocyte lysate system as described (Nicchitta and Blobel, 1989). Translations (20 μl) contained 8 μl of nucleasetreated rabbit reticulocyte lysate, 16 μCi of [35S] Pro-Mix (methionine/cysteine), 0.05 U/ml RNasin, 1 mM DTT, and 20 μM (−) methionine amino acid mix. Reactions were adjusted to 110 mM KOAc, 2.5 mM Mg(OAc)2. Rabbit reticulocyte lysate was prepared by the method of Jackson and Hunt (1983) and canine pancreas rough microsomes (RM) prepared by the method of Walter and Blobel (1983). Translations were performed for 30 min at 25°C.
Quantitation of Cell-Free Translation Products
The amount of free, nonradioactive methionine in the cell-free translation system was determined by isotope dilution. The addition of 1.4 μM nonradioactive methionine to the translation system decreased incorporation of radioactive methionine by 50%. Therefore, calculations of translation yield were based on an endogenous methionine concentration of 1.4 μM. The specific activity of the methionine pool, expressed as PSU units/nmol, was determined by phosphorimager based quantitation of a serial dilution series of the translation mix. The contribution of isotopically labeled cysteine to the total radioactivity of the translation products was <5% and was not included in the calculation.
Preparation of EDTA and KOAc Washed RM (EKRM)
EKRM were prepared by diluting 250 eq. of RM fourfold in buffer to yield final concentrations of 0.5 M KOAc, 10 mM EDTA, 25 mM K-Hepes, pH 7.2. After a 30-min incubation at 4°C, the membranes were collected by centrifugation for 10 min at 60,000 rpm in a TLA 100.2 rotor at 4°C (Beckman Instrs., Fullerton, CA). EKRM pellets were resuspended in RM buffer (0.25 M sucrose, 25 mM K-Hepes, pH 7.2, 25 mM KOAc), and stored at −80°C.
Reconstitution of SRα Activity (52 kD)
Isolation, partial purification, and reconstitution of the 52-kD fragment of SRα was performed as described in Nicchitta and Blobel (1989).
Sec61p Purification and Quantitation
Sec61p was purified by a modification of the procedures of Görlich and Rapoport (1993). 20 ml of RM, at a concentration of 1 eq./ml, were diluted 1:1 with a buffer consisting of 1 M KOAc, 10 mM EDTA. After a 30-min incubation on ice, RM were chromatographed, with upward flow, on a 170-ml Sepharose CL-2B column in 500 mM KOAc, 5 mM EDTA, 10 mM 2-mercaptoethanol, at a flow rate of 12 ml/h. The ribosome-stripped RM fractions were pooled and centrifuged for 1 h at 40 K in the Ti50.2 rotor. To remove the lumenal contents, pelleted membranes were resuspended in 0.1 M Na-CAPS, pH 10.5, 10 mM 2-mercaptoethanol, incubated on ice for 30 min, and recovered by centrifugation for 40 min at 45 K in the Ti50.2 rotor over a cushion of 0.5 M sucrose, 50 mM K-Hepes, pH 7.2 (Nicchitta and Blobel, 1993). The ribosome and lumenal protein depleted RM were resuspended in 15% glycerol, 750 mM NaCl, 25 mM K-Hepes (pH 7.2), 10 mM 2-mercaptoethanol (buffer A), and solubilized by addition of Nikkol to 1.5%. A high speed supernatant, containing soluble Sec61p, was obtained by centrifugation of the detergent/membrane mixture for 1 h at 45 K in the Ti50.2 rotor. The soluble fraction was subsequently depleted of glycoprotein components by chromatography on a 10-ml con A–Sepharose column, equilibrated in buffer A supplemented with 0.25 mg/ml egg yolk phosphatidylcholine (PC), at a flow rate of 1.5 ml/h. The flowthrough fraction, depleted of glycoproteins was then chromatographed on a Superdex 200/60 gel filtration column, equilibrated in buffer A adjusted to 500 mM NaCl, 0.5% Nikkol, 0.1 mg/ml PC at a flow rate of 0.5 ml/min. The Sec61p enriched fractions, identified by immunoblot with an NH2-terminal directed Sec61p antibody, were pooled and dialyzed overnight against buffer A adjusted to 50 mM NaCl, 0.25% Nikkol, and 0.1 mg/ml PC. After dialysis, the protein fraction was centrifuged for 30 min at 45 K in the Ti50.2 rotor, to remove aggregates, and the supernatant chromatographed on a 5-ml Q-Sepharose FF column, equilibrated in dialysis buffer. The flowthrough fractions were directly loaded onto a Mono S 10/10 column and eluted with a gradient of 50–500 mM NaCl in 25 mM K-Hepes, pH 7.4, 0.25% Nikkol, 0.1 mg/ml PC, 10 mM 2-mercaptoethanol. Peak Sec61p containing fractions were pooled and concentrated in a Centricon 30 ultrafiltration device. On the basis of Coomassie blue staining, Sec61p purifed by this protocol was ∼60% pure. Quantitation of the Sec61p content of pH 10.5 washed RM (Nicchitta and Blobel, 1993) was performed by quantitative immunoblot using purified Sec61p as standard and by densitometric analysis of Coomassie blue–stained gels, also with purified Sec61p as standard.
Purification of Reticulocyte Ribosomes
8 ml of nuclease-treated reticulocyte lysate was diluted to 12 ml using ribosome buffer (150 mM KCl, 25 mM K-Hepes, pH 7.2, 5 mM Mg(OAc)2). Lysate was then centrifuged for 35 min at 100,000 rpm in the TLA100.3 rotor (4°C). The supernatant was removed by aspiration, and the pellet resuspended in 1 ml of ribosome buffer by Dounce homogenization (B pestle) and agitation for 2 h at 4°C. Insoluble material was removed by centrifugation for 10 min at 10,000 g. Samples were then loaded on preparative 10–30% sucrose gradients and centrifuged at 40,000 rpm for 2 h at 25°C in the SW40.1 rotor. The lower 50% of each gradient was collected, combined, and centrifuged for 3 h at 45,000 rpm in the Ti50.2 rotor, 4°C. The supernatant was aspirated, the 80-S ribosomal pellets resuspended in ribosome buffer, and aliquots stored at −80°C. Ribosome concentrations were determined using the relationship 1A260 = 21.4 pmol 80S ribosomes (Martin et al., 1969)
NEM Treatment of RM
RM were diluted fivefold and treated with 1 mM NEM (200 mM stock in DMSO) for 20 min at 25°C. After treatment, DTT was added to a final concentration of 25 mM, and reactions incubated for an additional 10 min at 25°C. RM were layered over 0.5 M sucrose cushion, and collected by centrifugation (6 min, 60,000 rpm, TLA 100 rotor, 4°C). RM pellets were resuspended in RM buffer supplemented with 2 mM DTT.
Chemical Cross-linking
Chemical cross-linking of completed translation reactions were performed as follows. Translation reactions were chilled on ice and diluted fivefold with a physiological salt buffer consisting of 110 mM KOAc, 25 mM K-Hepes (pH 7.4), 2.5 mM Mg(OAc)2. Diluted reactions were overlayed onto a 1/3 vol cushion of 0.5 M sucrose, 25 mM K-Hepes, pH 7.4 and centrifuged for 10 min at 60,000 rpm in the TL100 rotor. Supernatant and cushion fractions were discarded and the membrane pellet resuspended in 0.25 M sucrose, 50 mM KOAc, 2.5 mM Mg(OAc)2. Cross-linking reactions (50 μl) were performed for the indicated time periods at 25°C, by addition of m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS) to a final concentration of 1 mM, from a 50-mM stock in dimethylformamide. Reactions were quenched by addition of 1 vol of PBS containing 50 mM dithiothreitol, 50 mM lysine, 1% SDS. Cross-linking reactions were precipitated by addition of TCA to 10% and processed for SDS-PAGE.
Results
Accessibility of ER Membrane Proteins to Proteolytic Degradation
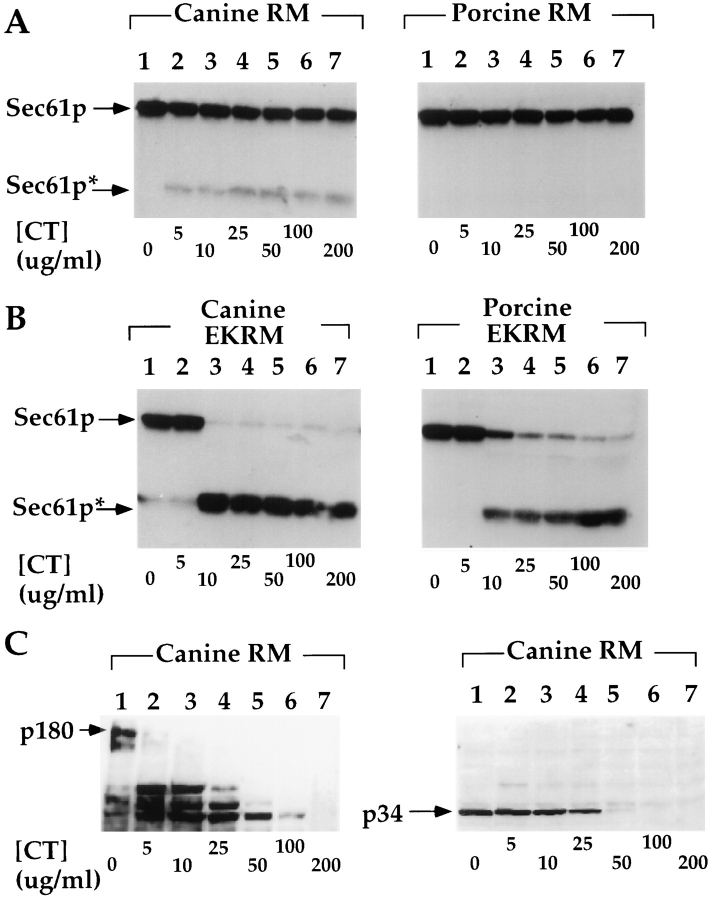
Consistent with its proposed role in ribosome binding, it has been reported that bound ribosomes protect Sec61p from digestion with exogenous proteases (Kalies et al., 1994). This observation was confirmed in experiments depicted in Fig. 1 A. In these experiments, RM were treated with increasing concentrations of chymotrypsin at 4°C, and immunoblotted with a polyclonal antibody directed against the NH2 terminus of Sec61p. In canine and porcine RM, Sec61p is insensitive to proteolytic degradation at chymotrypsin concentrations up to 200 μg/ml. In five independent experiments with native RM, canine Sec61p was >80% protected, while porcine Sec61p was >95% protected. Proteolytic cleavage of the Sec61p was assayed as the appearance of a limit digestion product, indicated by the asterisk, which maintains reactivity with the polyclonal antibody. To determine the contribution of bound ribosomes to the observed protection, ribosomes were extracted from RM by treatment with 15 mM EDTA and 0.5 M KOAc (EKRM). As depicted in Fig. 1 B, upon removal of bound ribosomes, the sensitivity of Sec61p to proteolytic digestion is dramatically enhanced, with degradation occurring at chymotrypsin concentration as low as 10 μg/ml. At higher chymotrypsin concentrations, >95% of Sec61p is degraded to the limit digestion product. These data suggest that in native RM, the vast majority of the Sec61p exists in association with bound ribosomes and by virtue of this association, is protected from proteolytic degradation by exogenous proteases.
Figure 1.
Protease protection of ER proteins in RM and EKRM. 4 eq. of either RM (A and C) or EKRM (B) were diluted to 20 μl in a buffer containing 25 mM K-Hepes, pH 7.2, 25 mM KOAc, and 2.5 mM Mg(OAc)2 at 4°C. Samples were treated with chymotrypsin (CT) at the indicated concentration for 30 min at 4°C and reactions quenched by addition of TCA to 10%. After centrifugation, samples were processed for SDS-PAGE and immunoblotted with antibodies directed against Sec61p, p180, and p34, as described in Materials and Methods. The migration of full-length Sec61p, p180, and p34 are indicated by arrows; a prominent limit digestion product of Sec61p is indicated by an asterisk.
Experiments were also performed to determine if bound ribosomes afforded protease protection to the ribosome receptors p180 and p34 (Savitz and Meyer, 1990; Tazawa et al., 1991; Ichimura et al., 1992; Savitz and Meyer, 1993, 1994; Wanker et al., 1995). The results of these studies are depicted in Fig. 1 C. In contrast to Sec61p, both p180 and p34 were sensitive to digestion by chymotrypsin in native RM. These data corroborate those of Kalies et al. (1994) and indicate that of the proposed ribosome receptors p180, p34, and Sec61p, only Sec61p is protected from proteolytic degradation by native, bound ribosomes.
Relationship between Ribosome Structure and Sec61p Accessibility
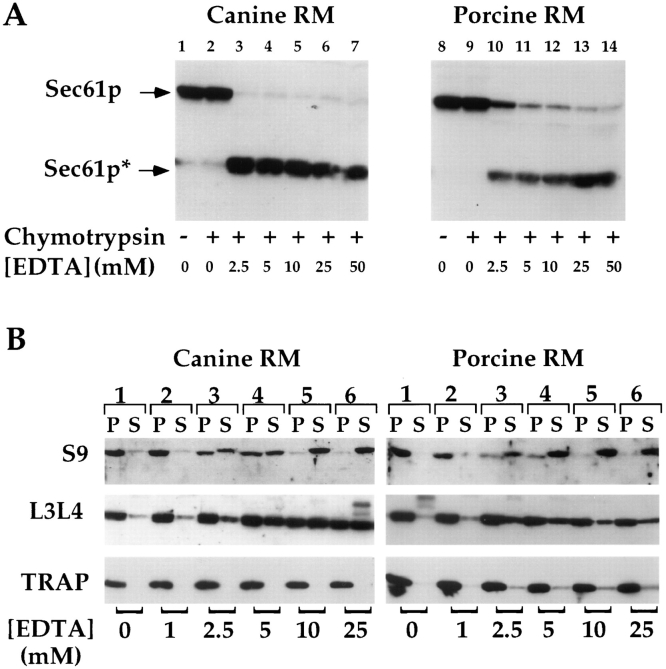
To further explore the correlation between membranebound ribosomes and the protease accessibility of Sec61p, canine and porcine RM were treated with increasing concentrations of EDTA, and subsequently assayed for the release of bound ribosomal subunits and Sec61p protease accessibility. It has been previously demonstrated that exposure of RM to increasing concentrations of EDTA yields preferential release of the small ribosomal subunit, and partial release of the large subunit (Sabatini et al., 1966). RM were incubated in the presence of EDTA, and either subjected to proteolysis with chymotrypsin, or centrifuged to separate membrane associated and free subunits. In the absence of EDTA treatment, >90% of the canine and porcine Sec61p was protected from proteolytic digestion (Fig. 2 A, lanes 2 and 9). Exposure to increasing concentrations of EDTA yielded a dramatic increase in the susceptibility of Sec61p to proteolytic digestion, an effect that was somewhat more pronounced in canine RM. An immunoblot analysis of the distribution of the large ribosomal subunit proteins L3 and L4 and the small subunit protein S9 is shown in Fig. 2 B. Samples were also immunoblotted with an antibody directed against the ER integral membrane protein TRAPα, to insure that centrifugation conditions yielded complete recovery of the microsomal membranes. Consistent with previous studies, treatment of RM with EDTA resulted in the preferential release of the small subunit and partial release of the large subunit (Fig. 2 B, lanes 2–6). From these data it is apparent that the protease accessibility of Sec61p appears coincident with the dissociation/denaturation of bound ribosomes.
Figure 2.
EDTA treatment of RM: effects on Sec61p susceptibility to protease digestion and release of bound 60 and 40 S ribosomal subunits. 4 eq. of either canine or porcine RM were diluted to 20 μl in buffer containing 25 mM K-Hepes pH 7.2, 25 mM KOAc at 4°C. After dilution, samples were treated with EDTA at the indicated concentration for 15 min at 4°C, and either treated with chymotrypsin (25 μg/ml) for 30 min at 4°C (A), or centrifuged to separate membraneassociated and free ribosomal subunits (B). (A) After chymotrypsin treatment and acid precipitation, samples were processed for SDSPAGE and immunoblotted with an antibody directed against Sec61p. As in Fig. 1, the migration of both full- length Sec61p and the limit digestion product are indicated. (B) After EDTA treatment, samples were diluted sevenfold in a physiological salts buffer, layered over a 0.5-M sucrose cushion, and centrifuged to separate bound from free ribosomal subunits (6 min, 60,000 rpm, TLA 100 rotor, 4°C). Pellet (P) and supernatant (S) samples were processed for SDSPAGE, and immunoblotted with antibodies directed against the 40-S subunit protein S9, 60 S subunit proteins L3 and L4 (L3L4), or TRAPα.
Sec61p-associated Ribosomes Do Not Release Upon Run-Off Translation
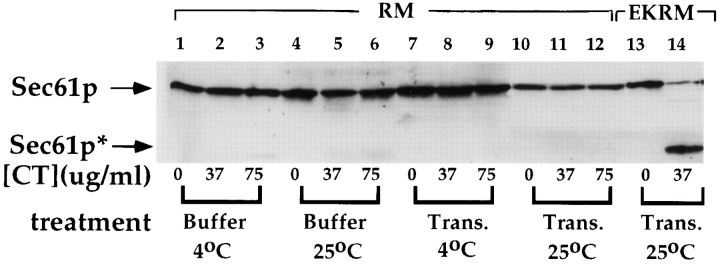
The data presented in Figs. 1 and 2 indicate that in native RM, bound ribosomes protect Sec61p from digestion with exogenous proteases and that release of bound ribosomes, by addition of EDTA, is accompanied by a dramatic increase in protease sensitivity. In vivo, this behavior is likely mimicked by the termination reaction, which yields the release of the nascent chain and the dissociation of the ribosome into its component subunits. To determine whether nascent chain termination on membrane-bound ribosomes results in ribosome release and an increase in Sec61p protease accessibility, the accessibility of Sec61p to proteolytic degradation was assessed following run-off translation. As depicted in Fig. 3, incubation of RM with reticulocyte lysate, either at 4°C (lanes 7–9) or at 25°C (lanes 10–12) did not alter the protease susceptibility of Sec61p, an observation consistent with a lack of ribosome release following runoff translation. Similar conclusions have been previously reported regarding rat liver RM (Sabatini and Blobel, 1970), where it has also been directly demonstrated that the large subunits of bound ribosomes do not undergo significant exchange with free large subunits (Borgese et al., 1973). When EKRM were treated under identical conditions, Sec61p remained highly sensitive to proteolytic digestion, indicating that the concentrations of ribosomes present in the reticulocyte lysate are insufficient to yield reprotection of the accessible Sec61p (Fig. 3, lanes 13 and 14).
Figure 3.
Run-off translation does not alter the accessibility of Sec61p to proteolytic digestion. RM were treated with buffer (25 mM K-Hepes, pH 7.2, 110 mM KOAc, 2.5 mM Mg(OAc)2) at 4°C or 25°C, or with reticulocyte translation mixture (see Materials and Methods) at 4°C or 2°C. Subsequently, samples were treated with the indicated concentration of chymotrypsin for 30 min at 4°C. Samples treated with buffer were processed for SDS-PAGE as described previously. Samples treated with reticulocyte translation mixture were diluted sevenfold in physiological salt buffer, overlaid onto a 0.5-M sucrose cushion, and centrifuged for 10 min at 60,000 rpm in a TLA 100 rotor, as described in the legend to Fig. 2. The pellet (RM) fraction was solubilized in SDS-PAGE sample buffer and processed for SDS-PAGE. After transfer to nitrocellulose, samples were immunoblotted with an antibody directed against Sec61p.
80 S Ribosomes Fail to Compete with Ribosome/pPl 86 for Binding to RM
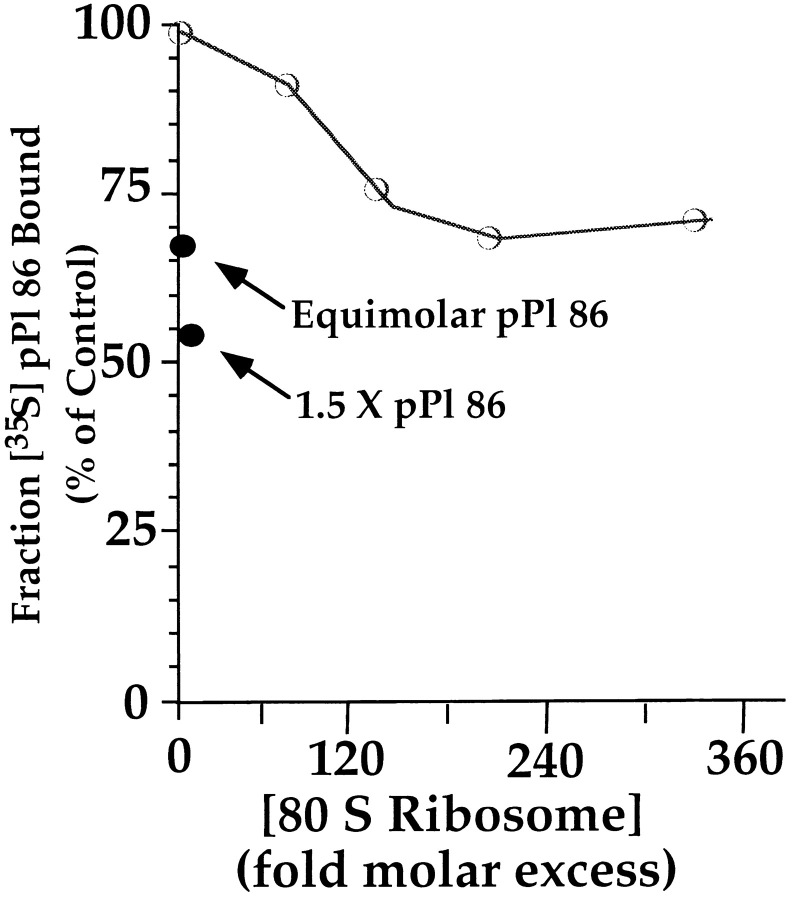
It has recently been reported that Sec61p displays nanomolar affinity for translationally inactive ribosomes at both low and physiological salt concentrations and thus represents the predominant site of ribosome-membrane interaction in RM (Kalies et al., 1994). While Sec61p appears to mediate the majority of ribosome binding (cf. Figs. 1–3), it has not been established whether Sec61p represents the sole site of ribosome-membrane association, particularly with regard to active, nascent chain bearing ribosomes. Thus, a competition study was performed to determine if free, inactive ribosomes compete with targeted, ribosome/ nascent chain complexes for binding sites on RM. In these experiments, 80 S ribosomes were purified from reticulocyte lysate, and used in competition binding studies with pPl 86, a truncated, ribosome-associated form of preprolactin that is fully active in posttranslational targeting assays (Connolly and Gilmore, 1986; Nicchitta and Blobel, 1989). Increasing amounts of purified 80 S ribosomes were added to RM, before the addition of ribosome/pPl 86 complexes and, following a 10-min incubation at 25°C, RM were separated from the translation mix by centrifugation. Samples were resolved by SDS-PAGE and the relative amount of bound and free nascent chains determined by phosphorimager analyses of the dried gels. All samples were normalized to a control sample lacking added ribosomes. In the experiment shown in Fig. 4, 50 fmol of ribosome/pPl 86 substrate were incubated with RM in the presence of increasing concentrations of free, inactive ribosomes (0–19,000 fmol). Clearly, purified 80 S ribosomes compete quite poorly with ribosome/pPl 86 complexes for membrane binding, with a 25% inhibition observed at a 160-fold excess of free ribosomes (Fig. 4). If there exist ribosome binding sites for active, nascent chain bearing ribosomes, however, it should be possible to compete radiolabeled ribosome/pPl 86 binding with an unlabeled ribosome/pPl 86 substrate. pPl 86 mRNA was thus translated in the presence and absence of radiolabeled methionine, and membrane binding of the labeled substrate assayed in the presence of increasing amounts of the unlabeled substrate. As shown in Fig. 4, unlabeled ribosome/pPl 86 was highly effective at competing for radiolabeled ribosome/pPl 86 binding. Addition of one translation equivalent of unlabeled substrate reduced binding of labeled substrate by ∼35% whereas a 1.5-fold excess of unlabeled precursor reduced binding by almost 50%. These results demonstrate that binding of ribosome/ pPl 86 complexes is saturable and specific and further suggest that there may exist, in addition to the previously identified Sec61p defined binding sites, additional ribosome binding sites in the ER membrane, possibly specific for nascent chain bearing ribosomes.
Figure 4.
Inactive ribososomes do not compete for binding with ribosome/nascent chain complexes. pPl 86 was translated in the absence of RM, and aliquots of the translation reaction (20 μl) added to 1 eq. of RM, subsequent to addition of the indicated quantity of 80 S reticulocyte lysate-derived ribosomes. Buffer conditions were adjusted such that the final KOAc concentration was 140 mM. After incubation at 25°C for 10 min, RM-bound pPl 86 was separated from unbound by centrifugation through a 0.5-M sucrose cushion as described in the legend to Fig. 2, and samples processed for SDS-PAGE. Quantitation of the [35S] pPl 86 was performed by phosphorimager analysis of the dried gels on a Fuji MacBAS 1000 phosphorimager.
EKRM Containing Proteolyzed Sec61p Support Ribosome/Nascent Chain Binding
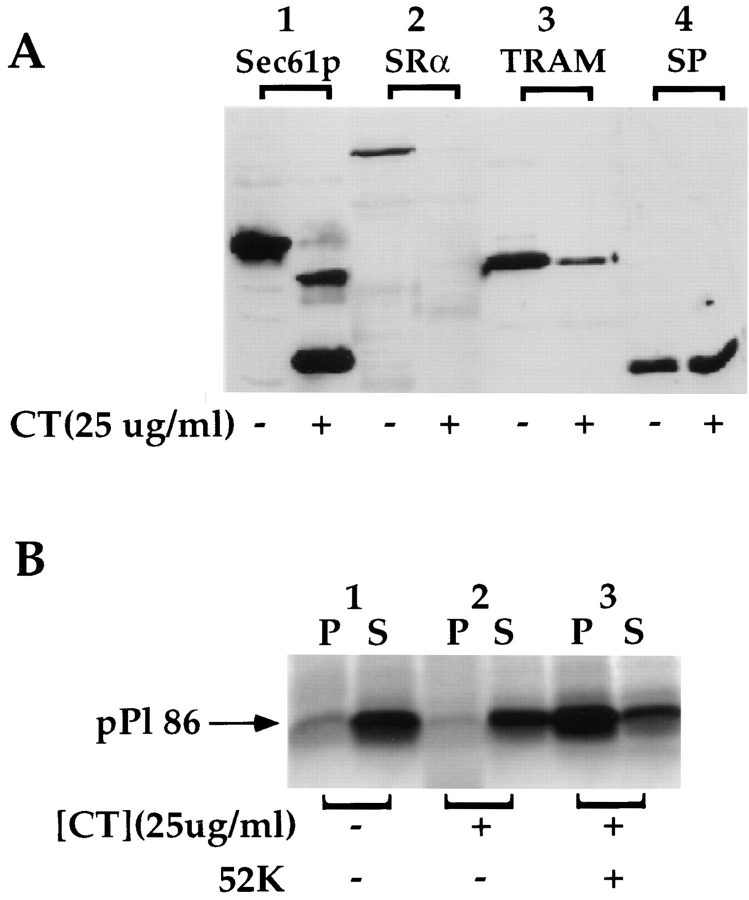
Having acquired evidence suggesting that ribosome/nascent chain binding to RM may involve multiple sites and/or mechanisms of association, we determined whether the binding of ribosome nascent chain complexes displayed a requirement for intact Sec61p. EKRM were treated with chymotrypsin, under conditions in which Sec61p is quantitatively clipped, and, following reconstitution of the membranes with the 52-kD fragment of the SRP receptor, used in binding reactions with pPl 86. To assess the structural state of various ER integral membrane proteins, chymotrypsintreated EKRM were immunoblotted for Sec61p, SRα, TRAM, and the 22/23-kD subunit of the signal peptidase complex (Fig. 5 A). From these data, it is apparent that under these conditions, Sec61p and SRα are fully degraded whereas TRAM is >85% proteolyzed. Signal peptidase (SPC) is a stable heterooligomeric complex and data with the SPC 22/23-kD subunit were included as a control for excessive proteolysis. As noted, digestion of EKRM under conditions which degrade Sec61p, also results in digestion of SRα. Therefore, in order to restore physiological targeting, the proteolyzed membranes were reconstituted with the cytoplasmic domain of SRα (Gilmore et al., 1982a ,b; Meyer et al., 1982). The activity of the reconstituted membranes in the targeting assay is shown in Fig. 5 B. In the absence of RM, or in the presence of proteolyzed EKRM, >90% of the pPl-86 is recovered in the supernatant fraction (Fig. 5 B, lanes 1 and 2). After reconstitution with the SRα receptor fragment, ribosome/pPl 86 binding was restored (Fig. 5 B, lane 3). We conclude from these data that intact Sec61p, and perhaps TRAM, are not required for the initial binding of active ribosome/nascent chain complexes to the ER membrane.
Figure 5.
Proteolysis of Sec61p does not alter ribosome/pPl 86 binding. (A) Untreated and chymotrypsin-treated EKRM were resolved on 12.5% SDS-PAGE gels, and immunoblotted with antibodies directed against Sec61p, SRα, TRAM, and the 22/23-kD subunit of the signal peptidase complex. (B) EKRM were treated with chymotrypsin at 25 μg/ml for 30 min at 4°C, conditions known to yield nearly quantitative digestion of Sec61p. After protease treatment, chymotrypsin-treated EKRM were tested for their ability to support binding of ribosome/pPl 86, either in the presence or absence of the 52-kD fragment of SRα. pPl 86 was translated in the absence of 1.0 eq. EKRM (lane 1), in the presence of 1.0 eq. chymotrypsin-treated EKRM (lane 2), and in the presence of 1.0 eq. chymotrypsin-treated EKRM supplemented with 52 kD SRα fragment (lane 3). The 52-kD SRα fragment was incubated with chymotrypsin-treated EKRM for 30 min at 4°C, before translation.
RM and EKRM Binding Capacity Does Not Correlate with Accessible Sec61p
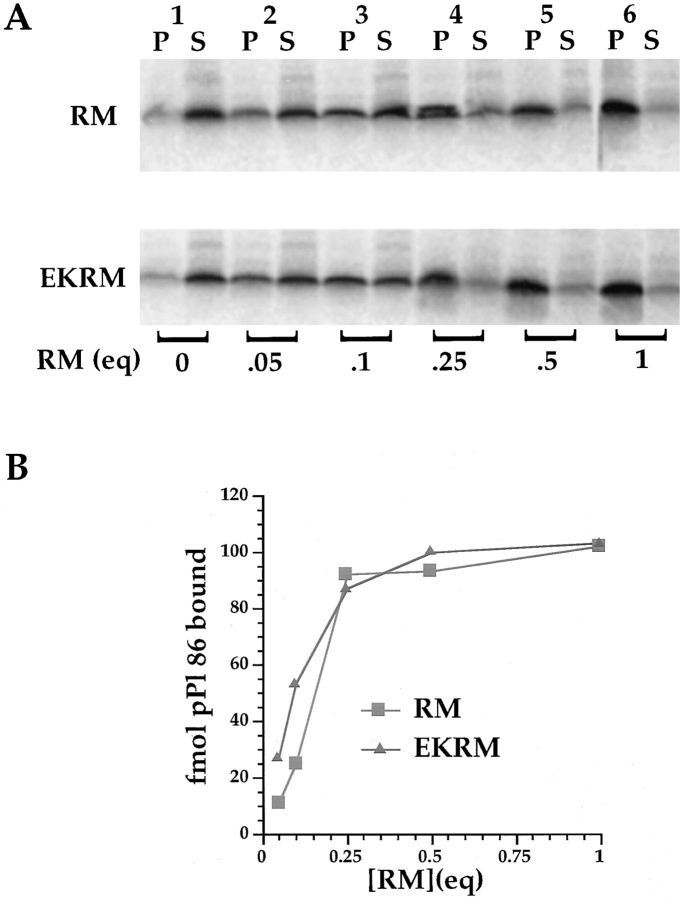
If Sec61p is the primary site of ribosome/nascent chain association with the ER membrane, the binding capacity of RM for targeted nascent chains should correlate with the concentration of free, or accessible, Sec61p. For example, from the protease protection data in Fig. 1 and Sec61p quantitative immunoblots (data not shown), there appear to be ∼200 fmol of available Sec61p per equivalent of canine RM (20% accessible, 1,000 fmol/eq. total) (Kalies et al., 1994), and ∼60 fmol of Sec61p per equivalent in porcine RM (5% accessible, 1,200 fmol/eq.). Also depicted in Fig. 1 is the observation that virtually 100% of Sec61p is protease accessible in both canine and porcine EKRM, suggesting (at a minimum stoichiometry of 1 ribosome: Sec61p complex) a binding capacity of ∼1,000 fmol of nascent chains/RM equivalent for EKRM. An experiment testing this hypothesis is depicted in Fig. 6 A. From determinations of the specific activity of the [35S]methionine pool and the incorporation of radiolabel into nascent chains, the molar concentration of newly synthesized nascent chains was calculated and, following isolation of the RM by centrifugation, the quantity of bound vs free ribosome/ nascent chain complexes determined. In Fig. 6 A, ∼150 fmol of ribosome/pPl 86 complex were incubated with quantities of RM or EKRM ranging from 0 eq. (lane 1) to 1 eq. (lane 6). For each condition depicted, and at the levels of pPl 86 translation in the reticulocyte lysate system, maximal binding (>75% bound) was seen using 0.25 eq./ reaction (lane 4), operationally defined as saturated binding. No further increase in binding was observed when 0.5 eq. (lane 5) or 1 eq. (lane 6) were used. The data are depicted graphically in Fig. 6 B. These results are evident from two significant points: First, and unexpectedly, EKRM, in which Sec61p is virtually 100% protease accessible, did not display an increased binding capacity for active ribosome/nascent chain complexes, relative to RM. Second, in the depicted experiment, 0.25 equivalents of RM bound ∼100 fmol of ribosome/nascent complexes, when, as determined by protease accessibility, only 15 fmol of Sec61p, were available. It does not appear that the binding capacity in excess of the available Sec61p can be attributed to bound ribosomes dissociating from Sec61p upon runoff translation, as the data in Fig. 3, as well as previous reports, indicate that this does not occur (Sabatini and Blobel, 1970; Borgese et al., 1973). Thus, the binding capacity of RM for targeted, ribosome-associated nascent chains is apparently insensitive to the ribosome occupancy state of Sec61p. These results suggest that either there are multiple sites of ribosome/nascent chain binding to the ER membrane or that the binding reaction is comprised of multiple stages, with ribosome/nascent chain-Sec61p association representing a stage subsequent to an initial binding event.
Figure 6.
Ribosome/pPl 86 binding capacity of RM and EKRM. pPl 86 was translated in the absence of microsomes and, following translation, 20-μl aliquots of the translation were added to the indicated quantities of either RM or EKRM and a binding reaction performed for 10 min at 25°C. Samples were diluted sevenfold using a buffer containing 25 mM K-Hepes, pH 7.2, 110 mM KOAc, and 2.5 mM Mg(OAc)2, and overlayed onto a 0.5 M sucrose cushion. RM were collected by centrifugation (6 min, 60,000 rpm, TLA 100 rotor, 4°C). Supernatants were fractionated by ammonium sulfate precipitation and prepared for SDS-PAGE as described in Materials and Methods. [35S] pPl 86 was quantitated using a Fuji MacBAS1000 phosphorimaging system. A digital image of the dried gels is depicted in A, and the data depicted graphically in B.
Characteristics of Ribosome/pPl 86 Binding
The results of the binding capacity experiments shown in Fig. 6 indicated that at limiting RM concentrations (0.25 eq.), the molar quantity of targeted and bound ribosome/ pPl 86 exceeded that of protease-accessible Sec61p. To further characterize the bound state observed under these conditions, four components of the binding reaction were examined; dependence on the SRP receptor (SRα) activity, protease accessibility of the nascent chain, salt sensitivity of ribosome/pPl 86 binding, and nascent chain proximity to Sec61p. It is well established that under standard assay conditions, membrane-associated pPl 86 is highly resistant to proteolytic digestion and remains associated with the RM following extraction with high salt (Connolly and Gilmore, 1986; Nicchitta and Blobel, 1989; Jungnickel and Rapoport, 1995). Conversely, binding of shorter ribosome/ pPl truncations (51-64 residues) has been shown to be salt and protease sensitive (Jungnickel and Rapoport, 1995).
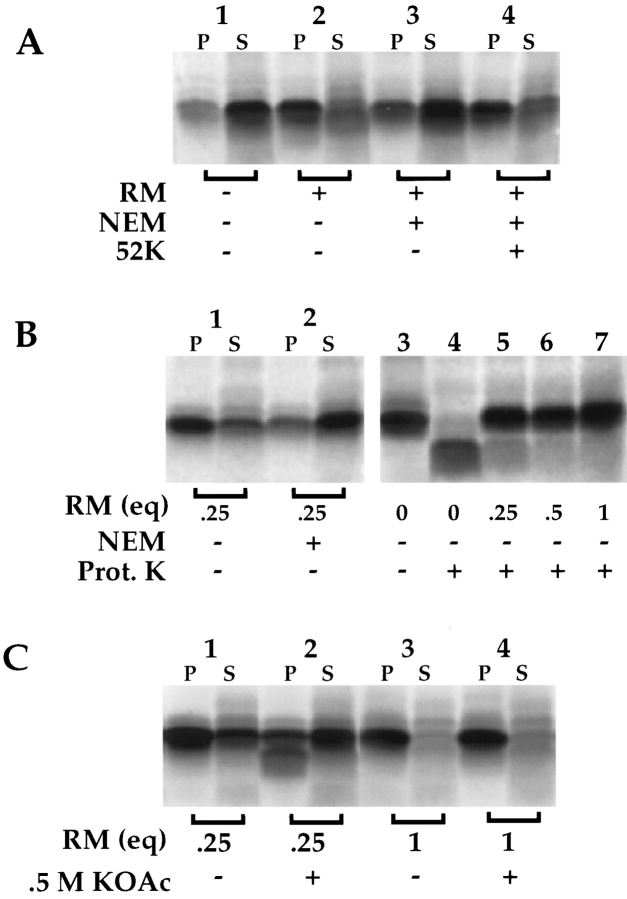
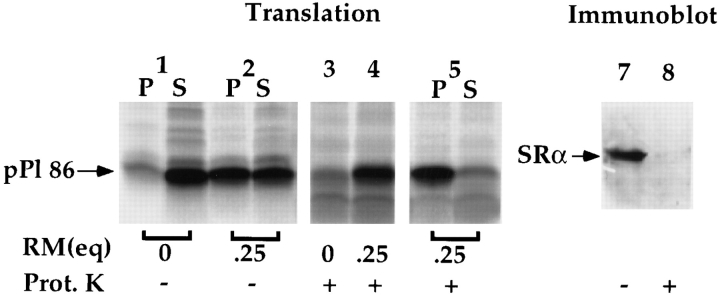
To determine whether the binding observed at saturating nascent chain/membrane ratios was dependent on SRα, and thus represented physiological targeting, RM were first treated with N-ethylmaleimide (NEM) to inactive SRα (Gilmore et al., 1982b ; Nicchitta and Blobel, 1989). After NEM treatment, RM were tested for their ability to support ribosome/nascent chain targeting and binding. As depicted in Fig. 7 A, in the presence of 1 eq. of RM, >80% of the pPl 86 is recovered with the RM in the pellet fraction (lane 2) whereas <15% of the pPl 86 is recovered in the membrane fraction when translation is performed in the presence of NEM-treated membranes (lane 3). Addition of the cytoplasmic domain of SRα to NEM-treated RM before targeting yielded reconstitution of binding activity (Fig. 7 A, lane 4). These data indicate that the pPl 86 binding observed in the reticulocyte lysate/canine RM system is strictly dependent upon the activity of SRα and is therefore specific.
Figure 7.
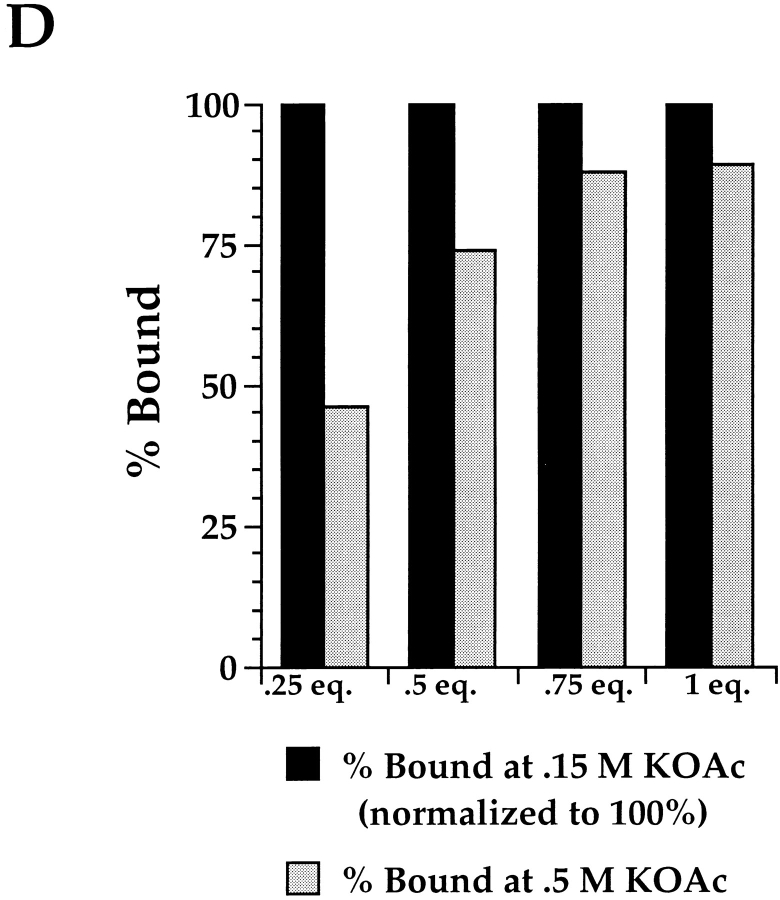
Characteristics of ribosome/pPl 86 binding. (A) Dependence on SRα activity. pPl 86 translations were performed in the absence of RM (lane 1) or the presence of RM (lane 2). An identical translation was done in the presence of NEM-treated RM (lanes 3 and 4) (see Materials and Methods). In lane 4, the NEMtreated membranes were reconstituted with the 52-kD fragment of SRα before translation. After translation, samples were diluted sevenfold, layered over 0.5 M sucrose, and processes by centrifugation, as described in the legend to Fig. 3. Pellet and supernatant samples were processed as previously described and resolved on SDS-PAGE gels. (B) Dependence on SRα activity at 0.25 eq. RM. pPl 86 translations were performed in the presence of 0.25 eq. RM (lane 1), or NEM-treated RM (lane 2). In lanes 3–7, pPl 86 was translated either in the absence of RM (lanes 3 and 4), or in the presence of 0.25 (lane 5), 0.5 (lane 6) or 1.0 eq. of RM (lane 7). Subsequent to translation, samples were either left untreated (lane 3) or digested with proteinase K (100 μg/ml) for 30 min at 4°C (lanes 4–7). Samples were resolved by SDS-PAGE (C) Saltextraction of bound translation products. pPl 86 was translated in the presence of 0.25 (lanes 1 and 2) or 1.0 eq. RM (lanes 3 and 4). After translation, samples 2 and 4 were diluted sevenfold in buffer yielding a final concentration of 0.5 M KOAc. After a 15-min incubation at 4°C, the reactions were fractionated by centrifugation, as described in the legend to Fig. 3, and pellet and supernatant samples processed for SDS-PAGE. Quantitation was performed by phosphorimager analysis; all translation products were included in the analyses. (D) Graph of data described in C, with the inclusion of samples containing 0.5 and 0.75 eq. RM. The percent bound at physiological salt (150 mM KOAc) has been normalized to 100%.
As noted, salt- and protease-resistant binding of the pPl 86 to RM is thought to reflect ribosome association with Sec61p and the insertion of the nascent chain into the translocation site, itself defined primarily by Sec61p (Connolly and Gilmore, 1986; Rapoport et al., 1996). Given the suggestion, derived from the binding studies depicted in Fig. 6, that Sec61p was not mediating the entirety of the pPl 86 binding occurring at 0.25 eq. of RM, the binding characteristics of the reaction performed under these conditions were determined. In the experiment shown in Fig. 7 B, lanes 3–7, pPl 86 was synthesized either in the absence of RM (lanes 3 and 4), or in the presence of 0.25 (lane 5), 0.5 (lane 6), or 1.0 eq. (lane 7) of RM. The efficiency of the binding reaction was then assayed by protease accessibility. In the absence of RM, virtually 100% of pPl 86 is sensitive to proteinase K (Fig. 7 B, lanes 3 and 4). The lower molecular weight, limit digestion product seen in lane 4 represents that portion of the pPl 86 which resides within the ribosome, and is thus inaccessible to protease (Malkin and Rich, 1967; Connolly and Gilmore, 1986; Nicchitta and Blobel, 1989). When translation was performed in the presence of 0.25 equivalents of RM, >75% of the pPl 86 was resistant to proteolytic digestion (Fig. 7 B, lane 5). This is equivalent to that fraction recovered in association with the membrane by sedimentation (Fig. 7 B, lane 2). Similarly, when translations were performed in the presence of 0.5 or 1.0 equivalent of RM, ∼90% of pPl 86 was resistant to protease digestion (Fig. 7 B, lanes 6 and 7), corresponding once again to the bound fraction in the sedimentation assay. Note that the binding reactions performed at 0.25 eq. were, as expected, dependent upon functional SRα (Fig. 7 B, lanes 1 and 2). From these data, it is clear that at both saturating (0.25 eq.) and nonsaturating conditions (0.5 eq. and 1.0 eq.), RM bound pPl 86 nascent chains are resistant to proteinase K digestion, a characteristic of membrane inserted nascent chains (Connolly and Gilmore, 1986; Nicchitta and Blobel, 1989; Jungnickel and Rapoport, 1995).
As a means of further characterizing the binding observed at saturation, pPl 86 was synthesized in the presence of either 0.25 or 1 eq. RM, treated with either 0.15 or 0.5 M KOAc, and centrifuged to separate membrane associated and membrane extracted nascent chains. Surprisingly, and as shown in Fig. 7 C, lanes 1 and 2, although >75% of the pPl 86 remains bound to 0.25 eq. RM following extraction with physiological salt, only 35% is membrane associated following extraction with 0.5 M KOAc (Fig. 5 C, lanes 1 and 2, and 5 D). In this experiment, the minor, faster migrating translation product observed in lane 2 P represents signal processed pPl 56-mer which is generated upon salt-dependent dissociation of the ribosomal subunits (Murphy III, E.C., and C.V. Nicchitta, unpublished observations). In contrast to these results, yet in complete agreement with previous studies, pPl 86 synthesized in the presence of 1.0 eq. of RM is completely resistant to extraction with 0.5 M KOAc (Fig. 7 C, lanes 3 and 4) (Connolly and Gilmore, 1986; Nicchitta and Blobel, 1989; Jungnickel and Rapoport, 1995). Thus, when translations are performed under conditions in which the membrane association reaction(s) is/are saturated, the majority of the nascent chains are bound to the membrane, as assayed by protease accessibility, yet are sensitive to extraction with 0.5 M KOAc. These observations define a novel state of ribosome/nascent chain association with the ER membrane.
pPl 86 Bound At Saturation Is a Posttargeting Intermediate
The kinetics and regulation of the transfer of the nascent chain from the SRP/SRα bound state to Sec61p have not been elucidated. In the absence of this information, it must be considered that at binding saturation, a substantial fraction of the bound ribosome/nascent chain complexes may exist in a stable complex with SRP and SRα, and thus comprise targeting, rather than membrane-bound intermediates. To test this hypothesis, translation reactions were performed at saturating ribosome/nascent chain ratios and the interactions between the ribosome/nascent chain complex and SRα investigated.
The bound state, whether obtained at limiting, or nonlimiting RM concentrations, is notable for the remarkable degree of protease resistance displayed by the nascent chain (cf. Fig. 7 B). If, under limiting RM concentrations, the ribosome/nascent chain complex is bound to the RM solely through physical association with SRα, it would be predicted that proteolytic degradation of SRα would result in release of the ribosome/nascent chain complex from the membrane. The experiment depicted in Fig. 8 was performed to test this prediction. pPl 86 was translated in the presence and absence of limiting RM concentrations and, consistent with previous data, was recovered in the pellet fraction when translations were performed in the the presence, but not the absence, of RM (Fig. 8, lanes 1 and 2). Also consistent with previous observations, membrane associated, but not free, pPl 86 was protected from digestion with exogenous proteases (Fig. 8, lanes 3 and 4). Significantly, the entire population of membrane bound nascent chains remained associated with the membrane following protease digestion (Fig. 8, lane 5). Under these conditions, SRα was completely degraded (Fig. 8, lanes 7 and 8). Thus, when pPl 86 is bound to RM at limiting RM concentrations, SRα is fully accessible to proteolytic digestion, yet the association of the ribosome/nascent chain complexes with the membrane is unaltered. These data indicate that the bound form of the pPl 86 obtained at limiting RM concentrations is, in fact, a posttargeting intermediate and does not arise through stable association with SRα.
Figure 8.
Analysis of ribosome/nascent chain-SRα complex formation. pPl 86 was translated in the presence or absence of 0.25 eq. of RM. Aliquots of the translation were removed and the membrane-bound translation products resolved by sedimentation analysis (lanes 1and 2), or subjected to proteolysis with 100 μg/ml proteinase K for 30 min at 4°C, before sedimentation (lanes 3–5). Fractions were processed as described in the legend to Fig. 3. Paired samples were processed in parallel for immunoblot analysis of SRα.
Analysis of pPl 86/Sec61p Interactions
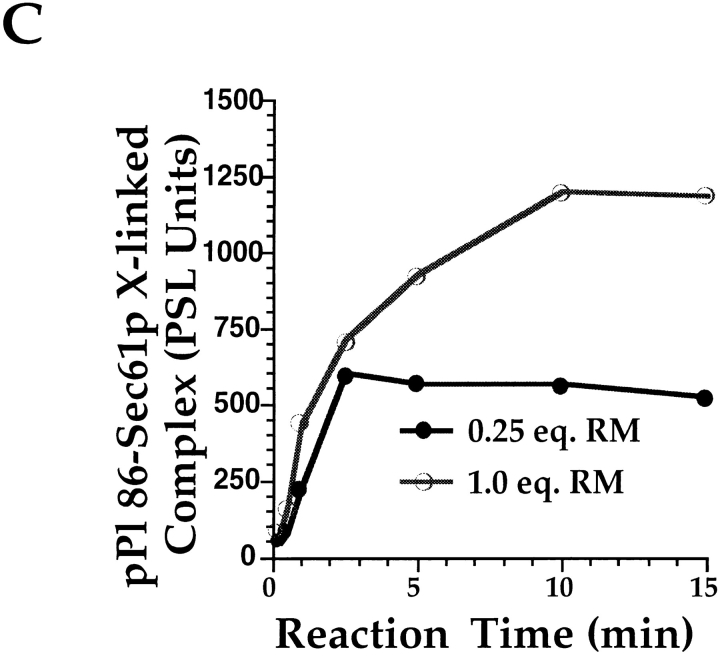
As an additional means of characterizing the binding observed at saturation, chemical cross-linking was employed to assess the molecular environment of the nascent chain. Equivalent amounts of pPl 86 were translated in the presence of 0.25 or 1.0 eq. of RM and chilled to 4°C. RM were recovered by centrifugation, resuspended, and treated with the hetero-bifunctional chemical cross-linker, m-maleimidobenzoyl-N-hydroxysuccinimide ester (MBS) for time periods ranging from 15 s–15 min. As depicted in Fig. 9, MBS treatment of ribosome/pPl 86 RM complexes at 0.25 eq. (saturation) and 1.0 eq. resulted in the formation of a 43-kD cross-linked species, previously demonstrated to be comprised predominantly of Sec61p (Nicchitta et al., 1995). This complex was seen at all time points assayed, at both saturating (Fig. 9 A) and nonsaturating (Fig. 9 B) conditions, and reached a maximum within the time frame of the experiment (Fig. 9 C). Quantitation of the pPl 86/Sec61p cross-linked complex formed under saturating and nonsaturating conditions revealed that twice as much pPl 86 became cross-linked to Sec61p under nonsaturating conditions (Fig. 8 C). These data suggest that when membrane binding sites are limiting, the topological relationship between the population of bound ribosome/pPl 86 complexes and Sec61p is significantly altered from that when membrane binding sites are in excess. At saturation, it appears that a significant fraction of the bound ribosome/ pPl 86 complexes are not in the physical proximity of Sec61p.
Figure 9.
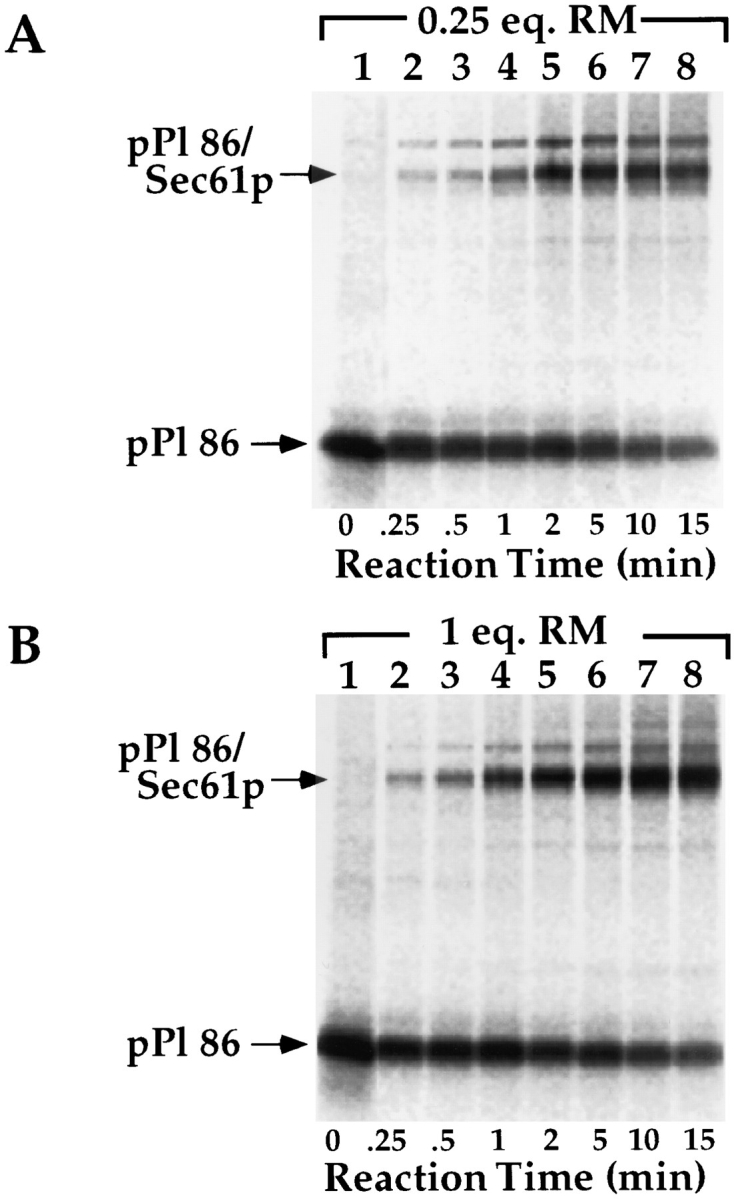
Cross-linking of bound pPl 86 to Sec61p at 0.25 eq. RM and 1.0 eq. RM. pPl 86 was translated either in the presence of 0.25 (A) or 1.0 eq. (B) of RM and processed for cross-linking with the heterobifunctional cross-linker MBS, as detailed in Materials and Methods. Cross-linking reaction times ranged from 15 s–15 min, and reactions were quenched by addition of 1 vol of PBS supplemented with 50 mM DTT, 50 mM lysine, 1% SDS. Samples were TCA precipitated and resolved on 12.5% SDS-PAGE gels. The positions of pPl 86 and the cross-linked pPl 86/Sec61p are indicated. Quantitation was by phosphorimager analysis (Fuji MacBAS 1000). (C) Graph of data derived from A and B.
Discussion
In this communication, we report the identification of a novel stage of ribosome/nascent chain binding to the ER membrane. Although independent of the apparent ribosome occupancy state of Sec61p, the described binding event yields a nascent chain which is protected from digestion with exogenous proteases, an established characteristic of translocation competent, membrane-bound ribosome/nascent chain complexes (Connolly and Gilmore, 1986; Nicchitta and Blobel, 1989; Jungnickel and Rapoport, 1995). The identification of this binding reaction was dependent upon two experimental manipulations. One, and in contrast to most previous studies, ribosome binding was assayed as the functional association of targeted ribosome/ nascent chain complexes with the ER membrane, rather than binding of translationally inactive ribosomes (Borgese et al., 1974; Savitz and Meyer, 1990; Kalies et al., 1994). Secondly, binding reactions were performed as a function of membrane concentration, which, under conditions where membrane binding sites were limiting, revealed a novel, protease resistant, salt-sensitive association of the nascent chain with the ER membrane.
The primary objective of these studies was to characterize ribosome/nascent chain binding using translationally active, nascent chain bearing ribosomes and native rough microsomes, an approach distinct from previous ribosomes binding studies (Borgese et al., 1974; Savitz and Meyer, 1990; Kalies et al., 1994). Two experimental observations warranted this approach. Foremost, binding of nascent chainbearing ribosomes to RM is known to require a specific targeting event, which limits ribosome binding to those ribosomes synthesizing signal or topogenic sequence bearing nascent chains. Furthermore, native RM, although richly endowed with bound ribosomes, are fully functional in vitro, and thus must contain binding sites relevant to the translocation reaction. In addition to these experimental considerations, the observation that translocation in the mammalian ER is strictly cotranslational indicates that the mechanism and regulation of ribosome binding to the ER membrane is intimately related to the mechanism and regulation of protein translocation (Blobel and Dobberstein, 1975).
Current models of the mechanism of protein translocation into the ER indicate that vectorial translocation is a consequence of topological restriction. In these models, a tight junction between the ribosomal nascent chain exit site and components of the protein conducting channel functions to restrict transit of the nascent chain to the ER lumen (Walter and Johnson, 1994; Rapoport et al., 1996). This model is founded upon established experimental observations. For example, translocation-competent binding of truncated preprolactin precursors is accompanied by conversion of the nascent chain from a protease-sensitive, to a protease-insensitive state, data interpreted to be indicative of a tight junction (Connolly and Gilmore, 1986; Nicchitta and Blobel, 1989; Jungnickel and Rapoport, 1995). Subsequently, it was observed that the resident ER membrane protein Sec61p, is highly enriched in the ribosome fraction of solubilized ER membranes and is thus a likely candidate for the ribosome receptor (Görlich et al., 1992). Sec61p has also reported to reside in close physical proximity to translocating secretory and membrane protein precursors (Thrift et al., 1991; Görlich et al., 1992; High et al., 1993; Mothes et al., 1994; Do et al., 1995; Jungnickel and Rapoport, 1995; Nicchitta et al., 1995). In addition, Kalies et al. (1994) observed high affinity binding of inactive ribosomes to Sec61p and demonstrated that inactive ribosomes, in binding to Sec61p, protect it from proteolytic degradation (Kalies et al., 1994). Direct evidence for tight ribosomemembrane coupling was reported in studies of the quenching kinetics of nascent chains bearing fluorescent reporter groups (Crowley et al., 1993, 1994). In these studies, the fluoresence of membrane-bound short nascent chains bearing reporter groups were demonstrated to be insensitive to collisional quenching by exogenous iodide ions, indicating that the ribosome-membrane junction is essentially contiguous. These data support a model in which the ribosome is tightly coupled to the ER membrane through direct physical interactions with Sec61p (Walter and Johnson, 1994; Rapoport et al., 1996). While it is clear that binding of ribosome/nascent chain complexes culminates in a tight interaction with Sec61p, the mechanism by which this stage is ultimately achieved is not. The answer to this question bears significant ramifications on the mechanism and function of ribosome binding to the ER membrane.
We observed that in both canine and porcine RM, Sec61p was largely protected from protease digestion by bound ribosomes. Conversely, treatment of canine and porcine RM with EDTA and 0.5 M KOAc, conditions known to dissociate ribosomes from the ER membrane, resulted in a dramatic increase in the sensitivity of Sec61p to proteolytic digestion. However, comparison of the binding capacity of RM and EKRM for ribosome/nascent chain complexes indicated that the binding capacity did not correlate with the ribosome occupancy state of Sec61p. This was surprising, as we had expected that removal of bound ribosomes from Sec61p would increase the total ribosome/nascent chain binding capacity. These data could be reconciled, however, if ribosome/nascent chain binding were comprised of sequential Sec61p independent and Sec61p dependent stages. Such a proposal would also relieve the stoichiometric constraints imposed by the observations, from recent imaging studies of the ribosome/Sec61p complex, that a single ribosome binds three Sec61p complexes (Hanein et al., 1996).
Because it has been reported that inactive ribosomes bind to Sec61p with nanomolar affinity (Kalies et al., 1994), we reasoned that if the described binding stage was independent of Sec61p, isolated ribosomes would not compete for binding of targeted ribosome/nascent chain complexes. In support of this hypothesis, addition of up to a 350-fold molar excess of purified 80 S ribosomes failed to yield substantial competition for binding. In contrast, addition of a 1.5-fold molar excess of unlabeled ribosome/pPl 86 resulted in efficient binding competition. These results lend credence to a model in which the initial binding event can occur independent of interactions with Sec61p. Most significantly, and in context of current models of the mechanism of protein translocation, these data suggest that formation of a protease-resistant ribosome-membrane junction can precede physical interaction of the ribosome with Sec61p. We have been as yet unable to identify a role for the ribosome receptors p180 and p34 in the binding of active, ribosome/nascent chain complexes. However, as both p180 and p34 were identified in binding studies using inactive ribosomes, it is reasonable to speculate that the binding of active and inactive ribosomes are independently regulated. In support of this, we have observed that under conditions in which ribosome/nascent chain binding is saturated, there is no apparent decrease in the relative protease sensitivity of p180 or p34 (Murphy, E.C., and C.V. Nicchitta, unpublished observations).
To further characterize this novel binding event, binding reactions were performed as a function of membrane concentration and the characteristics of the bound nascent chain analyzed by established criteria. As has been shown in previous reports, bound ribosome/pPl 86 was highly protease and salt resistant when binding was performed in the presence of 1 eq. of RM (Connolly and Gilmore, 1986; Nicchitta and Blobel, 1989; Jungnickel et al., 1995). However, when the same criteria were applied to samples containing 0.25 eq. RM, it was found that, while protease resistant, the majority of bound ribosome/pPl 86 was salt extractable. That the binding obtained at 0.25 eq. is dependent upon physiological targeting is supported by the observation that bound nascent chains are protease resistant, and that binding is blocked by treatment of the RM with NEM, which, at the concentrations used, is known to inactivate the SRP receptor α subunit (Gilmore et al., 1982; Nicchitta and Blobel, 1989).
Could this novel binding stage represent a continued interaction of the ribosome/nascent chain/SRP complex with SRα? All available evidence indicates that the interaction of SRα with nascent chain–bound SRP results in SRP release (Gilmore et al., 1982a ,b; Miller et al., 1993; Bacher et al., 1996). Furthermore, it is not clear how such a continued interaction would yield a protease resistant form of the precursor, given that in the absence of RM, the nascent chain, with bound SRP, is readily digested by exogenous proteases (Connolly and Gilmore, 1986; Nicchitta and Blobel, 1989; Jungnickel and Rapoport, 1995; Fig. 5). In addition, we observed that at saturation, SRα can be readily and fully degraded by exogenous proteases, yet the ribosome/nascent chain complexes remain bound to the RM. Given these considerations, we favor a model in which the initial targeting reaction places the nascent chain in a protease resistant, yet salt extractable, environment. We postulate that at this stage, the ribosome is binding to factors other than Sec61p, and that the signal sequence is associated with components of the ER membrane through hydrophobic and electrostatic interactions, perhaps directly involving the lipid bilayer. At a subsequent stage, occurring coincident with the interaction of the ribosome with Sec61p, the nascent chain is stably inserted into the translocon and assumes a translocation competent conformation. This model predicts an alternative binding site on the ER membrane for targeted ribosome-nascent chain complexes. The identity of this site is currently under investigation.
We have provided direct evidence for the existence of a novel stage of ribosome association with the ER membrane. The identification of this stage has significant ramifications for proposed mechanisms of protein translocation. First, it provides a means by which the translocon can distinguish between cytosolic, free ribosomes and ribosomes engaged in the synthesis of secretory or membrane protein substrates. The translocon can, in effect, select ribosomes which are membrane associated via this novel stage, thereby excluding those ribosomes which are cytosolic. In the absence of such a mechanism, one would expect competition between free ribosomes and ribosomes synthesizing signal-bearing nascent chains (Nicchitta, 1996). Furthermore, we postulate that this novel stage provides the nascent chain the opportunity to attain a translocation competent topology, perhaps through recruitment of translocon components in a manner first proposed in the signal hypothesis (Blobel and Dobberstein, 1975). In this scenario, only those precursor proteins that have properly assembled into the ER membrane are available for subsequent translocation by the translocon.
Acknowledgments
We thank Drs. Edwin C. Murphy Jr., T. Bell, M. Likker, and S. Dias for critical comments, advice, and support through the course of these studies. We also wish to express our gratitude to Dr. David I. Meyer, for providing antibodies directed against p180 and helpful advice, and Dr. Stephen High, for providing antibodies to p34.
This work was supported by National Institutes of Health grant DK47897 (CVN).
Abbreviations used in this paper
- EKRM
KOAc and EDTA washed RM
- NAC
nascent chain–associated protein complex
- NEM
N-ethyl maleimide
- RM
rough microsome
- SRP
signal recognition particle
References
- Adelman MR, Sabatini DD, Blobel G. Ribosome-membrane interaction: I. Non-destructive disassembly of rat liver rough microsomes into ribosomal and membranous components. J Cell Biol. 1973;56:206–229. doi: 10.1083/jcb.56.1.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacher G, Lutcke H, Jungnickel B, Rapoport TA, Dobberstein B. Regulation by the ribosome of the GTPase of the signal recognition particle during protein targeting. Nature (Lond) 1996;381:248–251. doi: 10.1038/381248a0. [DOI] [PubMed] [Google Scholar]
- Blobel G, Dobberstein B. Transfer of proteins across membranes II. Reconstitution of functional rough microsomes from heterologous components. J Cell Biol. 1975;67:852–862. doi: 10.1083/jcb.67.3.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgese D, Blobel G, Sabatini DD. In vitro exchange of ribosomal subunits between free and membrane-bound ribosomes. J Mol Biol. 1973;74:415–438. doi: 10.1016/0022-2836(73)90037-5. [DOI] [PubMed] [Google Scholar]
- Borgese N, Mok W, Kreibich G, Sabatini DD. Ribosomal-membrane interaction: in vitro binding of ribosomes to microsomal membranes. J Mol Biol. 1974;88:559–580. doi: 10.1016/0022-2836(74)90408-2. [DOI] [PubMed] [Google Scholar]
- Brundage L, Hendrick JP, Schiebel E, Driessen AJM, Wickner W. The purified E.coliintegral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- Collins PG, Gilmore R. Ribosome binding to the endoplasmic reticulum-a 180 kD protein identified by cross-linking to membrane-bound ribosomes is not required for ribosome binding activity. J Cell Biol. 1991;114:639–649. doi: 10.1083/jcb.114.4.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly T, Gilmore R. Formation of a functional ribosome-membrane junction during translocation requires the participation of a GTPbinding protein. J Cell Biol. 1986;103:2253–2261. doi: 10.1083/jcb.103.6.2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley KS, Reinhart GD, Johnson AE. The signal sequence moves through a ribosomal tunnel into a noncytoplasmic aqueous environment at the ER membrane early in translocation. Cell. 1993;73:1101–1115. doi: 10.1016/0092-8674(93)90640-c. [DOI] [PubMed] [Google Scholar]
- Crowley KS, Liao S, Worrell VE, Reinhart GD, Johnson AE. Secretory proteins move through the endoplasmic reticulum membrane via an aqueous, gated pore. Cell. 1994;78:461–471. doi: 10.1016/0092-8674(94)90424-3. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Schekman R. A yeast mutant defective at an early stage in import of secretory protein precursors into endoplasmic reticulum. J Cell Biol. 1987;105:633–645. doi: 10.1083/jcb.105.2.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do H, Falcone D, Lin J, Andrews KW, Johnson AE. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- Florini JR, Breuer CB. Amino acid incorporation into protein by cell-free systems from rat skeletal muscle. V. Effects of pituitary growth hormone on activity of ribosomes and ribonucleic acid polymerase in hypophysectomized rates. Biochemistry. 1966;5:1870–1876. doi: 10.1021/bi00870a013. [DOI] [PubMed] [Google Scholar]
- Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. JCell Biol. 1982a;95:461–469. doi: 10.1083/jcb.95.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmore R, Walter P, Blobel G. Protein translocation across the endoplasmic reticulum. II. Isolation and characterization of the signal recognition particle receptor. J Cell Biol. 1982b;95:470–477. doi: 10.1083/jcb.95.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görlich D, Rapoport TA. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell. 1993;75:615–630. doi: 10.1016/0092-8674(93)90483-7. [DOI] [PubMed] [Google Scholar]
- Görlich D, Prehn S, Hartmann E, Kalies KU, Rapoport TA. A mammalian homolog of SEC61p and SECYp is associated with ribosomes and nascent polypeptides during translocation. Cell. 1992;71:489–503. doi: 10.1016/0092-8674(92)90517-g. [DOI] [PubMed] [Google Scholar]
- Hanein D, Matlack KES, Jungnickel B, Plath K, Kalies K-U, Miller KR, Rapoport TA, Akey CW. Oligomeric rings of the Sec61p complex induced by ligands required for protein translocation. Cell. 1996;87:721–732. doi: 10.1016/s0092-8674(00)81391-4. [DOI] [PubMed] [Google Scholar]
- High S, Andersen SS, Görlich D, Hartmann E, Prehn S, Rapoport TA, Dobberstein B. Sec61p is adjacent to nascent type I and type II signal-anchor proteins during their membrane insertion. J Cell Biol. 1993a;121:743–750. doi: 10.1083/jcb.121.4.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High S, Martoglio B, Görlich D, Andersen SS, Ashford AJ, Giner A, Hartmann E, Prehn S, Rapoport TA, Dobberstein B. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J Biol Chem. 1993b;268:26745–26751. [PubMed] [Google Scholar]
- Ichimura T, Ohsumi T, Shindo Y, Ohwada T, Yagame H, Momose Y, Omata S, Sugano H. Isolation and some properties of a 34 kD membrane protein that may be essential for ribosome binding in rat liver rough microsomes. FEBS Lett. 1992;296:7–10. doi: 10.1016/0014-5793(92)80391-s. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Hunt T. Preparation and use of nuclease-treated rabbit reticulocyte lysates for the translation of eukaryotic messenger RNA. Methods Enzymol. 1983;96:50–74. doi: 10.1016/s0076-6879(83)96008-1. [DOI] [PubMed] [Google Scholar]
- Jungnickel B, Rapoport TA. A posttargeting signal sequence recognition event in the endoplasmic reticulum. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Kalies KU, Görlich D, Rapoport TA. Binding of ribosomes to the rough endoplasmic reticulum mediated by the Sec61p-complex. J Cell Biol. 1994;126:925–934. doi: 10.1083/jcb.126.4.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreibich G, Ulrich BC, Sabatini DD. Proteins of rough microsomal membranes related to ribosome binding. I. Identification of ribophorins I and II, membrane proteins characteristic of rough microsomes. J Cell Biol. 1978;77:464–487. doi: 10.1083/jcb.77.2.464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring B, Kreibich G, Wiedmann M. The intrinsic ability of ribosomes to bind to endoplasmic reticulum membranes is regulated by signal recognition particle and nascent-polypeptide-associated complex. Proc Natl Acad Sci USA. 1995a;92:9435–9439. doi: 10.1073/pnas.92.21.9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring B, Sakai H, Kreibich G, Wiedmann M. Nascent polypeptide-associated complex prevents mistargeting of nascent chains to the endoplasmic reticulum. Proc Natl Acad Sci USA. 1995b;92:5411–5415. doi: 10.1073/pnas.92.12.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malkin LI, Rich A. Partial resistance of nascent polypeptide chains to proteolytic digestion due to ribosome shielding. J Mol Biol. 1967;26:329–346. doi: 10.1016/0022-2836(67)90301-4. [DOI] [PubMed] [Google Scholar]
- Martin TE, Rolleston FS, Low RB, Wool IG. Dissociation and reassociation of skeletal muscle ribosomes. J Mol Biol. 1969;43:135–149. doi: 10.1016/0022-2836(69)90084-9. [DOI] [PubMed] [Google Scholar]
- Martoglio B, Hofmann MW, Brunner J, Dobberstein B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell. 1995;81:201–214. doi: 10.1016/0092-8674(95)90330-5. [DOI] [PubMed] [Google Scholar]
- Meyer DI, Krause E, Dobberstein B. Secretory protein translocation across membranes-the role of the “docking protein.” . Nature (Lond) 1982;297:647–650. doi: 10.1038/297647a0. [DOI] [PubMed] [Google Scholar]
- Miller JD, Wilhelm H, Gierasch L, Gilmore R, Walter P. GTP binding and hydrolysis by the signal recognition particle during initiation of protein translocation. Nature (Lond) 1993;366:351–354. doi: 10.1038/366351a0. [DOI] [PubMed] [Google Scholar]
- Mothes W, Prehn S, Rapoport TA. Systematic probing of the environment of a translocating secretory protein during translocation through the ER membrane. EMBO (Eur Mol Biol Organ) J. 1994;13:3973–3982. doi: 10.1002/j.1460-2075.1994.tb06713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta CV. Protein translocation in the endoplasmic reticulum: the search for a unified molecular mechanism. Sem Cell Dev Biol. 1996;7:497–503. [Google Scholar]
- Nicchitta CV, Blobel G. Nascent chain binding and translocation are distinct processes: differentiation by chemical alkylation. J Cell Biol. 1989;108:789–795. doi: 10.1083/jcb.108.3.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicchitta CV, Blobel G. Lumenal proteins of the mammalian endoplasmic reticulum are required to complete protein translocation. Cell. 1993;73:989–998. doi: 10.1016/0092-8674(93)90276-v. [DOI] [PubMed] [Google Scholar]
- Nicchitta CV, Murphy EC, III, Haynes R, Shelness GS. Stage and ribosome-specific alterations in nascent chain-Sec61p interactions accompany translocation across the ER membrane. J Cell Biol. 1995;129:957–970. doi: 10.1083/jcb.129.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari JM, Zimmerman DL, Ogg SC, Walter P. Characterization of the rough endoplasmic reticulum ribosome-binding activity. Nature (Lond) 1991;352:638–640. doi: 10.1038/352638a0. [DOI] [PubMed] [Google Scholar]
- Rapoport TA, Jungnickel B, Katay U. Protein transport across eukaryotic endoplasmic reticulum and bacterial inner membranes. Annu Rev Biochem. 1996;65:271–303. doi: 10.1146/annurev.bi.65.070196.001415. [DOI] [PubMed] [Google Scholar]
- Rothblatt JA, Deshaies RJ, Sanders SL, Daum G, Schekman R. Multiple genes are required for proper insertion of secretory proteins into the endoplasmic reticulum in yeast. J Cell Biol. 1989;72:61–68. doi: 10.1083/jcb.109.6.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DD, Blobel G. Controlled proteolysis of nascent polypeptides in rat liver cell fractions. II. Location of the polypeptides within ribosomes. J Cell Biol. 1970;45:130–145. doi: 10.1083/jcb.45.1.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini DD, Tashiro Y, Palade GE. On the attachment of ribosomes to microsomal membranes. J Mol Biol. 1966;19:503–524. doi: 10.1016/s0022-2836(66)80019-0. [DOI] [PubMed] [Google Scholar]
- Savitz AJ, Meyer DI. Identification of a ribosome receptor in the rough endoplasmic reticulum. Nature (Lond) 1990;346:540–544. doi: 10.1038/346540a0. [DOI] [PubMed] [Google Scholar]
- Savitz AJ, Meyer DI. 180-kD ribosome receptor is essential for both ribosome binding and protein translocation. J Cell Biol . 1993;120:853–863. doi: 10.1083/jcb.120.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirling CJ, Rothblatt J, Hosobuchi M, Deshaies RJ, Schekman R. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol Biol Cell. 1992;3:129–142. doi: 10.1091/mbc.3.2.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tazawa S, Unuma M, Tondokoro Y, Asano T, Ohsumi Y, Ichimura T, Sugano H. Identification of a membrane protein responsible for ribosome binding in rough microsomal membranes. J Biochem (Tokyo) 1991;109:89–98. [PubMed] [Google Scholar]
- Thrift RN, Andrews DW, Walter P, Johnson AE. A nascent membrane protein is located adjacent to ER membrane proteins throughout its integration and translation. J Cell Biol. 1991;112:809–821. doi: 10.1083/jcb.112.5.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G. Preparation of microsomal membranes for cotranslational protein translocation. Methods Enzymol. 1983;96:84–93. doi: 10.1016/s0076-6879(83)96010-x. [DOI] [PubMed] [Google Scholar]
- Walter P, Johnson AE. Signal sequence recognition and protein targeting to the endoplasmic reticulum membrane. Annu Rev Cell Biol. 1994;10:87–119. doi: 10.1146/annurev.cb.10.110194.000511. [DOI] [PubMed] [Google Scholar]
- Wanker EE, Sun Y, Savitz AJ, Meyer DI. Functional characterization of the 180-kD ribosome receptor in vivo. J Cell Biol. 1995;130:29–39. doi: 10.1083/jcb.130.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weitzmann CJ, Cunningham PR, Ofengand J. Cloning in vitro transcription and biological activity of Eschericia coli23S ribosomal RNA. Nucleic Acids Res. 1990;18:3515–3520. doi: 10.1093/nar/18.12.3515. [DOI] [PMC free article] [PubMed] [Google Scholar]