Abstract
The nuclear lamina is a fibrous structure that lies at the interface between the nuclear envelope and the nucleoplasm. The major proteins comprising the lamina, the nuclear lamins, are also found in foci in the nucleoplasm, distinct from the peripheral lamina. The nuclear lamins have been associated with a number of processes in the nucleus, including DNA replication. To further characterize the specific role of lamins in DNA replication, we have used a truncated human lamin as a dominant negative mutant to perturb lamin organization. This protein disrupts the lamin organization of nuclei when microinjected into mammalian cells and also disrupts the lamin organization of in vitro assembled nuclei when added to Xenopus laevis interphase egg extracts. In both cases, the lamina appears to be completely absent, and instead the endogenous lamins and the mutant lamin protein are found in nucleoplasmic aggregates. Coincident with the disruption of lamin organization, there is a dramatic reduction in DNA replication. As a consequence of this disruption, the distributions of PCNA and the large subunit of the RFC complex, proteins required for the elongation phase of DNA replication, are altered such that they are found within the intranucleoplasmic lamin aggregates. In contrast, the distribution of XMCM3, XORC2, and DNA polymerase α, proteins required for the initiation stage of DNA replication, remains unaltered. The data presented demonstrate that the nuclear lamins may be required for the elongation phase of DNA replication.
The nuclear lamin proteins form a fibrous structure, termed the nuclear lamina, which is concentrated at the nucleoplasmic face of the nuclear envelope (40). The lamins are also found in nucleoplasmic foci, the distribution of which is related to the cell cycle (7, 18, 32, 39, 48). The lamins are highly conserved proteins that are closely related to cytoplasmic intermediate filament (IF)1 proteins and, as such, are classified as type V IF (51). In vertebrates, as many as five lamin proteins have been reported. These are divided into two types, A and B, based on criteria such as expression patterns and exon positions (53). B-type lamins are expressed in all cells, while the A-type lamins are expressed in differentiated cells (40). In Xenopus laevis, there are five or more lamins, also showing cell type–specific expression patterns (3, 40, 54). As is the case with cytoplasmic IF, the lamins have a central rod domain that forms an α helix, composed of heptad repeats. The rod domains are primarily responsible for the higher order lamin–lamin interactions that govern lamin assembly (20). The two non–α-helical end domains are also involved in assembly and may interact with other nuclear structures (see 40, for detailed discussion).
In addition to providing mechanical support to the nucleus and influencing its shape and volume, the lamins appear to interact with other nuclear components and thereby may influence a number of nuclear processes (40). For example, the lamins interact with chromatin in vitro and probably in vivo (2, 16, 22, 47). Through this interaction, the lamins may be involved in DNA replication. For example, lamin B is associated with replicating chromatin in mammalian cells (39). During S phase, lamin B appears not only at the nuclear periphery, but also in nucleoplasmic foci that frequently coincide with sites of bromodeoxyuridine incorporation and the location of the DNA replication factor, PCNA (39). It has also been suggested on morphological grounds that a filamentous network of lamins acts as a scaffold for “DNA replication factories” (24, 25).
Further evidence for a role of the nuclear lamins in DNA replication comes from the nuclear assembly system prepared from Xenopus laevis eggs (3, 4, 30). In this system, nuclei rapidly assemble when DNA or chromatin is added to interphase egg extracts. These nuclei carry out processes such as nuclear import, lamin assembly, and DNA replication. The presence of highly concentrated, soluble, nuclear components in the extract makes the Xenopus system particularly useful for biochemical manipulations. Proteins from a wide range of species, including yeast and humans, have been added to interphase and mitotic extracts to examine their function in the regulation of the cell cycle (41). In addition, immunodepletion of specific proteins from these extracts has been used to determine their involvement in nuclear functions. For example, when the major endogenous lamin (lamin B3) is immunodepleted from interphase extracts, nuclei form, but they cannot replicate their DNA (34, 45). Furthermore, when the eluted lamin B3 is added back to depleted extracts, nuclear DNA replication is restored (17). These results suggest that nuclear lamins play a role in DNA replication, although it is unclear how or at what stage DNA synthesis is blocked under these experimental conditions.
The Xenopus nuclear assembly system has also been useful in characterizing other proteins involved in regulating DNA replication. Immunodepletion experiments involving the removal of XMCM3 and XORC2, as well as a number of other factors, have demonstrated that they are essential for DNA replication. XMCM3, for example, is a putative component of the licensing factor that is thought to limit replication to one round for each cell cycle. It binds to chromatin early in the process of nuclear assembly, before the nuclear membrane forms (10, 29, 33). XORC2 is the Xenopus homologue of the yeast protein, ORC2 (9). In yeast, this protein is required to initiate DNA synthesis and is part of a complex that binds to origins of replication (55). In Xenopus, XORC2 binds to chromatin before nuclear envelope formation and appears to be involved in the initiation of DNA synthesis (9). Other proteins involved in replication have also been identified as constituents of the Xenopus system, including PCNA (26), a required cofactor of DNA polymerase δ. This polymerase is responsible for the elongation phase of DNA replication (59).
The immunodepletion approach has been extremely valuable in defining roles for the lamins as well as for other proteins in DNA synthesis, but this method has several limitations. For example, in the case of the nuclear lamins, it is difficult to completely immunodeplete lamin proteins (31). In addition, any effects seen after immunodepletion may not be due to the removal of targeted antigenic components, but rather to the coimmunoprecipitation of bound, associated proteins. Immunodepletions of XMCM3, XORC2, and lamin B3 all result in the specific removal of other proteins, in addition to the antigen targeted by the antibody (9, 10, 17, 33).
To define more precisely the functions of nuclear lamins in nuclear assembly and DNA replication, we have developed a method that avoids some of the pitfalls inherent in the immunodepletion techniques. Our approach uses a human lamin mutant as a dominant negative disrupter of the nuclear lamin organization in two experimental systems. When this mutant protein is microinjected into mammalian cells or added to the Xenopus nuclear assembly extract, the lamin organization is disrupted. The mutant protein as well as the endogenous lamins appear to colocalize in nucleoplasmic aggregates, with little or no detectable lamin protein at the nuclear periphery. We have concentrated our efforts on defining the effects of this disruption in nuclei assembled in Xenopus extracts, since more nuclei can be studied under conditions permitting coordinated biochemical and morphological assays. As in the case for Xenopus nuclei assembled after lamin B3 immunodepletion (34, 45), the disrupted nuclei described in this study cannot complete DNA replication. However, because the lamins are retained within the nucleus, we are able to examine their distribution relative to other DNA replication markers. We find that, as a consequence of the disruption, there is an altered distribution of proteins specifically involved in the elongation phase, but not in the initiation phase of DNA replication.
Materials and Methods
Expression of Human Lamins in Escherichia coli
Full-length human lamin A (LA) and ΔNLA were cloned in a pET-derived vector and expressed in the NovaBlue (DE3) strain of E. coli (Novagen, Madison, WI). ΔNLA lacks the first 33 amino acids of human lamin A. Both of the expressed proteins were purified by ion-exchange chromatography as described previously (38). The protein in column buffer (6 M urea, 25 mM Tris, 2 mM EDTA, and 1 mM DTT) was dialyzed against PB (300 mM NaCl, 25 mM Tris Base, pH 9, and 1 mM DTT). After dialysis, SDS-PAGE analysis of the resulting protein solution showed the presence of one major band of protein. In the case of ΔNLA, the protein had a molecular mass of ∼69 kD (Fig. 1 a). This 69-kD protein reacted with a rabbit polyclonal antibody directed against human lamins A/C as demonstrated by immunoblotting (Fig. 1 b). SDS-PAGE and blotting analyses were carried out as described in (39). The relatively minor bands seen in the immunoblot (Fig. 1 b) are due to a small amount of proteolysis that is present in all preparations of nuclear lamins (38). The same gel profiles are seen for the wild-type LA protein, but the apparent molecular mass is 72 kD (not shown). The protein solutions were aliquoted and stored at −80°C at a final concentration of 2 mg/ml. Before use in microinjection experiments and nuclear assembly assays, samples were centrifuged at 20,000 g for 10 min at room temperature to remove insoluble material.
Figure 1.
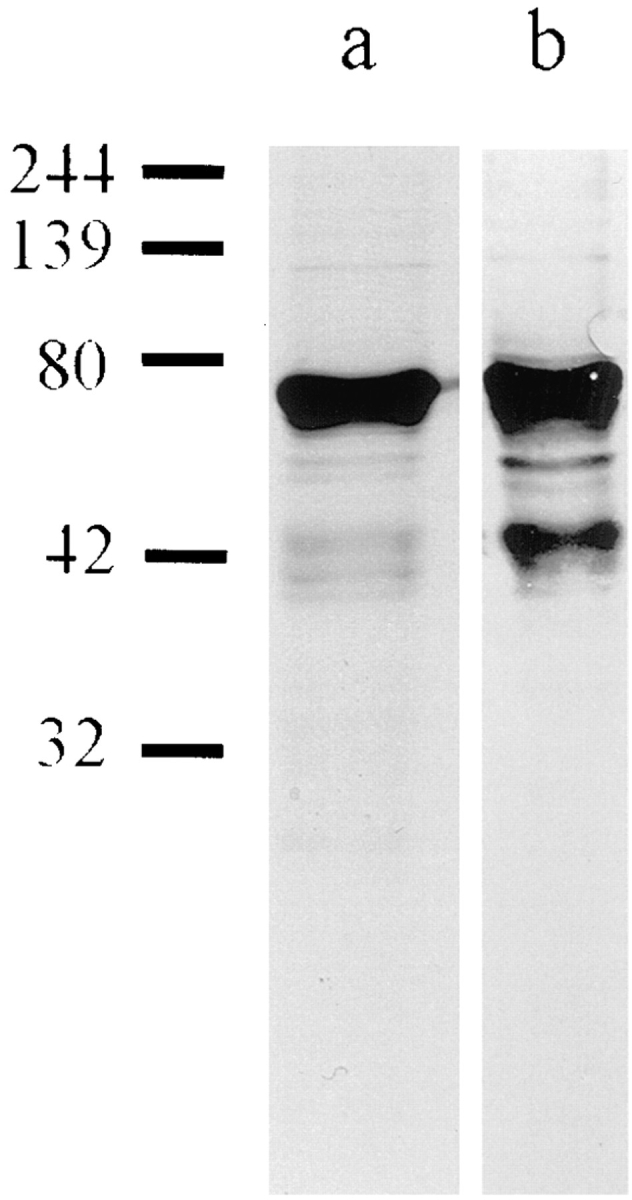
(a) SDS-PAGE of purified ΔNLA and (b) corresponding Western blot using a polyclonal lamin A/C antibody (39). The major band runs at 69 kD, the predicted molecular mass of ΔNLA. There are also minor bands, representing the proteolytic fragments seen in nuclear lamin preparations expressed in E. coli (38). Numbers on the left represent molecular mass size markers in kD.
Microinjection of Mammalian Cells and Analysis by Immunofluorescence
BHK-21 cells were cultured as described elsewhere (18). Single cells were injected with ΔNLA at a concentration of 1 mg/ml in PB (18). Controls consisted of the injection of cells with PB. Cells were fixed in methanol 2 h after microinjection and processed for immunofluorescence as described previously (18). A rat polyclonal antibody directed against human lamins A and C (39) or a rabbit polyclonal antibody directed against human lamin B (39) was diluted 1:100 in PBS for use as a primary antibody for immunofluorescence. Secondary antibodies were diluted 1:50 in PBS and included FITC-labeled goat anti–rat IgG (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA), tetramethyl rhodamine isothiocyanate–labeled goat anti–rabbit IgG, and lissamine rhodamine-labeled donkey anti–rabbit IgG (Kirkegaard & Perry Laboratories, Inc., Gaithersburg, MD).
Xenopus Interphase Extracts and In Vitro Nuclear Assembly Reactions
Xenopus laevis egg interphase extracts were prepared as described in (43). The extract was frozen in liquid nitrogen in 70-μl aliquots. Demembranated chromatin from Xenopus sperm was prepared as described in (42). For all nuclear assembly reactions, aliquots of interphase extract were thawed rapidly and brought to 15 mM Hepes, pH 7.4. For ATP generation, the extract was made 1 mM ATP (Sigma Chemical Co., St. Louis, MO), 10 mM phosphocreatine (Sigma Chemical Co.), and 50 μg/ml creatine phosphokinase (Sigma Chemical Co.) (see 43). Stock solutions of bacterially expressed human LA or ΔNLA in PB were added to the assembly reaction to a final concentration of 200 μg/ml. Controls consisted of the addition of an equal volume of PB to parallel assembly reactions. The volume of protein solution or PB alone added to an assembly reaction was maintained at ⩽10% of the final volume of the reaction mixture. 15 min after the addition of protein or PB to an assembly reaction, demembranated sperm chromatin was added to initiate nuclear assembly. Sufficient sperm chromatin was added to achieve a final concentration of 1,000 nuclei per μl. Unless otherwise specified, nuclei were either fixed for immunofluorescence or prepared for electrophoretic analysis at 90–120 min after initiating the assembly reaction as described below.
In some experiments, assembled nuclei were removed from one assembly reaction and transferred to another. To accomplish this, the assembly reaction mixture was diluted 50-fold with NWB (200 mM sucrose, 15 mM Hepes, pH 7.4, 50 mM NaCl, 2.5 mM MgCl2, and 1 mM DTT), and the nuclei were recovered by centrifugation for 3 min at 1,600 g (5). The resulting pellets were then resuspended in fresh nuclear assembly reaction mixture.
The effects of ΔNLA on assembled nuclei were studied by adding ΔNLA (200 μg/ml final concentration) at a time interval of 90 min after the addition of sperm chromatin to a nuclear assembly reaction. After an additional 45 min, nuclei were fixed and processed for immunofluorescence as described below.
In Vitro DNA Replication Assays
DNA replication in the in vitro assembled nuclei was assayed with fluorescence microscopy by adding 1 mM bio-11-dUTP (Enzo Diagnostics, Farmingdale, NY) to the nuclear assembly reaction to achieve a final concentration of 10 μM (6). The incorporated nucleotides were detected with fluorochrome-tagged streptavidin as described below. DNA synthesis was also assayed by adding 1.5 μCi of [32P]dCTP (6,000 Ci/mM; Amersham Corp., Arlington Heights, IL) to a 15-μl nuclear assembly reaction before the addition of sperm chromatin. DNA replication was measured as described in (12). Briefly, the reactions were stopped, treated with proteinase K, and resolved by electrophoresis on a Tris borate–buffered, 0.8% agarose gel. The gel was dried and exposed for autoradiography and phosphoimaging with a FUJIX BAS 2000 (Fuji Photo Film Co., Tokyo, Japan) to quantitate the amount of [32P]dCTP incorporation.
Preparation of Nuclear Matrices
Nuclear matrices were prepared from the nuclei assembled in vitro as described in (11). Nuclei were assembled in a 100-μl nuclear assembly reaction, and the reaction mixture was diluted with 650 μl of NWB containing 0.5% Triton X-100. DNase I (DPRF; Worthington Biochemical Corp., Freehold, NJ) was added to a final concentration of 8.3 μg/ml, and, after a 10min incubation at 20°C, an additional 750 μl of 4 M NaCl, 20 mM Hepes, pH 7.4, 20 mM EDTA, and 1 mM DTT was added. After 10 min at 20°C, the matrices were fixed at the same temperature by adding 160 μl of 10 mM ethylene bis [succinimidyl succinate] (Pierce Chemical Co., Rockford, IL). The fixation was stopped after 7 min by the addition of 42 μl of 1 M Tris HCl, pH 7.4.
Microscopic Analyses of Xenopus Nuclei Assembled In Vitro
Nuclei assembled in vitro were fixed for 10 min at 20°C by diluting the assembly reaction mixture 10-fold in NWB and adding 0.1 vol of 100 mM ethylene glycol bis [succinimidylsuccinate] in DMSO (35). Fixation was stopped by adding 1 M Tris HCl, pH 7.4, to achieve a final concentration of 25 mM. Alternatively, nuclei were fixed with 4% paraformaldehyde in NWB for staining with XMCM3 (33). Subsequently, nuclei were pelleted onto coverslips as described elsewhere (37). After fixation, coverslips were placed in 0.1% NP-40 or 0.1% Triton X-100 in PBS for 2 min, and then rinsed twice for 2 min in PBS at room temperature. 30-μl aliquots of primary antibodies, diluted 1:20 in PBS, were overlaid onto coverslips. After a 30-min incubation at 37°C, coverslips were washed four times with PBS and incubated for an additional 30 min at 37°C with a 1:50 dilution of the appropriate fluorochrome-labeled secondary antibody. The coverslips were then washed five times in PBS and mounted in 50 mM Tris Base, pH 9.0, 50% glycerol, and 2 mg/ml of p-phenylenediamine (Sigma Chemical Co.).
The rabbit polyclonal sera used for these studies were directed against human lamins A and C (39), XORC2 (9; a gift from William Dunphy, California Institute of Technology, Pasadena), and XMCM3 (33; a gift from Ronald Laskey, Cambridge University, UK). Monoclonal ascites and supernatants used included L6-5D5 directed against Xenopus lamin B3 (52), CRL 1640 directed against DNA polymerase α (57; American Type Culture Collection, Rockville, MD), α RFC 11 directed against the large subunit of replication factor C (8; a gift from Bruce Stillman, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), and PC10 directed against PCNA (Boehringer Mannheim Biochemicals, Indianapolis, IN). The myc 9E10 epitope antibody was also used (13; American Type Culture Collection). The antibodies directed against PCNA, DNA polymerase α, XMCM3, and XORC2 have all been shown to react only with their targeted antigens in Xenopus extracts ( 9, 26, 27, 33). The antibody directed against human lamin A does not cross-react with Xenopus lamin B3 in immunofluorescence assays (data not shown). The secondary antibodies used were FITC-labeled donkey anti–mouse IgG, tetramethyl rhodamine isothiocyanate–labeled donkey anti–rabbit IgG, and lissamine rhodamine-labeled donkey anti–rabbit IgG (Jackson ImmunoResearch Laboratories, Inc.). Detection of bio-11-dUTP incorporation involved the incubation of fixed nuclei for 30 min at 37°C in Texas red– or FITC-labeled streptadivin (Amersham Corp.) at a 1:500 dilution in the presence of 12.5 μg/ml of RNase A (4; Sigma Chemical Co.). Nuclear membranes were stained with the lipophilic dye dihexyloxacarbocyanine (DIOC6) (Molecular Probes, Eugene, OR) at 2.5 μg/ml during the secondary antibody incubation and at 0.25 μg/ml in the mounting medium (44). Nuclear pore proteins were stained by adding FITC-labeled WGA (Sigma Chemical Co.) at 50 μg/ml during secondary antibody incubation (14). DNA was visualized by adding Hoechst dye (Molecular Probes) at 1 μg/ml to the mounting medium. Microscopic observations were carried out on an Axiophot (Carl Zeiss, Inc., Thornwood, NY) equipped with a 35-mm camera or an LSM 410 confocal microscope (Carl Zeiss, Inc.) equipped with an argon/krypton laser. Confocal micrographs were stored on optical disks, and micrographs were printed on an UP-D8800 video printer (Sony Corp., Park Ridge, NJ).
Results
ΔNLA Disrupts Nuclear Lamin Organization In Mammalian Nuclei
To determine the function of nuclear lamins, we sought a method to perturb lamin organization. In previous studies, we found that microinjection of bacterially expressed human LA into mammalian cells resulted in its incorporation into the endogenous nuclear lamin structures (18). This technique allowed us to follow the pathway of nuclear lamin assembly in situ (18). We used this same technique to introduce a mutant lamin protein into BHK-21 cells. The mutant, ΔNLA, lacks the NH2-terminal nonhelical domain (33 amino acids) of LA, but it contains the entire α-helical rod and COOH terminus. Bacterially expressed ΔNLA cannot form typical LA paracrystals in vitro, showing that the NH2 terminus is required for higher order lamin assemblies (21, 38). To determine the effects of ΔNLA in vivo, a solution of this mutant protein (1 mg/ml; see Materials and Methods) was injected into the cytoplasm of BHK cells. Injected cells were fixed after 2 h and then processed for immunofluorescence. As can be seen in Fig. 2, there is a dramatic alteration in the distribution and organization of the nuclear lamins. Instead of producing the typical rim pattern associated with the nuclear envelope as well as nucleoplasmic foci (Fig. 2, a and b), lamins A/C and B colocalize almost exclusively in large nucleoplasmic aggregates in injected cells (Fig. 2, c and d). These results show that ΔNLA acts as a dominant negative mutant that can induce the rapid disruption of the endogenous lamin structures that are composed of both A- and B-type lamins.
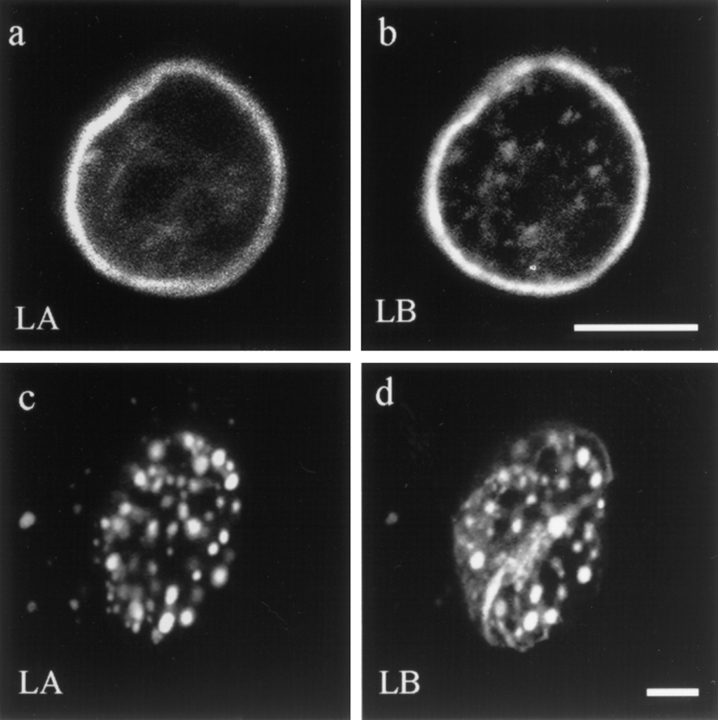
Figure 2.
Double label immunofluorescence showing nuclear lamin patterns in a normal interphase BHK cell and in a cell injected with ΔNLA. Nuclei were stained for lamins A/C (LA) (a and c) or lamin B (LB) (b and d). In the uninjected cell, there is a distinctive lamin rim as well as less intense nucleoplasmic foci (a and b), as previously described (39). In cells fixed 2 h after injection, the lamin rim is no longer obvious, and the lamin staining for both lamins A/C and B appears mainly in the same foci (c and d). Confocal optics showing sections through the mid-region of the nucleus. Bar, 5 μm.
Human Lamin A Is Incorporated into the Endogenous Lamin Structures of Xenopus Nuclei Assembled In Vitro
While microinjection of ΔNLA can disrupt the lamina of individual BHK-21 cells, such studies are not amenable to detailed biochemical and structural analyses, as only a limited number of cells can be injected, and it is difficult to control the amount of protein injected (for a discussion of this latter point, see 19). Similarly, others have shown that transfection of lamin cDNA mutants into mammalian cells can also result in similar nucleoplasmic structures and apparent disruption of the endogenous lamin organization, but the amount of protein expressed per cell and the percentage of cells expressing the protein are quite variable (23). In light of these limitations, we have developed a hybrid system in which controlled concentrations of mutated and wild-type human lamins are added to nuclear assembly extracts prepared from Xenopus laevis eggs (4, 30, 43). One of the major advantages of this in vitro nuclear assembly system includes the ability to precisely control the amount of protein added to the extract. In addition, large numbers of nuclei can be prepared for morphological and biochemical studies. We have used the human lamins in this system because species-specific antibodies allow us to distinguish between the endogenous Xenopus lamin B3 and the mutant human lamin.
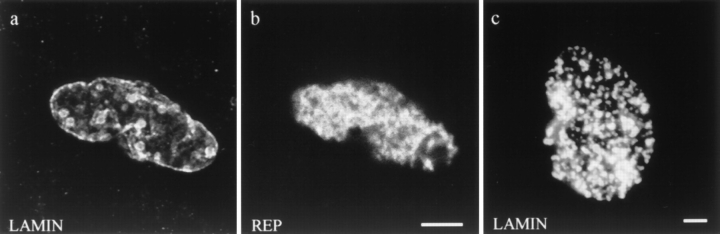
As a control for the use of the Xenopus system, wildtype LA was added to the nuclear assembly reactions. Nuclei assembled under these conditions contain LA, which colocalizes with the endogenous Xenopus lamin B3 (LB3) (Fig. 3, a and b). Staining is present at the nuclear periphery and elsewhere through the nucleus when viewed by conventional immunofluorescence. The LB3 staining pattern is similar to that seen in nuclei assembled without LA (compare Fig. 3 b with Fig. 6 a). Furthermore, nuclei assembled in the presence of LA contain a normal DNA staining pattern as indicated with Hoechst dye (Fig. 3 f). They also possess a normal distribution of nuclear membrane, as shown by staining with the membrane intercalating dye DIOC6 (data not shown), and a nuclear envelope with nuclear pore complexes, as suggested by the presence of a typical WGA staining pattern (Fig. 4, c and d). This organization of human LA in a heterologous system is consistent with results obtained in other laboratories. For example, when human lamin RNA is expressed in Xenopus oocytes, the expressed protein is localized to the lamina of the germinal vesicle (28). Similarly, Xenopus lamins expressed in mammalian cells integrate normally into the lamina (15).
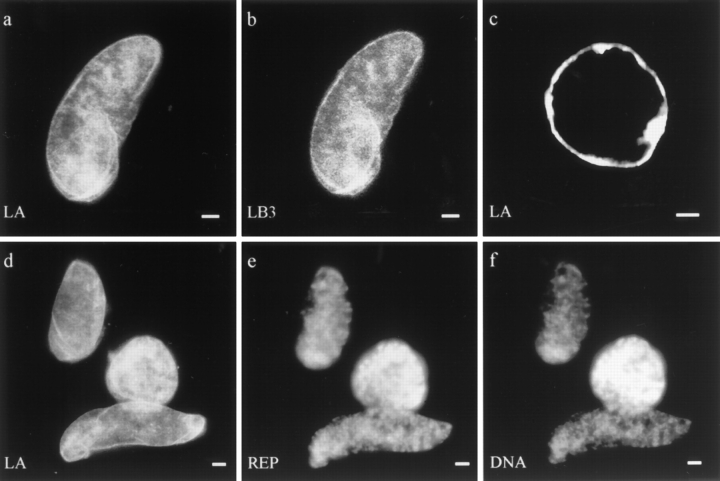
Figure 3.
Characterization of nuclei assembled in reactions containing human LA. (a and b) Nucleus stained for (a) human lamin A and (b) Xenopus LB3. The human LA does not disrupt the lamin network and appears to colocalize with the endogenous LB3. (c) Confocal microscopic image of a nucleus after nuclear matrix extraction and staining for human LA. LA staining is retained in the peripheral lamina after the matrix extraction protocol. (d–f) Fluorescence images from a triple label preparation of the same nuclei. (d) Nucleus stained for human lamin A; (e) biotinylated dUTP incorporation pattern as shown by Texas red–conjugated streptavidin (REP); (f) Hoechst dye showing the location of DNA. The incorporation of lamin A does not appear to alter DNA replication in the extract. Bar, 5 μm.
Figure 6.
The addition of ΔNLA to nuclear assembly reactions inhibits DNA replication as shown by the reduced incorporation of biotinylated dUTP. Fluorescence images of the LB3 pattern and biotinylated dUTP incorporation in (a and b) control nuclei and (c–f) nuclei formed in the presence of ΔNLA. (a, c, and e) Immunofluorescence using the antibody against Xenopus LB3; (b, d, and f) Texas red–conjugated streptavidin shows the patterns of incorporation of biotinylated dUTP. Disruption of lamin organization greatly inhibited the incorporation of biotinylated nucleotide when compared with control nuclei. However, all of the lamin-disrupted nuclei do contain a faint punctate nucleoplasmic pattern of labeled nucleotide incorporation that is readily resolved by confocal microscopy. (a–d) Conventional optics (see f). (e and f) Confocal optics. Bar, 5 μm.
Figure 4.
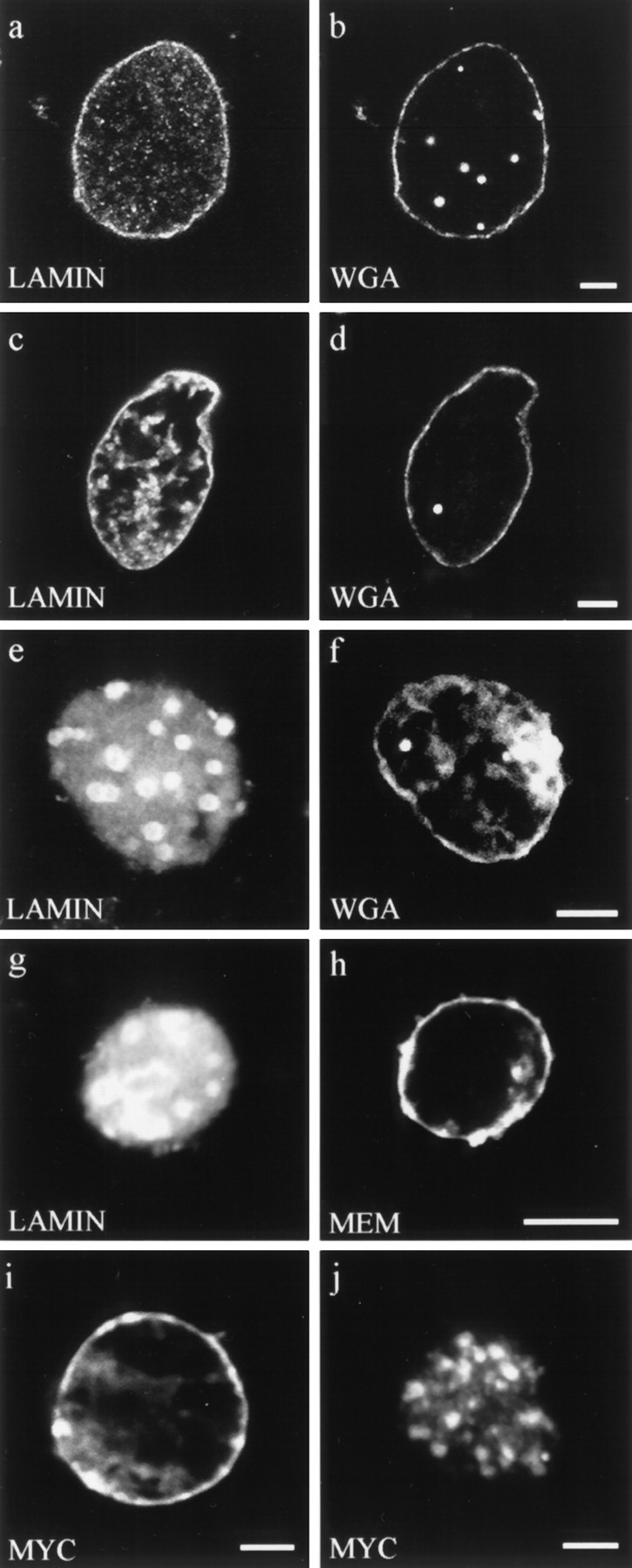
Double label fluorescence observations of nuclei stained for different aspects of nuclear envelope structure and function. (a–f) Nuclei were assembled in interphase extracts containing: (a and b) buffer control, (c and d) lamin A, and (e and f) ΔNLA. Nuclei were stained for (a) lamin B3 or (c and e) human lamin A, and (b, d, and f) the nuclear pore WGA binding proteins using fluorescently tagged WGA. Nuclei assembled under all three conditions appear to have essentially normal distributions of WGA binding proteins at the nuclear periphery. (g and h) Nucleus assembled in the presence of ΔNLA and stained for (g) ΔNLA and (h) the membrane dye DIOC6 (MEM). The nucleus contains a disrupted lamin organization but retains normal membrane staining. Bar, 5 μm. (i and j) Import of wild-type lamin A into (i) buffer control and (j) ΔNLA-disrupted nuclei. The wildtype lamin A was detected using the myc 9E10 epitope antibody (13). Nuclei were assembled with or without ΔNLA, and 90 min after the initiation of assembly, myc-tagged human lamin A was added to the reaction. The nuclei were fixed 20 min later and stained with the myc antibody. Both (i) control and (j) ΔNLAdisrupted nuclei show prominent myc staining, demonstrating that the disrupted nuclei retain the ability to import protein. The majority of the imported protein localizes to the characteristic foci of ΔNLA-disrupted nuclei. Confocal optics showing sections through the mid-region of nuclei. Bar, 5 μm.
To determine if LA is stably incorporated into the endogenous nuclear lamina, nuclear matrices were prepared from nuclei assembled in the presence of LA. Nuclei were first treated with DNase I, and subsequently extracted with 2 M NaCl and 0.1% Triton X-100 (see Materials and Methods). The nuclear lamina is resistant to this digestion and extraction procedure that has been shown to remove ∼90% of the nuclear proteins and DNA (11). Indirect immunofluorescence shows that the LA within the lamina is resistant to extraction and is therefore incorporated into the lamina of these nuclei (Fig. 3 c).
Xenopus nuclei assembled in vitro replicate their DNA once per cell cycle in a semiconservative manner (4, 30, 43). To determine whether the incorporation of LA affects DNA replication, biotinylated dUTP was added to nuclear assembly reactions (see Materials and Methods) containing LA. After fixation and staining with Texas red– streptavidin, fluorescence microscopic observations show that this nucleotide is incorporated in a fashion indistinguishable from control nuclei (compare Figs. 3, d and e, and 6, a and b).
ΔNLA Disrupts Nuclear Lamin Organization in Nuclei Assembled In Vitro and Inhibits DNA Replication
When the mutant human lamin ΔNLA is added to the nuclear assembly reaction at the same concentration as that used for the wild-type LA (200 μg/ml final concentration; see Materials and Methods), normal lamin assembly is altered in >90% of the nuclei. Instead of producing a typical nuclear lamin staining pattern, antibodies directed against Xenopus LB3 and LA stain large nucleoplasmic spheroidal bodies (Fig. 5, a and b). Confocal microscopic analysis of nuclei assembled with ΔNLA shows that both Xenopus LB3 and ΔNLA colocalize within these nucleoplasmic aggregates and that there is no obvious staining of the nuclear periphery (Fig. 5, d and e). However, the DNA of these lamin-disrupted nuclei appears to be distributed normally as indicated by Hoechst dye (Fig. 5 c). Furthermore, nuclear pore complexes, as indicated by WGA staining (Fig. 4, e and f), and the nuclear membranes, as indicated by DIOC6 staining, (Fig. 4, g and h) appear to be normal.
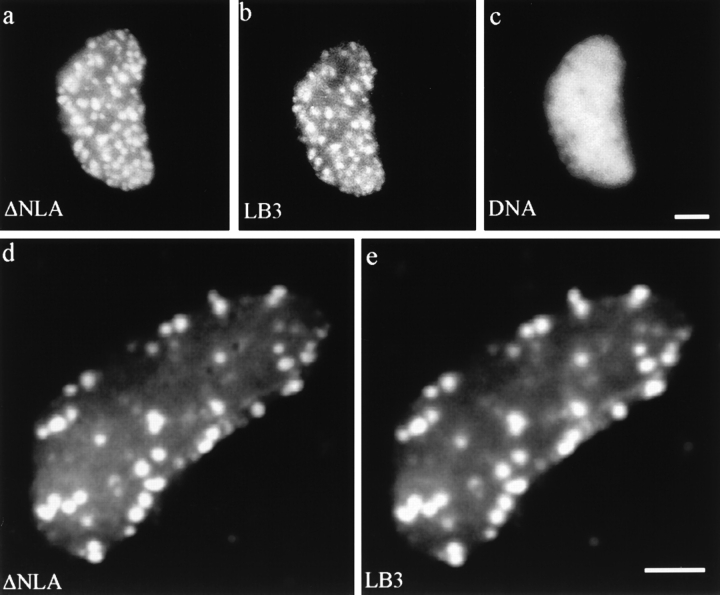
Figure 5.
Nuclei assembled in an interphase extract containing ΔNLA. (a–c) Conventional fluorescence images of lamin and DNA patterns of a nucleus stained for (a) human LA, (b) Xenopus LB3, and (c) DNA. (d and e) Confocal images of a disrupted nucleus stained for (d) human LA and (e) Xenopus LB3. The endogenous lamin structure has been disrupted and LB3 appears in foci colocalizing with ΔNLA. Bar, 5 μm.
We also determined if nuclear protein import could take place in disrupted nuclei. This involved the use of nuclei that were assembled in the presence of ΔNLA for 90 min (see Materials and Methods). Under these conditions, 100% (n = 84), of the in vitro assembled nuclei were disrupted. Subsequently myc-tagged wild-type LA was added to the same extract at a concentration equivalent to 25% of the mutant protein. 30 min later, the reactions were stopped and immunofluorescence assays demonstrated that the wild-type lamin A was imported into 100% (n = 80) of the disrupted nuclei (Fig. 4, i and j). These observations demonstrate that the disrupted nuclei are able to import nuclear proteins. Furthermore, under the experimental conditions used, the nuclei maintained their disrupted phenotype throughout the transport process. In contrast with the control nuclei, the myc-tagged lamin A colocalized with ΔNLA in the nucleoplasmic aggregates in disrupted nuclei (Fig. 4, i and j).
Interestingly, the disrupted nuclei are smaller than those formed in control assembly reactions or in reactions containing LA (e.g., compare Fig. 6 a and 6 b with 6 c and 6 d). Nuclei assembled in the presence of ΔNLA also seem more fragile than nuclei formed in either control or LAcontaining assembly reactions, as indicated by their increased tendency to break open when centrifuged at low forces (see Materials and Methods; data not shown).
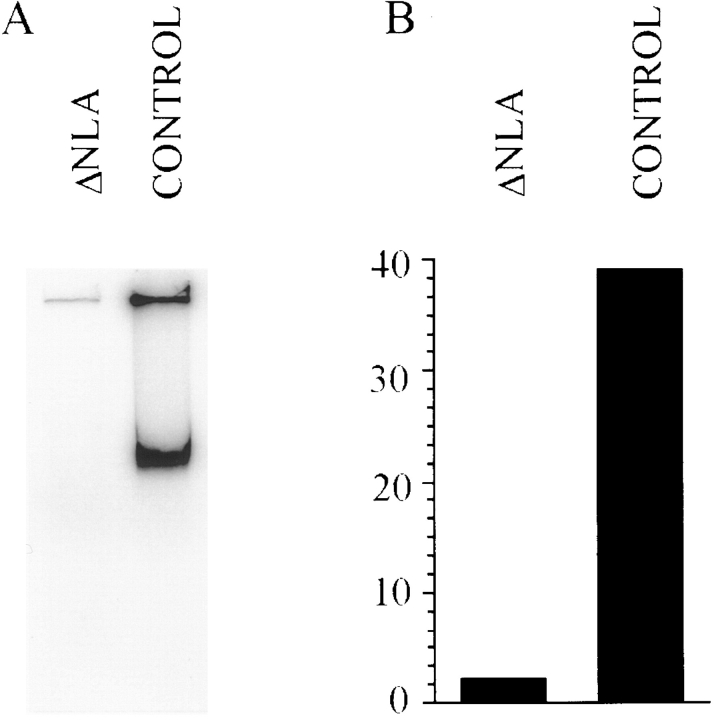
In contrast with the results obtained with the buffer control (Fig. 6, a and b) or with LA (see Fig. 3 e), the addition of ΔNLA to nuclear assembly reactions greatly inhibits DNA replication. When examined with conventional fluorescence optics, the disrupted nuclei show greatly reduced or no detectable incorporation of biotinylated dUTP after 90- and 180-min incubations (Fig. 6 d). However, confocal microscopy demonstrates that the majority of these nuclei do exhibit a faint punctate nucleoplasmic pattern of biotinylated dUTP incorporation (Fig. 6 f). To obtain a more quantitative measure of the inhibition of DNA synthesis, 32P-labeled dCTP was added to nuclear assembly reactions, in the presence or absence of ΔNLA. After a 90-min incubation, the DNA was isolated and resolved by gel electrophoresis (see Materials and Methods). Autoradiographic (Fig. 7 A) and phosphoimage analysis (Fig. 7 B) demonstrates that ΔNLA reduces the level of 32P-incorporation by 94% in disrupted nuclei.
Figure 7.
The addition of ΔNLA to nuclear assembly reactions reduces [32P]dCTP incorporation by ∼95%. (A) Autoradiogram of an agarose gel showing the incorporation of 32P-labeled dCTP into the DNA of nuclei formed in the presence of buffer control or ΔNLA. After nuclear assembly, the samples were treated as described in Materials and Methods and resolved on an 0.8% agarose gel. The upper band is at the origin of the gel. (B) Quantitation of replication assays shown in A. The radioactive signal of the dried gel was quantitated with a FUJIX BAS 2000 phosphoimager. The sum of the signal intensity/area value for both bands in each lane was used to measure the total incorporation of radioactivity into DNA. The average value for four replicate assays was plotted in a bar graph, where the vertical axis represents the signal/area values (in thousands) determined by the imager. The average value for samples containing ΔNLA was 2,292, with values ranging from 2,035–2,513. The average value for the control samples was 39,030, with samples ranging from 34,514–47,443. The addition of ΔNLA to the nuclear assembly reaction reduced the incorporation of 32P-labeled dCTP to ∼5% of that found in control reactions.
Disruption of Lamin Organization Alters the Distribution of Factors Required for the Elongation Phase of DNA Synthesis
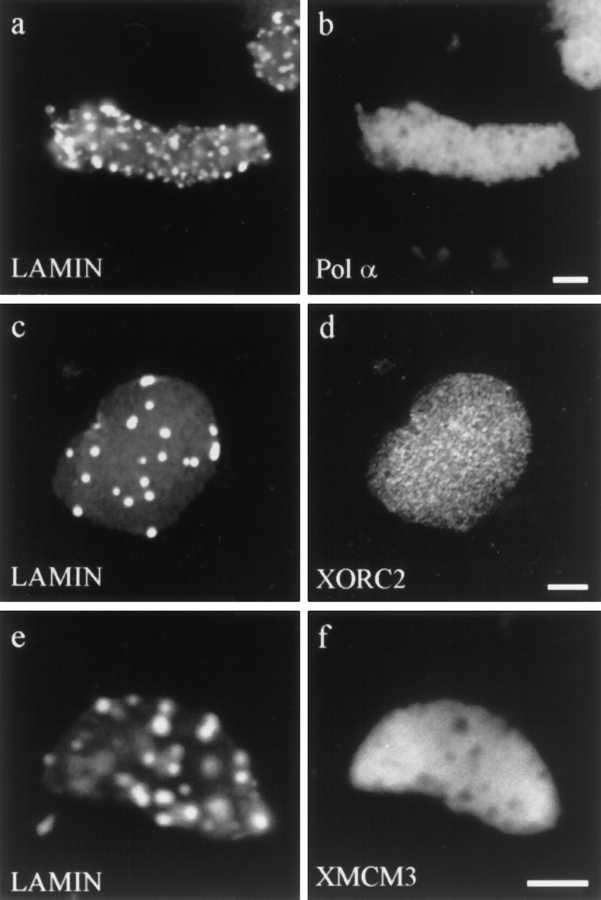
To examine the effects of altered lamin organization on the major steps of DNA replication, the distributions of five proteins known to be involved in either the initiation or the elongation phases of DNA replication were examined. The organization of factors involved in the initiation phase of DNA replication was studied with antibodies directed against DNA polymerase α, XORC2, and XMCM3. DNA polymerase α is believed to catalyze the formation of primers at origins of replication (55). XORC2 has been shown to be essential for DNA replication in the Xenopus nuclear assembly extracts, and, as a result of its sequence homology to the yeast ORC2, it is most likely involved in the initiation of DNA replication (9). XMCM3 has been characterized as a component of the licensing factor for DNA replication in Xenopus nuclear assembly extracts, and it is thought to be required for the initiation of DNA replication (10, 29, 33).
The DNA polymerase α and XORC2 staining patterns were unaffected in ΔNLA-disrupted nuclei (Fig. 8, a–d), compared with nuclei formed in the presence of buffer (data not shown). In both disrupted and control nuclei, staining with these antibodies indicated that they colocalized with chromatin (not shown). This is in agreement with previous studies (9, 26). These results suggest that the disruption of the lamin network does not affect the early stages of replication. The XMCM3 staining pattern of ΔNLA-disrupted nuclei was also coincident with chromatin (Fig. 8, e and f). This was true of 45-, 90-, and 180-min nuclear assembly reactions. In addition, we noticed that, in nuclei assembled in the presence of buffer, the XMCM3 staining colocalizes with chromatin at 45 min, but the fluorescence intensity decreases over time so that at 90 min it is not detectable (data not shown). This loss of XMCM3 signal is identical to the results obtained by other groups and is believed to represent a displacement of the protein from chromatin as replication proceeds (10, 29, 33). The disruption of lamin organization apparently prevents this XMCM3 displacement.
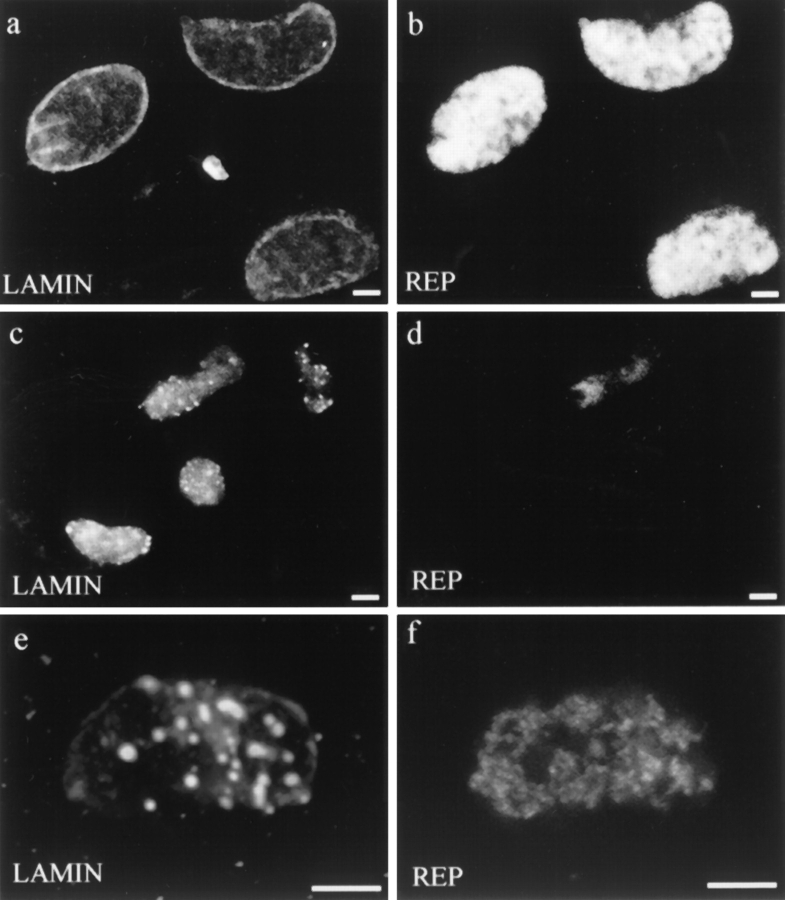
Figure 8.
Nuclei formed in the presence of ΔNLA, and then subsequently stained for ΔNLA or lamin B3 and one of several early DNA replication markers. (a and b) Nucleus stained for (a) ΔNLA and (b) DNA polymerase α. (c and d) Nucleus stained for (c) LB3 and (d) XORC2. (e and f) Nucleus stained for (e) LB3 and (f) XMCM3. The distribution of DNA polymerase α, XORC2, and XMCM3 is not altered by the disruption of the lamin structure. Confocal microscopic images showing sections through the middle of the nuclei. Bar, 5 μm.
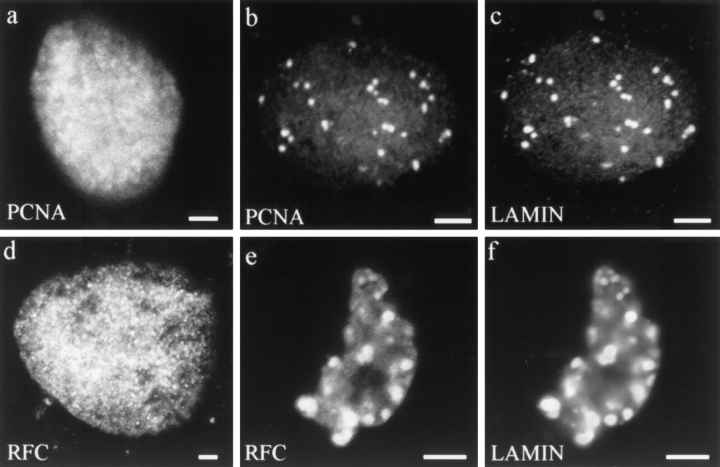
In contrast, the staining patterns produced by the antibodies directed against the elongation factors, PCNA and the large subunit of RFC, were dramatically altered in nuclei assembled in the presence of ΔNLA. Both of these proteins are required cofactors for DNA polymerase δ, the polymerase known to be responsible for chain elongation during DNA synthesis (55, 59). As assayed by indirect immunofluorescence, these two factors are associated with chromatin in nuclei assembled in control reactions (Fig. 9, a and d) or in assembly reactions containing LA (data not shown). However, when nuclei are assembled in the presence of ΔNLA, PCNA and RFC colocalize with the lamin aggregates (Fig. 9, b, c, e, and f). These observations suggest that the inhibition of DNA replication resulting from the disruption of nuclear lamin organization may be caused by alterations in the localization and/or targeting of the components of the DNA replication machinery responsible for chain elongation.
Figure 9.
Staining patterns of lamin and DNA replication factors involved in elongation in nuclei assembled in the presence of (a and d) buffer or (b, c, e, and f) ΔNLA. (a) Control nucleus stained for PCNA. (b and c) Nucleus assembled in the presence of ΔNLA stained for (b) PCNA and (c) ΔNLA. (d) Control nucleus stained for RFC. (e and f) Nucleus assembled in the presence of ΔNLA stained for (e) RFC and (f) ΔNLA. PCNA and RFC distributions are altered from the control as a consequence of lamin disruption such that PCNA and RFC colocalize with lamin aggregates in these nuclei. Confocal microscope showing sections through the middle of the nuclei. Bar, 5 μm.
Disruption of Nuclear Lamin Assembly Is Reversible
To determine if the ΔNLA-induced alterations of nuclear structure and function are reversible, nuclei were assembled in the presence of ΔNLA for 90 min. The disrupted nuclei were then removed from nuclear assembly reactions containing ΔNLA by centrifugation. These nuclei were resuspended in fresh extract to which no ΔNLA was added and were assayed for DNA replication. The nuclei were fixed 90 min later for immunofluorescence or processed for autoradiography (see Materials and Methods). As seen by both biotinylated dUTP incorporation (Fig. 10, a and b) and [32P]dCTP incorporation (data not shown), nuclear DNA synthesis was “rescued” when the disrupted nuclei were transferred to normal nuclear assembly reactions. Interestingly, although a few of the lamin aggregates remained in these nuclei, apparently normal nuclear lamina staining was reestablished (Fig. 10 a).
Figure 10.
(a and b) Lamin and biotinylated dUTP incorporation in a nucleus formed in the presence of ΔNLA, and subsequently transferred to an interphase extract containing biotinylated dUTP but lacking ΔNLA (see text). Confocal micrographs showing sections through the middle of the nucleus. (a) Nucleus stained for Xenopus LB3. (b) Pattern of biotinylated dUTP incorporation as shown by binding of Texas red–conjugated streptavidin. The disrupted nuclei were transferred to a nuclear assembly reaction lacking ΔNLA, where they form a lamin rim and replicate DNA. However, some lamin foci remain. (c) Postassembly disruption of the lamin structure of an in vitro assembled nucleus. ΔNLA was added 90 min after the onset of nuclear formation, a point at which the nuclei have normal lamin organization and have largely completed DNA replication. The addition of ΔNLA disrupts the assembled LB3 staining pattern. Bar, 5 μm.
Disruption of Nuclear Lamin Organization after Nuclear Assembly In Vitro
The microinjection of ΔNLA into cultured mammalian cells demonstrates the capacity of this truncated protein to disrupt endogenous lamin organization in fully formed interphase nuclei. At 90 min, nuclei formed in the Xenopus nuclear assembly reactions under normal conditions contain a normal lamin organization as indicated by lamin antibody staining (see, e.g., Fig. 4 a). DNA replication is complete or nearly complete, as indicated by the low level of incorporation of a 5-min pulse of biotinylated dUTP at this time interval (data not shown). To determine if ΔNLA can disrupt the lamin organization once it is established, ΔNLA was added to the nuclear assembly reaction 90 min after the initiation of nuclear assembly. The reaction was allowed to continue for an additional 45 min (see Materials and Methods). We found that the addition of ΔNLA under these conditions induced a dramatic disruption of the endogenous lamina in >90% of the nuclei observed. This resulted in the formation of nucleoplasmic aggregates containing LB3 (Fig. 10 c) and ΔNLA (data not shown). The addition of ΔNLA at both earlier and later time points (45, 120, and 150 min) after the initiation of nuclear assembly also resulted in the formation of nucleoplasmic aggregates indistinguishable from those seen in Fig. 10 c. In all cases, these aggregates appear to be structurally identical to those formed when nuclei are assembled in the presence of ΔNLA (see Figs. 3–6). However, it should be noted that nuclei disrupted 90 min after the initiation of assembly are larger than nuclei formed in the presence of ΔNLA.
Discussion
In this study we describe the use of a dominant negative mutant human lamin to disrupt the organization of the nuclear lamin assemblies both in vivo by microinjection into mammalian cells and in in vitro assembled Xenopus nuclei. We further demonstrate that a normal distribution of nuclear lamins is required for DNA synthesis. Specifically, the addition of ΔNLA, a mutant human lamin, to the Xenopus laevis nuclear assembly system blocks the formation of a normal lamina at the nuclear periphery. Instead, the endogenous LB3 and ΔNLA are found as constituents of the same large aggregates dispersed throughout the nucleoplasm. Under these conditions, DNA synthesis is dramatically reduced to ∼5% of its normal level. These results are consistent with those obtained from immunodepletion studies demonstrating that LB3 is required for DNA synthesis in in vitro assembled nuclei (17, 34, 45). The nuclei formed in LB3-immunodepleted extracts or ΔNLA-containing extracts described in this study have other features in common, including the fact that nuclear membranes and pores appear to assemble in a relatively normal fashion (34, 45). However, the dominant negative approach introduced in this study avoids one of the major problems inherent in the immunodepletion experiments: that the block in DNA replication could be caused by the removal of other proteins associated with lamin B3 in the assembly extract. In the experiments presented in this study, no components are removed from the nuclear assembly system.
The disruption of lamin organization alters the distribution of both PCNA and the large subunit of RFC, two essential cofactors for DNA polymerase δ during the elongation phase of replication (55). Normally both the large subunit of RFC (see Fig. 9) and PCNA (26) are distributed along chromatin. In ΔNLA-disrupted nuclei, these cofactors are reorganized, along with the nuclear lamins, to form nucleoplasmic aggregates. These aggregates are not obviously associated with chromatin, and this in turn may have an inhibitory effect(s) on the assembly and function of the elongation machinery. Alternatively, disruption of lamin organization may also alter an aspect of the initiation process itself that we have been unable to detect.
At the present time, we believe that our results support an effect on the elongation phase of DNA replication for several reasons. Evidence suggesting that the initiation of DNA synthesis does not rely on an intact lamin organization comes from our immunofluorescence studies of XORC2, XMCM3, and DNA polymerase α. These three proteins are thought to be involved in the initiation and primer formation steps of DNA replication (9, 10, 29, 33, 55). The disruption of the nuclear lamin structure does not detectably alter the distribution of DNA polymerase α and XORC2 or the initial distribution of XMCM3, and all three of these factors remain associated with chromatin. Interestingly, during nuclear assembly, it appears that XORC2 and XMCM3 along with two other initiation factors, RFA and FFA, bind to chromatin before the assembly of either higher order lamin structures or the nuclear membrane (1, 9, 10, 29, 33, 60, 61). These findings suggest that both the sites of initiation of DNA replication, as well as the organization of initiation cofactors at these sites, take place early in the process of nuclear assembly. In addition, the results reported here indicate that the location, organization, and function of these origins of replication may be independent of the presence of normal lamin organization.
Evidence indicating that the initiation of DNA synthesis occurs in disrupted nuclei comes from the observations that ΔNLA dramatically reduces but does not eliminate incorporation of biotinylated dUTP or [32P]dCTP. The very faint punctate pattern of biotinylated nucleotide incorporation in disrupted nuclei (Fig. 6 f) is reminiscent of the centers of DNA synthesis observed in normal nuclei at the onset of replication (24, 25, 37). Therefore, it is possible that the pattern and low levels of nucleotide incorporation seen in the disrupted nuclei described in this study may result from sites of primer and initial strand synthesis. This is also consistent with the unaltered distribution of DNA polymerase α in the lamin-disrupted nuclei. However, at this point, we cannot eliminate the possibility that the low level of incorporation of biotinylated nucleotide detected in disrupted nuclei may be due to other processes such as DNA repair.
Further clues for the role of the nuclear lamins are derived from the immunofluorescence pattern of XMCM3 in ΔNLA-disrupted nuclei. Normally, in control nuclei, XMCM3 is displaced from chromatin as replication progresses and a dramatic decrease in fluorescence intensity is observed (10, 29, 33). However, in the ΔNLA-disrupted nuclei reported in this study, the fluorescence intensity of XMCM3 staining appeared unchanged throughout the entire assembly reaction. This suggests that the disruption of the lamin structure prevents the dissociation of XMCM3 from chromatin, presumably by arresting replication before the normal displacement of this factor. Interestingly, it has been proposed that XMCM3 may be involved in the switch between the initiation and elongation phases of replication (60). Taken together, these results suggest that disruption of lamin organization blocks replication after the initiation of DNA synthesis and prevents the switch from the initiation to the elongation phase of DNA replication. However, the precise elucidation of the point at which lamin disruption blocks DNA synthesis requires a more complete understanding of the steps involved in DNA replication.
Most of our understanding of the biochemistry of DNA replication has come from the coupling of genetic studies in yeast with in vitro studies of SV-40 replication (55). The mechanisms involved in the regulation of DNA replication in more complex genomes remain largely unknown. However, it appears from the data presented in this study and elsewhere (17, 34, 44, 55) that, in higher eukaryotic cells, the nuclear lamins play a vital role in this process. We have previously reported that lamin B colocalizes with PCNA in mammalian cells during S phase (39). In this report we find that the disruption of nuclear lamin organization also alters the normal organization of RFC and PCNA, such that all three proteins are found in the same aggregates. These results suggest that lamins interact with the components of the strand elongation complexes. This interaction could be direct, with lamins binding to a component of the replication machinery, or indirect through unknown nuclear proteins or structural entities. This proposed interaction of PCNA with nuclear lamins is also supported by the finding that PCNA is readily extracted from nuclei formed in lamin B3–depleted assembly reactions, but is resistant to extraction in nuclei assembled in control reactions (27).
Traditionally, lamins have been thought to be located exclusively at the nuclear periphery, which makes it difficult to model lamin involvement in DNA synthesis, since much of the replication process takes place deep in the nucleoplasm. However, the presence of lamins in the nucleoplasm is supported by a number of studies (7, 18, 39), and we have found lamin B3 within the nucleoplasm of control Xenopus nuclei (Fig. 3 a). This lamin staining may be part of a dynamic nucleoplasmic lamin network. Such a network could form a scaffold (24, 25) upon which replication factors are assembled into functional units that facilitate the formation of active elongation complexes and/or stabilize such complexes once they are formed. Such an organization would explain why perturbations in nuclear lamin organization can block DNA replication and cause the abnormal distribution of RFC and PCNA.
The reduced size of nuclei assembled in the presence of ΔNLA provides evidence that the nuclear lamins are involved in the growth of the nucleus after its initial assembly. In addition, the increased fragility of the nuclei assembled in the presence of ΔNLA supports the idea that the nuclear lamin structure also provides a mechanical support system for the nucleus. These findings are consistent with previous reports of the small size and increased fragility of nuclei assembled in the absence of LB3 (34, 45).
The ΔNLA-induced disruption of lamin structure is most likely related to the dynamic characteristics of lamins in vivo. These properties of nuclear lamins are very similar to those found for other types of intermediate filament systems (36, 46, 50, 56, 58). Specifically, it has been shown that during interphase the nuclear lamins do not form a static polymer in vivo, but rather they are in a state of dynamic equilibrium between subunits and polymer. For example, microinjected nuclear lamins are rapidly incorporated into endogenous lamin polymers (18). Similarly, the results of fluorescence recovery after photobleaching experiments demonstrate that lamin assemblies undergo continuous subunit exchange in living cells (49).
Since it is known from in vitro studies that the central α-helical rod domain is required for normal lamin–lamin interactions, it is probable that ΔNLA and LB3 interact through their highly conserved rod domains to form heterocomplexes. However, the in vitro assembly of higher order lamin structures such as tetramers and larger oligomeric complexes requires the NH2-terminal domain that is missing in ΔNLA (20, 38). Therefore, the ΔNLA/LB3 heterocomplexes most likely cannot be incorporated into higher order lamin complexes. In the presence of excess ΔNLA, the normal process of subunit exchange could produce a large pool of heterocomplexes. In turn, this could drive the equilibrium in the direction of disassembly, ultimately resulting in the disruption of the endogenous lamin structure described in this study.
In summary, the addition of exogenous normal and mutated lamins to the Xenopus nuclear assembly system has provided evidence that a normal nuclear lamin organization is required to proceed from the initiation to the elongation phase of DNA replication. The assays used are relatively simple and should continue to provide further structural and biochemical information about the role of nuclear lamins in DNA replication. Furthermore, the availability of a soluble pool of nuclear components should allow us to fractionate the interphase extract and to determine whether lamin proteins are interacting directly with elongation factors at replication forks or indirectly through other unidentified nuclear components. This use of Xenopus extracts has already proven to be very important in identifying and characterizing the interactions of factors involved in the initiation of DNA replication within nuclei (1, 9, 10, 29, 33, 61,). We believe such an approach will also help to elucidate the role of nuclear lamins in other processes such as postmitotic nuclear assembly, nuclear growth, and the maintenance of the overall shape and structural integrity of the nucleus.
Acknowledgments
We thank Ms. Satya Khuon for help in preparing some of the micrographs and Ms. Laura Davis for help in manuscript preparation.
Abbreviations used in this paper
- IF
intermediate filament
- LA
lamin A
- LB3
lamin B3
- DIOC6
dihexyloxacarbocyanine
Footnotes
This work is supported by grant CA31760 from the National Cancer Institute.
T.P. Spann and R.D. Moir contributed equally to this work.
References
- 1.Adachi Y, Laemmli UK. Identification of nuclear pre-replication centers poised for DNA synthesis in Xenopusegg extracts: immunolocalization study of replication protein A. J Cell Biol. 1992;119:1–15. doi: 10.1083/jcb.119.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Belmont AS, Zhai Y, Thilenius A. Lamin B distribution and association with peripheral chromatin revealed by optical sectioning and electron microscopy tomography. J Cell Biol. 1993;123:1671–1685. doi: 10.1083/jcb.123.6.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benavente R, Krohne G, Franke WW. Cell-type specific expression of nuclear lamina proteins during development of Xenopus laevis. . Cell. 1985;41:177–190. doi: 10.1016/0092-8674(85)90072-8. [DOI] [PubMed] [Google Scholar]
- 4.Blow JJ, Laskey RA. Initiation of DNA replication in nuclei and purified DNA by a cell-free extract of Xenopuseggs. Cell. 1986;47:577–587. doi: 10.1016/0092-8674(86)90622-7. [DOI] [PubMed] [Google Scholar]
- 5.Blow JJ, Laskey RA. A role for the nuclear envelope in controlling DNA replication within the cell cycle. Nature (Lond) 1988;332:546–548. doi: 10.1038/332546a0. [DOI] [PubMed] [Google Scholar]
- 6.Blow JJ, Watson JV. Nuclei act as independent and integrated units of replication in a Xenopuscell-free DNA replication system. EMBO (Eur Mol Biol Organ) J. 1987;7:1997–2002. doi: 10.1002/j.1460-2075.1987.tb02463.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridger JM, Kill IR, O'Farrell M, Hutchison CJ. Internal lamin structures within G1 nuclei of human dermal fibroblasts. J Cell Sci. 1993;104:297–306. doi: 10.1242/jcs.104.2.297. [DOI] [PubMed] [Google Scholar]
- 8.Bunz F, Kobayashi R, Stillman B. cDNAs encoding the large subunit of human replication factor C. Proc Natl Acad Sci USA. 1993;90:11014–11018. doi: 10.1073/pnas.90.23.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter PB, Mueller PR, Dunphy WG. Role for a XenopusORC2-related protein in controlling DNA replication. Nature (Lond) 1996;379:357–360. doi: 10.1038/379357a0. [DOI] [PubMed] [Google Scholar]
- 10.Chong JP, Mahbubani HM, Khoo CY, Blow JJ. Purification of an MCM-containing complex as a component of the DNA replication licensing system. Nature (Lond) 1995;375:418–421. doi: 10.1038/375418a0. [DOI] [PubMed] [Google Scholar]
- 11.Cockerill PN, Garrard WT. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell. 1986;44:273–282. doi: 10.1016/0092-8674(86)90761-0. [DOI] [PubMed] [Google Scholar]
- 12.Dasso M, Newport JW. Completion of DNA replication is monitored by a feedback system that controls the initiation of mitosis in vitro: studies in Xenopus. . Cell. 1990;61:811–823. doi: 10.1016/0092-8674(90)90191-g. [DOI] [PubMed] [Google Scholar]
- 13.Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finlay DR, Forbes DJ. Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell. 1990;60:17–29. doi: 10.1016/0092-8674(90)90712-n. [DOI] [PubMed] [Google Scholar]
- 15.Firmbach-Kraft I, Stick R. The role of CaaX-dependent modifications in membrane association of Xenopusnuclear lamin B3 during meiosis and the fate of B3 in transfected mitotic cells. J Cell Biol. 1993;123:1661–1670. doi: 10.1083/jcb.123.6.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glass CA, Glass JR, Taniura H, Hasel KW, Blevitt JM, Gerace L. The α-helical rod domain of human lamins A and C contains a chromatin binding site. EMBO (Eur Mol Biol Organ) J. 1993;12:4413–4424. doi: 10.1002/j.1460-2075.1993.tb06126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg M, Jenkins H, Allen T, Whitfield WG, Hutchison CJ. Xenopuslamin B3 has a direct role in the assembly of a replication competent nucleus: evidence from cell-free egg extracts. J Cell Sci. 1995;108:3451–3461. doi: 10.1242/jcs.108.11.3451. [DOI] [PubMed] [Google Scholar]
- 18.Goldman AE, Moir RD, Montag-Lowy M, Stewart M, Goldman RD. Pathway of incorporation of microinjected lamin a into the nuclear envelope. J Cell Biol. 1992;119:725–735. doi: 10.1083/jcb.119.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman RD, Khuon S, Chou YH, Opal P, Steinert PM. The function of intermediate filaments in cell shape and cytoskeletal integrity. J Cell Biol. 1996;134:971–983. doi: 10.1083/jcb.134.4.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heins S, Aebi U. Making heads and tails of intermediate filament assembly, dynamics and networks. Curr Opin Cell Biol. 1994;6:25–33. doi: 10.1016/0955-0674(94)90112-0. [DOI] [PubMed] [Google Scholar]
- 21.Heitlinger E, Peter M, Lustig A, Villiger W, Nigg EA, Aebi U. The role of the head and tail domain in lamin structure and assembly: analysis of bacterially expressed chicken lamin A and truncated B2 lamins. J Struct Biol. 1992;108:74–89. doi: 10.1016/1047-8477(92)90009-y. [DOI] [PubMed] [Google Scholar]
- 22.Höger TH, Krohne G, Kleinschmidt JA. Interaction of Xenopus lamins A and LII with chromatin in vitromediated by a sequence element in the carboxy-terminal domain. Exp Cell Res. 1991;197:280–289. doi: 10.1016/0014-4827(91)90434-v. [DOI] [PubMed] [Google Scholar]
- 23.Holtz D, Tanaka RA, Hartwig J, McKeon F. The CaaX motif of lamin A functions in conjunction with the nuclear localization signal to target assembly to the nuclear envelope. Cell. 1989;59:969–977. doi: 10.1016/0092-8674(89)90753-8. [DOI] [PubMed] [Google Scholar]
- 24.Hozak P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- 25.Hozak P, Jackson DA, Cook PR. Replication factories and nuclear bodies: the ultrastructural characterization of replication sites during the cell cycle. J Cell Sci. 1994;107:2191–2202. doi: 10.1242/jcs.107.8.2191. [DOI] [PubMed] [Google Scholar]
- 26.Hutchison C, Kill I. Changes in the nuclear distribution of DNA polymerase alpha and PCNA/cyclin during the progress of the cell cycle, in a cell-free extract of Xenopuseggs. J Cell Sci. 1989;93:605–613. doi: 10.1242/jcs.93.4.605. [DOI] [PubMed] [Google Scholar]
- 27.Jenkins H, Holman T, Lyon C, Lane B, Stick R, Hutchison C. Nuclei which lack a lamina accumulate karyophilic proteins and assemble a nuclear matrix. J Cell Sci. 1993;106:275–285. doi: 10.1242/jcs.106.1.275. [DOI] [PubMed] [Google Scholar]
- 28.Krohne G, Waizenegger I, Höger TH. The conserved carboxy-terminal cysteine of nuclear lamins is essential for lamin association with the nuclear envelope. J Cell Biol. 1989;109:2003–2011. doi: 10.1083/jcb.109.5.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubota Y, Mimura S, Nishimoto S, Takisawa H, Nojima H. Identification of the yeast MCM3-related protein as a component of XenopusDNA replication licensing factor. Cell. 1995;81:601–609. doi: 10.1016/0092-8674(95)90081-0. [DOI] [PubMed] [Google Scholar]
- 30.Lohka MJ, Masui Y. Formation in vitroof sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science (Wash DC) 1983;220:719–721. doi: 10.1126/science.6601299. [DOI] [PubMed] [Google Scholar]
- 31.Lourim D, Krohne G. Membrane-associated lamins in Xenopusegg extracts: identification of two vesicle populations. J Cell Biol. 1993;123:501–512. doi: 10.1083/jcb.123.3.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lutz RJ, Trujillo MA, Denham KS, Wenger L, Sinensky M. Nucleoplasmic localization of pre-lamin A: implications for prenylationdependent lamin A assembly into the nuclear lamina. Proc Natl Acad Sci USA. 1992;89:3000–3004. doi: 10.1073/pnas.89.7.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madine MA, Khoo CY, Mills AD, Laskey RA. MCM3 complex required for cell cycle regulation of DNA replication in vertebrate cells. Nature (Lond) 1995;375:421–424. doi: 10.1038/375421a0. [DOI] [PubMed] [Google Scholar]
- 34.Meier J, Campbell KH, Ford CC, Stick R, Hutchison CJ. The role of lamin LIII in nuclear assembly and DNA replication, in cellfree extracts of Xenopuseggs. J Cell Sci. 1991;98:271–279. doi: 10.1242/jcs.98.3.271. [DOI] [PubMed] [Google Scholar]
- 35.Miake-Lye R, Kirschner MW. Induction of early mitotic events in a cell-free system. Cell. 1985;41:165–175. doi: 10.1016/0092-8674(85)90071-6. [DOI] [PubMed] [Google Scholar]
- 36.Miller RK, Khuon S, Goldman RD. Dynamics of keratin assembly: exogenous type I keratin rapidly associates with type II keratin in vivo. J Cell Biol. 1993;122:123–135. doi: 10.1083/jcb.122.1.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mills AD, Blow JJ, White JG, Amos WB, Wilcock D, Laskey RA. Replication occurs at discrete foci spaced throughout nuclei replicating in vitro. . J Cell Sci. 1989;94:471–477. doi: 10.1242/jcs.94.3.471. [DOI] [PubMed] [Google Scholar]
- 38.Moir RD, Donaldson AD, Stewart M. Expression in Escherichia coliof human lamins A and C: influence of head and tail domains on assembly properties and paracrystal formation. J Cell Sci. 1991;99:363–372. doi: 10.1242/jcs.99.2.363. [DOI] [PubMed] [Google Scholar]
- 39.Moir RD, Montag-Lowy M, Goldman RD. Dynamic properties of nuclear lamins: lamin B is associated with sites of DNA replication. J Cell Biol. 1994;125:1201–1212. doi: 10.1083/jcb.125.6.1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moir RD, Spann TP, Goldman RD. The dynamic properties and possible functions of nuclear lamins. Int Rev Cytol. 1995;162B:141–182. doi: 10.1016/s0074-7696(08)62616-9. [DOI] [PubMed] [Google Scholar]
- 41.Murray, A.W., and T. Hunt. 1993. The Cell Cycle. First edition. W.H. Freeman Co., New York. 251 pp.
- 42.Newmeyer DD, Wilson KL. Egg extracts for nuclear import and nuclear assembly reactions. Methods Cell Biol. 1991;36:607–634. doi: 10.1016/s0091-679x(08)60299-x. [DOI] [PubMed] [Google Scholar]
- 43.Newport J. Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell. 1987;48:205–217. doi: 10.1016/0092-8674(87)90424-7. [DOI] [PubMed] [Google Scholar]
- 44.Newport J, Spann T. Disassembly of the nucleus in mitotic extracts: membrane vesicularization, lamin disassembly, and chromosome condensation are independent processes. Cell. 1987;48:219–230. doi: 10.1016/0092-8674(87)90425-9. [DOI] [PubMed] [Google Scholar]
- 45.Newport JW, Wilson KL, Dunphy WG. A lamin-independent pathway for nuclear envelope assembly. J Cell Biol. 1990;111:2247–2259. doi: 10.1083/jcb.111.6.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Okabe S, Miyasaka H, Hirokawa N. Dynamics of the neuronal intermediate filaments. J Cell Biol. 1993;121:375–386. doi: 10.1083/jcb.121.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paddy MR, Belmont AS, Saumweber H, Agard DA, Sedat JW. Interphase nuclear envelope lamins form a discontinuous network that interacts with only a fraction of the chromatin in the nuclear periphery. Cell. 1990;62:89–106. doi: 10.1016/0092-8674(90)90243-8. [DOI] [PubMed] [Google Scholar]
- 48.Sasseville AM, Raymond Y. Lamin A precursor is localized to intranuclear foci. J Cell Sci. 1995;108:273–285. doi: 10.1242/jcs.108.1.273. [DOI] [PubMed] [Google Scholar]
- 49.Schmidt M, Tschodrich-Rotter M, Peters R, Krohne G. Properties of fluorescently labeled Xenopus lamin A in vivo. . Eur J Cell Biol. 1994;65:70–81. [PubMed] [Google Scholar]
- 50.Skalli O, Chou Y-H, Goldman RD. Intermediate filaments: not so tough after all. Trends Cell Biol. 1992;2:308–312. doi: 10.1016/0962-8924(92)90121-3. [DOI] [PubMed] [Google Scholar]
- 51.Stewart M. Intermediate filament structure and assembly. Curr Opin Cell Biol. 1993;5:3–11. doi: 10.1016/s0955-0674(05)80002-x. [DOI] [PubMed] [Google Scholar]
- 52.Stick R. CDNA cloning of the developmentally regulated lamin LIII of Xenopus laevis. . EMBO (Eur Mol Biol Organ) J. 1988;7:3189–3197. doi: 10.1002/j.1460-2075.1988.tb03186.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stick, R. 1995. Nuclear lamins and the nucleoskeleton. In The Cytoskeleton. I.I.F Pryme and J.E. Hesketh, editors. JAI Press, Greenwich, CT. 257–296.
- 54.Stick R, Hausen P. Changes in the nuclear lamina composition during early development of Xenopus laevis. . Cell. 1985;41:191–200. doi: 10.1016/0092-8674(85)90073-x. [DOI] [PubMed] [Google Scholar]
- 55.Stillman B. Smart machines at the DNA replication fork. Cell. 1994;78:725–728. doi: 10.1016/s0092-8674(94)90362-x. [DOI] [PubMed] [Google Scholar]
- 56.Straube-West K, Loomis PA, Opal P, Goldman RD. Alterations in neural intermediate filament organization: functional implications and the induction of pathological changes related to motor neuron disease. J Cell Sci. 1996;109:2319–2329. doi: 10.1242/jcs.109.9.2319. [DOI] [PubMed] [Google Scholar]
- 57.Tanaka S, Hu SZ, Wang TS, Korn D. Preparation and preliminary characterization of monoclonal antibodies against human DNA polymerase α. J Biol Chem. 1982;257:8386–8390. [PubMed] [Google Scholar]
- 58.Vikstrom KL, Lim SS, Goldman RD, Borisy GG. Steady state dynamics of intermediate filament networks. J Cell Biol. 1992;118:121–129. doi: 10.1083/jcb.118.1.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Waga S, Stillman B. Anatomy of a DNA replication fork revealed by reconstitution of SV40 DNA replication in vitro. . Nature (Lond) 1994;369:207–212. doi: 10.1038/369207a0. [DOI] [PubMed] [Google Scholar]
- 60.Yan H, Newport J. An analysis of the regulation of DNA synthesis by cdk2, cip1, and licensing factor. J Cell Biol. 1995;129:1–15. doi: 10.1083/jcb.129.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yan H, Newport J. FFA-1, a protein that promotes the formation of replication centers within nuclei. Science (Wash DC) 1995;269:1883–1885. doi: 10.1126/science.7569932. [DOI] [PubMed] [Google Scholar]