Abstract
Amphiphysin (amphiphysin I), a dominant autoantigen in paraneoplastic Stiff-man syndrome, is a neuronal protein highly concentrated in nerve terminals, where it has a putative role in endocytosis. The yeast homologue of amphiphysin, Rvs167, has pleiotropic functions, including a role in endocytosis and in actin dynamics, suggesting that amphiphysin may also be implicated in the function of the presynaptic actin cytoskeleton. We report here the characterization of a second mammalian amphiphysin gene, amphiphysin II (SH3P9; BIN1), which encodes products primarily expressed in skeletal muscle and brain, as differentially spliced isoforms. In skeletal muscle, amphiphysin II is concentrated around T tubules, while in brain it is concentrated in the cytomatrix beneath the plasmamembrane of axon initial segments and nodes of Ranvier. In both these locations, amphiphysin II is colocalized with splice variants of ankyrin3 (ankyrinG), a component of the actin cytomatrix. In the same regions, the presence of clathrin has been reported. These findings support the hypothesis that, even in mammalian cells, amphiphysin/Rvs family members have a role both in endocytosis and in actin function and suggest that distinct amphiphysin isoforms contribute to define distinct domains of the cortical cytoplasm. Since amphiphysin II (BIN1) was reported to interact with Myc, it may also be implicated in a signaling pathway linking the cortical cytoplasm to nuclear function.
Amphiphysin I, a human autoantigen in Stiff-man syndrome associated with breast cancer (De Camilli et al., 1993; Folli et al., 1993), is a neuronal protein highly concentrated in the cortical cytomatrix of nerve terminals where it has a putative role in synaptic vesicle endocytosis (Lichte et al., 1992; David et al., 1994, 1996; Shupliakov et al., 1997). It comprises an NH2-terminal region, which is predicted to form coiled coil structures, a COOHterminal SH3 domain, and a proline-rich linker region between these two domains that is poorly conserved evolutionarily (David et al., 1994). Biochemical studies, complemented by colocalization and coimmunoprecipitation experiments, have strongly suggested that the two main physiological ligands for the SH3 domain of amphiphysin I are the GTPase dynamin I (David et al., 1996; Grabs et al., 1997) and the inositol–5-phosphatase synaptojanin (McPherson et al., 1996). Dynamin I participates in synaptic vesicle recycling via its critical role in the fission of clathrincoated vesicles from the nerve terminal plasmalemma (Kosaka and Ikeda, 1983; Koenig and Ikeda, 1989; Shpetner and Vallee, 1989; Takei et al., 1995), and synaptojanin is thought to function in a closely related step (McPherson et al., 1996). In addition, amphiphysin I interacts in vitro, via a region distinct from its SH3 domain, with the appendage domain of the α subunit of the clathrin adaptor AP2 (Wang et al., 1995; David et al., 1996). It has, therefore, been suggested that one of the functions of amphiphysin I is to recruit dynamin I and synaptojanin at the clathrin coat of synaptic vesicles (David et al., 1996). Consistent with this hypothesis, disruption of amphiphysin SH3 domain interactions in living nerve terminals produces a potent block of synaptic vesicle endocytosis at the stage of deeply invaginated clathrin coated pits (Shupliakov et al., 1997).
Amphiphysin I shares substantial primary sequence similarity and a similar domain structure, with the yeast protein Rvs167. Furthermore, the NH2-terminal portion of both proteins is similar to the yeast protein Rvs161 (Bauer et al., 1993; David et al., 1994; Sivadon et al., 1995). Mutations in either RVS161 or RVS167 block receptor-mediated and fluid phase endocytosis in yeast, strongly supporting a role of the Amphiphysin/Rvs family in endocytic processes (Munn et al., 1995). In addition, RVS161 and RVS167 mutants also exhibit defects in the function of the actin cytoskeleton, in agreement with the general link between actin and endocytosis that has emerged from yeast studies (Munn et al., 1995; Sivadon et al., 1995). A corresponding link between amphiphysin I and the function of the actin cytoskeleton has been suggested by studies in cultured hippocampal neurons (Mundigl O., C. Ochoa, C. David, A.K. Kabanov, and P. De Camilli. Mol. Biol. Cell (Suppl.). 7:84a.). Finally, rvs mutants impair the ability of yeast cells to enter stationary phase upon exposure to nutrient starvation (reduced viability upon starvation) suggesting an indirect role of the RVS genes in controlling cell proliferation (Crouzet et al., 1991; David et al., 1994).
Amphiphysin I is expressed at a high concentration in brain and testis, at a lower concentration in neuroendocrine tissues (De Camilli et al., 1993; Folli et al., 1993; Lichte et al., 1992), and at only much lower levels in most other tissues (Butler, M.H., S. Floyd, and P. De Camilli, unpublished results). We have characterized here the product of a second amphiphysin gene, which we refer to as amphiphysin II. Amphiphysin II is localized primarily in specialized regions of the cortical cytoplasm of axons and muscle cells. These observations add further evidence for a general connection between proteins of the amphiphysin/Rvs family and the function of the cortical cell cytoskeleton.
Materials and Methods
Antibodies
The following affinity purified rabbit polyclonal antibodies were generated in our laboratory. CD5 antibodies specific for amphiphysin I (raised against a GST-fusion protein comprising the full length amphiphysin I protein [David et al., 1994]); CD7 and CD8 antibodies specific for amphiphysin II (raised against a GST-fusion protein comprising the last 68 amino acids of amphiphysin II; in this region, the amino acid identity between amphiphysin I and II is 55%); CD9 antibodies directed against both amphiphysin I and II (raised against a synthetic peptide corresponding to amino acids 26 to 40 of amphiphysin I, which is 100% conserved in amphiphysin II). A mouse polyclonal serum specific for amphiphysin I was raised against the polyhistidine-tagged full length amphiphysin I protein (David et al., 1996). Rabbit polyclonal antibodies directed against synaptojanin were previously described (McPherson et al., 1996). DG1 antibodies specific for dynamin I were generated against a dynamin I GST-fusion protein lacking the proline-rich domain. Antibodies directed against desmin, MAP2, and myelin basic protein (MBP) were purchased from Immunon (Pittsburgh, PA), Boehringer Mannheim Corp. (Indianapolis, IN), and Sternberger Monoclonals (Baltimore, MD), respectively. Polyclonal anti-ankyrin3 antibodies and anti-glut4 antibodies were kind gifts of J. Morrow (Yale University, New Haven, CT; Devarajan et al., 1996) and D. James (University of Queensland, Australia), respectively. A monoclonal anti-clathrin antibody (X22) was a kind gift of F. Brodsky (University of California, San Francisco, CA; Brodsky, 1985). A mouse monoclonal antibody (GAD6) against glutamic acid decarboxylase (GAD) was a kind gift of D. Gottlieb (Washington University, St. Louis, MO; Chang and Gottlieb, 1988).
DNA Cloning
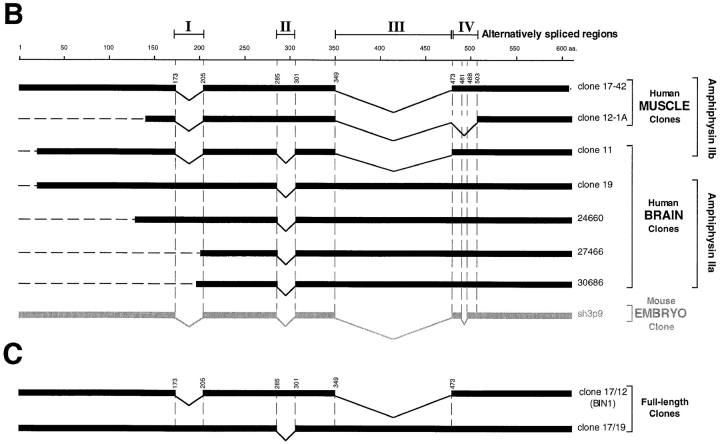
By searching the database for amphiphysin I homologues, a sequence of 289 bp was identified from a human muscle library with 76% identity to the COOH-terminal region of human amphiphysin I. (These sequence data are available from Genbank/EMBL/DDBJ under accession Z24784.) A probe corresponding to the first 251 bp of this sequence was amplified by PCR (forward primer 5′-cccaagcacgactacacggc-3′; reverse primer 5′ggaggaggtgttcttcacacgc-3′) from a human skeletal muscle cDNA library constructed in λZAPII phage (Stratagene, La Jolla, CA). The probe was then radioactively labeled by primer-direct labeling (Bogue et al., 1994) and used to screen 2 × 106 plaques of the same library. The two longest clones isolated by the screen (clones 17-42 and 12-1A) were partially characterized by restriction mapping and found to overlap extensively. Both clones were fully sequenced, and clone 17-42 was found to encode a nearly full length protein, missing only three amino acids at its COOH-terminal end. Subsequent searches of the database revealed additional expressed sequence tag (EST) sequences from a human infant brain library that were identical to portions of our clone 17-42. Clones 24660, 30686, and 27466 (Genbank/EMBL/DDBJ T80281, R18250 and R12992, respectively) were obtained through the IMAGE Consortium (Research Genetics Inc., Huntsville, AL) and fully sequenced. A human brain cDNA library constructed in λgt11 (Clontech, Palo Alto, CA) was then screened with a 220bp probe amplified by PCR (forward primer 5′-cttggggagggtggccccg-3′; reverse primer 5′-agcaagctcaaccagaacc-3′) and labeled by primer-direct labeling. 26 positive plaques out of 1 × 106 were identified, and the two longest clones (clone 11 and clone 19) were fully sequenced. These clones were identical to the IMAGE cDNA clones mentioned above, except for some additional alternative splicings (see Fig. 1 B). Since none of the clones isolated encoded an entire reading frame, the full length clones were assembled as follows starting from the two longest clones. Clone 17/12 (see Fig. 1 C) was assembled from clone 17-42 by replacing its COOH terminus with that of clone 12-1A using the SapI restriction site. Clone 17/19 (see Fig. 1 C) was assembled from clone 19 (Genbank/EMBL/DDBJ U87558) by replacing its NH2-terminal region with that of clone 17-42 at the unique restriction site BsaAI. Nucleotide sequences were analyzed by Blast and Fasta and aligned by Bestfit and Pileup (Genetics Computer Group, Madison, WI). Chromatograms from sequencing analysis were assembled by Seqman (DNASTAR, Inc., Madison, WI). Coiled coil structure identification was performed using Coils 2.2 (Lupas, 1996).
Figure 1.
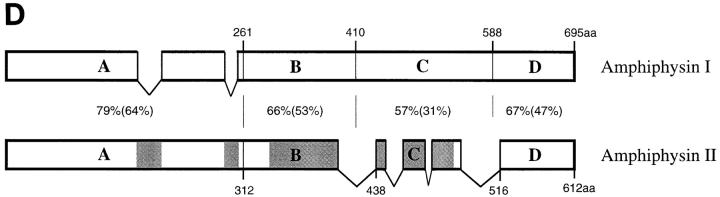
(A) Human amphiphysin II contiguous sequence obtained from a Pileup analysis of the human clones shown in B. Alternatively spliced regions are depicted by shaded amino acid residues. (B) Schematic representation of the human amphiphysin II clones analyzed in this study and of the mouse homologue of amphiphysin II previously reported (Sparks et al., 1996). The calibration bar (top) indicates number of amino acid residues. Alternatively spliced regions are indicated by roman numerals (I–IV). (C) Schematic representation of full length amphiphysin II clones assembled from clones 17-42 and 12-1A and from clones 19 and 1742, respectively. Clone 17/12 is identical to the recently reported BIN1 clone with the exception of a K→ E difference at position 434 of BIN1 and the corresponding position 591 of the contiguous sequence shown in Fig. 1 A (Sakamuro et al., 1996). (D) Domain diagram of human amphiphysins I and II showing the homology between the two genes. The boundaries of the A–D domains are delineated by amino acid numbers. A–D domains were previously defined as follows based on comparisons among human and chicken amphiphysin I and yeast Rvs proteins (David et al., 1994). The A, B, and D domains are the regions most highly conserved between chicken and human amphiphysin, while the C domain is poorly conserved. The A domain, within the A and B region, is defined by the yeast proteins Rvs161, which comprises this domain only. The percent similarity and identity (in parenthesis) for each domain is given. The shaded areas in the amphiphysin II gene represent the alternatively spliced regions outlined in Fig. 1 B.
Northern Blot Analysis
Northern blot analysis of amphiphysin II expression was performed on a human multiple tissue RNA filter (Clontech) containing 2 μg of Poly A+ RNA on each lane. The full length amphiphysin II cDNA (clone 17/12) and an oligonucleotide of 120 bp corresponding to amino acid 351–390 of the contiguous sequence shown in Fig. 1 A, were labeled by random priming (Boehringer Mannheim Corp.; 2 × 106 cpm/ml) and hybridized at 42°C in 50% formamide, 6× SSC, 0.1% SDS, 2× Denhardt's, 100 μg/ml salmon sperm DNA. An actin probe was used as a control to assess gel loading. Filters were washed twice for 20 min at high stringent conditions in 0.2× SSC, 0.1% SDS at 50°C.
Cell Transfection
COS-7 cells were transiently transfected with cDNAs corresponding to either clone 17/12 or clone 17/19 (see Fig. 1 C). The two cDNAs were subcloned in pcDNA3 (Invitrogen, San Diego, CA) and then purified with a Maxiprep kit (Qiagen, Chatsworth, CA). COS-7 cells (American Type Culture Collection, Rockville, MD) were transfected with lipofectamine (GIBCO BRL, Gaithersburg, MD), according to standard procedures (Chen and Okayama, 1987). Triton X-100 extracts of transfected cells and untransfected COS-7 cells were harvested after 24 h and analyzed by SDSPAGE and Western blotting.
Immunocytochemistry
Light microscopy.
Rat brains were fixed and frozen sectioned as described (De Camilli et al., 1983). Small fragments of rat soleus skeletal muscle were incubated in relaxing media (100 mM Hepes, 100 mM potassium propionate, 3 mM MgCl, 5 mM EGTA, 15 mM phosphocreatine, 2 mM NaATP; Kaufman et al., 1990) at room temperature for 10 min. Muscles were then stretched, fixed in 4% paraformaldehyde/0.1 M phosphate buffer, pH 7.4, and semithin sectioned (0.5 mm) on an ultramicrotome (Ultracut FCS; Reichert, Vienna, Austria). Rat muscle and brain sections were stained for indirect immunofluorescence according to De Camilli et al. (1983). Pictures were recorded on black and white films (T-MAX 100; Kodak, Rochester, NY) with a microscope (Axiophot; Zeiss Inc., Thornwood, NY) equipped for epifluorescence microscopy.
Electron microscopy.
Small pieces of rat soleus skeletal muscle were rapidly excised and fixed by immersion in 4% paraformaldehyde in 0.12 M sodium phosphate buffer, pH 7.4. The samples were then infiltrated with polyvinylpyrrolidone/sucrose for 2 h. Ultrathin frozen sections were cut onto an ultramicrotome (Reichert) with FCS attachment and immediately processed for immunogold labeling (10-nm gold) as previously described (Keller et al., 1984; Tokuyasu et al., 1989).
Miscellaneous Procedures
SDS-PAGE and Western blotting were performed essentially as described by Laemmli (1970) and Towbin et al. (1979), respectively. Immunoreactive bands were detected by either alkaline phosphatase conjugated secondary antibodies (Bio Rad, Hercules, CA) or 125I-protein A (105 cpm/ ml; Dupont/NEN, Boston, MA).
Results
Cloning of Amphiphysin II
Human adult skeletal muscle and brain cDNA libraries were screened with probes corresponding to human partial cDNA sequences present in the database with significant homology to human amphiphysin I. Four clones identified by this screening (muscle clones 17-42 and 12-1A and brain clones 11 and 19) and three human infant brain clones (clones 24660, 27466, and 30686) obtained from the IMAGE Consortium were fully sequenced.
A comparative analysis of all these clones suggests that they are derived by alternative splicing from a single gene. The products of this gene will be collectively referred to as amphiphysin II, because of their strong sequence similarity to amphiphysin I. The contiguous amino acid sequence derived from the analysis of the individual human clones is shown in Fig. 1 A. Fig. 1 B is a schematic alignment of the human clones with a mouse sequence (SH3P9) that represents the murine homologue of amphiphysin II and which was obtained during a screen for SH3 domain-containing proteins (Sparks et al., 1996). The partial clones depicted in Fig. 1 B were used to assemble the full length clones 17/12 and 17/19, as shown in Fig. 1 C (see Materials and Methods). These full length clones correspond to two alternative splicing variants of amphiphysin II. Clone 17/12 is identical to BIN1 (with the exception of a single amino acid; see Fig. 1 C, legend), a protein recently identified in a two hybrid screen for MYC-interacting proteins (Sakamuro et al., 1996).
The percentages of similarity and identity between the contiguous sequence of human amphiphysin II (as defined by Fig. 1 A) and human amphiphysin I (David et al., 1994) are 71 and 55%, respectively. Fig. 1 D shows a schematic alignment of the two sequences, as well as the boundaries of the A–D domains as defined previously on the basis of blocks of similarity between the Rvs yeast proteins and human and chicken amphiphysins (David et al., 1994; Fig. 1, legend).
Note that alternatively spliced fragments I and II (Fig. 1 B) of amphiphysin II coincide with gaps in the amphiphysin I sequence, raising the possibility that even the amphiphysin I gene may undergo a similar alternative splicing in this region.
Amphiphysin II Is Primarily Expressed in Brain and Muscle
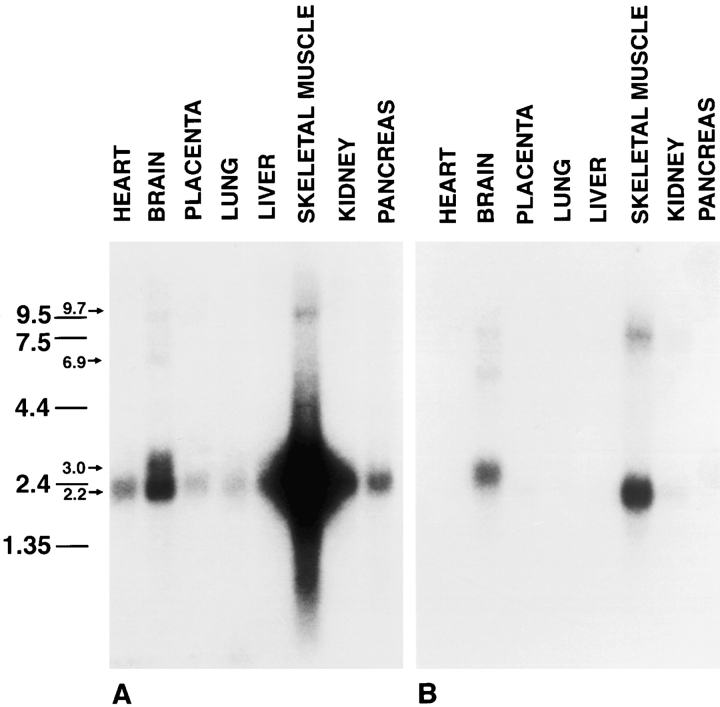
Northern blot analysis of different tissues with the full length amphiphysin II cDNA (clone 17/12) revealed that skeletal muscle is, by far, the major site of expression of this gene (Fig. 2 A). A major band centered at 2.2 kb was present in this tissue. The same band was present at much lower levels in brain and at an even lower concentration in several other tissues (Fig. 2 A). Several minor transcripts were also visible, including a band of 3 kb in brain. Since the putative alternatively spliced sequence III was only detected in brain clones (Fig. 1 B), we probed a blot identical to that of Fig. 2 A with constructs generated by PCR and corresponding either to this entire sequence (Fig. 1 A, amino acids 350–472) or to its 5′ portion (Fig. 1 A, amino acids 350–390). Both probes produced an identical pattern (Fig. 2 B and data not shown) and labeled bands with similar mobility as those labeled by the full length probe (clone 17/12) but with different relative intensities. The most striking difference is a strong labeling of the 3-kb band in brain and the weaker labeling of transcripts migrating at the 2.2-kb region. These observations confirm the preferential inclusion of splice segment III in brain amphiphysin II. No cross-reactivity with amphiphysin I mRNA (major transcript at 4.5 kb [David et al., 1994]) was observed in the high stringency conditions at which the Northern blot analysis was performed.
Figure 2.
Northern blot analysis of human tissues demonstrating patterns of expression of amphiphysin II mRNAs. Two identical blots containing Poly(A+) RNA from a variety of tissues were probed with clone 17/12 (A) and a probe corresponding to alternatively spliced segment III (B). Note the different labeling patterns produced by the two probes. Amphiphysin II is expressed primarily in skeletal muscle and brain. Numbers at left indicate molecular weights (kb).
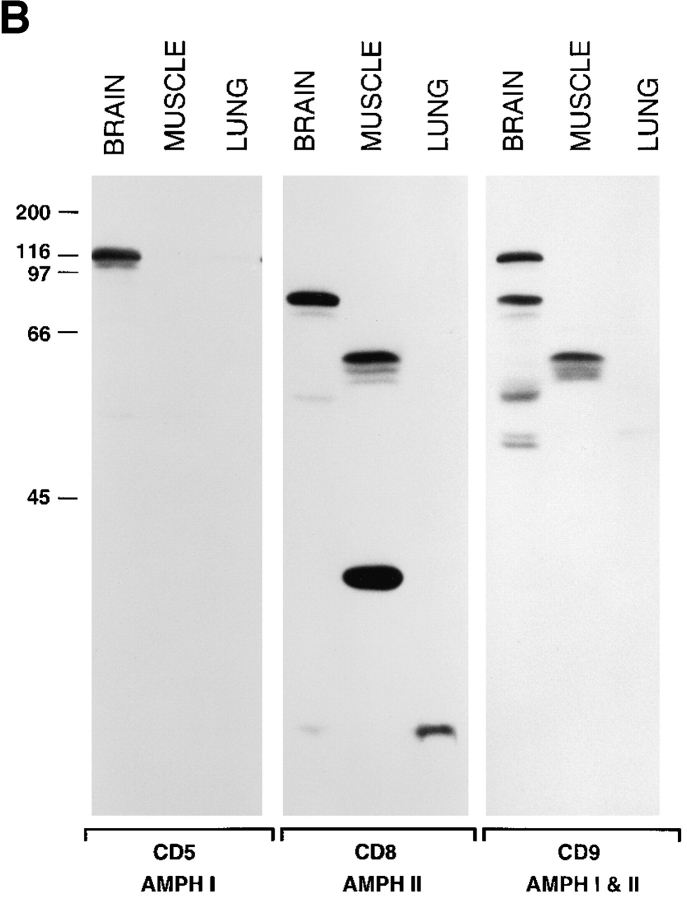
To determine the electrophoretic mobility and the tissue distribution of amphiphysin II, two rabbit antibodies (CD7 and CD8) were raised against the COOH-terminal 68 amino acids of amphiphysin II and tested by Western blotting against a variety of tissues (Fig. 3 A and data not shown). These antibodies specifically recognized very strongly a cluster of bands around 85 kD in brain and around 60 kD in skeletal muscle (Fig. 3 A) and did not react with amphiphysin I. Bands of similar molecular weight were seen in other tissues (primarily lung) only after very prolonged autoradiographic exposures (not shown). In these long exposures, the 85-kD band was also detectable in skeletal muscle in agreement with the Northern blot data of Fig. 2 B. In addition, a prominent band of 35 kD was visible in skeletal muscle (Fig. 3 A). As seen by a comparison of Figs. 2 and 3 A, there is a discrepancy between overall levels of amphiphysin II proteins and amphiphysin II mRNAs detected in muscle and brain. This discrepancy may be partially explained by a less efficient extractability of amphiphysin II from skeletal muscle. Alternatively, it may be due to the occurrence of major differences in amphiphysin II mRNA translation efficiency and/or in protein turnover in the two tissues.
Figure 3.
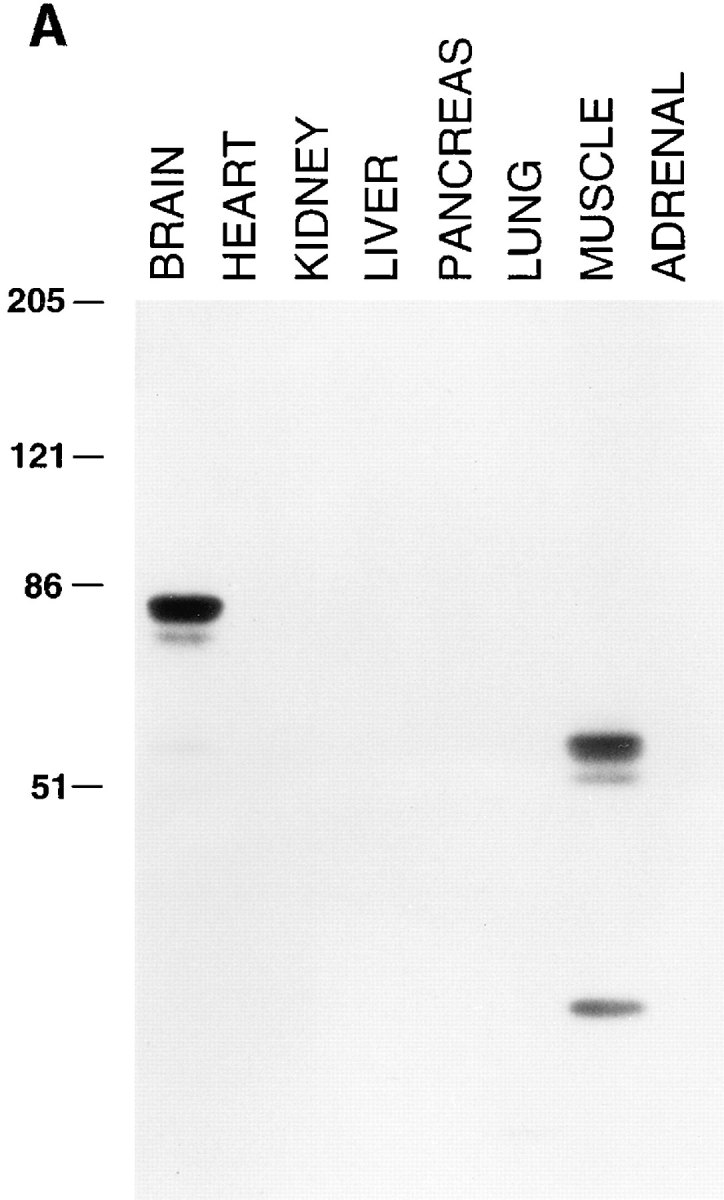
(A) Tissue distribution of amphiphysin II as demonstrated by Western blotting. Amphiphysin II is expressed primarily in brain and skeletal muscle. Equal protein amounts of post nuclear supernatants prepared from rat tissues were loaded in each lane and probed with the CD8 polyclonal rabbit serum specific for amphiphysin II. Bound antibodies were detected by 125Iprotein A. (B) Comparison of amphiphysin I and amphiphysin II expression in rat brain, skeletal muscle, and lung. Extracts of the three tissues were probed with an antibody specific for amphiphysin I (CD5), for amphiphysin II (CD8), and with an antibody that recognizes both amphiphysin I and II (CD9). Bound antibodies were detected by 125I-protein A. The low molecular weight bands labeled by the CD8 antibody are not visible in the brain and lung lanes of Fig. 3 A because they had migrated at the gel front. Numbers at left indicate molecular weights (kD).
To corroborate the identification of the brain and muscle bands as amphiphysin II, a distinct antiserum (CD9) was raised against a 15-mer peptide corresponding to amino acids 26–40 of amphiphysin I, which is identical to the corresponding region of amphiphysin II. This antibody recognized both the 128-kD brain amphiphysin I band (as the CD5 antibody does) and the bands immunoreactive with the amphiphysin II-specific antibody CD8 (Fig. 3 B). The CD9 antibody also recognized a few other bands in brain, suggesting the existence of additional amphiphysin isoforms. The CD7/CD8-immunoreactive 35-kD protein band of skeletal muscle was not recognized by the CD9 antibody and may therefore represent a variant of amphiphysin II that does not include its NH2-terminal domain. (Henceforth, we will refer to the brain 85- and muscle 60kD bands as amphiphysin IIa and IIb, respectively.)
Both amphiphysin IIa and IIb were recognized by antibodies directed against either NH2-terminal (CD 9) or COOH-terminal (CD7 and CD8) epitopes. Their different mobility suggests therefore internal alternative splicing. Most likely, this difference reflects the presence of splice fragment III (127 amino acids) in amphiphysin IIa. This hypothesis was supported by the transfection of COS-7 cells with clones 17/12 and 17/19. Amphiphysin II immunoreactivity induced by these transfections comigrated with amphiphysin IIa and IIb, respectively (Fig. 4). The difference of ∼25 kD between amphiphysin IIa and IIb is more than the difference expected by the inclusion of 127 amino acids. However, it was previously shown that amphiphysin I has an aberrant mobility in SDS-PAGE, migrating significantly slower (∼128 kD) than predicted by its amino acid sequence (76 kD; Lichte et al., 1992; David et al., 1994). This aberrant mobility is primarily due to a region (David et al., 1994) that strikingly corresponds to the alternatively spliced region III in amphiphysin II.
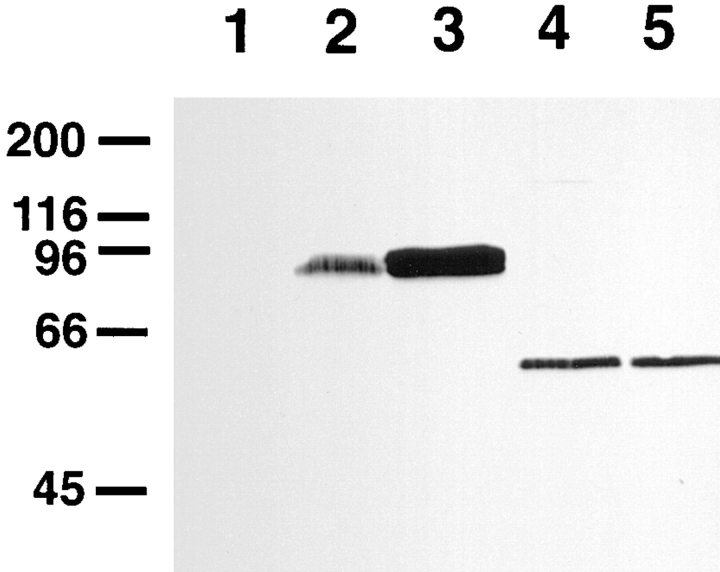
Figure 4.
Comparison of the electrophoretic mobility of amphiphysin II expressed in COS-7 cells with the electrophoretic mobilities of muscle and brain amphiphysin II. Triton X-100 extracts of tissues and COS-7 cells were probed by Western blotting with the amphiphysin II specific antibody, CD7. Lanes are as follows: 1, control untransfected COS-7 cells; 2, rat brain; 3, COS-7 cells transfected with clone 17/19; 4, COS-7 cells transfected with clone 17/12; 5, skeletal muscle. Immunoreactive bands were detected by using alkaline phosphatase-conjugated anti–rabbit IgG. Numbers at left indicate molecular weights (kD).
To determine whether the SH3 domains of amphiphysin I and II have similar properties, we carried out parallel affinity-purification experiments of brain extracts on GST fusion proteins comprising either one of the two SH3 domains. Both amphiphysin SH3 domains were equally effective in binding dynamin I, but the SH3 domain of amphiphysin II bound synaptojanin less effectively (not shown). Thus, amphiphysin I and amphiphysin II's SH3 domains have similar but not identical binding properties.
Localization of Amphiphysin II in the Nervous System
Considering their significant primary sequence similarity, amphiphysin I and II may have overlapping functions in brain. Therefore, we investigated whether these two proteins have a similar subcellular distribution by double immunofluorescence of rat brain frozen sections. Fig. 5 shows a comparison of the localization of amphiphysin I and II in three different gray matter regions of the brain: the cerebral cortex, the hippocampus, and the cerebellum. In all regions, amphiphysin I immunoreactivity (Fig. 5, B, D, and F) has the punctate distribution typical of nerve terminal staining. In contrast, amphiphysin II immunoreactivity occurs in the shape of short segments emerging from neuronal perikarya (Fig. 5, A, C, and E). In each region, the site of emergence of these processes and their shape is consistent with their identification as axon initial segments (Peters et al., 1991). High magnification views indicate that amphiphysin II is strictly confined to the cortical cytoplasm (Fig. 5 A, insets).
Figure 5.
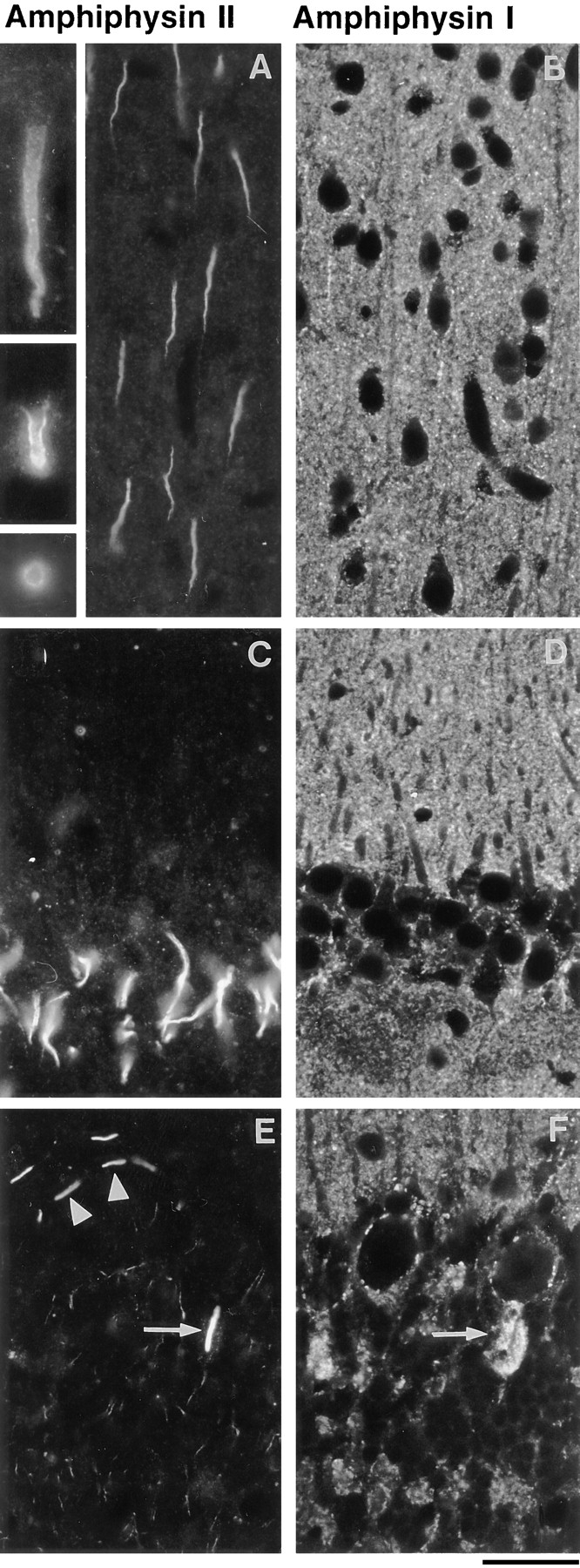
Comparison of the localization of amphiphysin I (mouse polyclonal serum) and amphiphysin II (rabbit antibody CD8) in rat brain. Double immunofluorescence micrographs. In all fields, amphiphysin I immunoreactivity (B, D, and F) has a typical nerve terminal pattern represented by small puncta throughout the gray matter. Amphiphysin II (A, C, and E) is primarily localized at initial axon segments. (A and B) cerebral cortex. The inset of A shows high power views of two longitudinal sections and one transverse section of initial axon segments. Note the concentration of immunoreactivity in the cortical region of the cytoplasm. (C and D) CA1 region of the hippocampus demonstrating in C the initial axon segments of pyramidal neurons visible in D as negative images. (E and F) Cerebellar cortex. Arrows point to the amphiphysin II positive initial segment of a Purkinje cell axon, which is surrounded by amphiphysin I positive nerve terminals of basket cells. Arrowheads in E point to initial axon segments of stellate cells. Bar, 63 μm; inset, 126 μm.
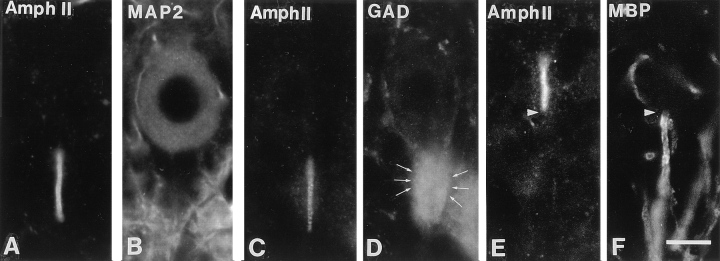
The specific localization of amphiphysin II at axon initial segments is further demonstrated in the high power views of cerebellar sections double stained for amphiphysin II and other protein markers (Fig. 6). Labeling for MAP2, a marker of perikarya and dendrites (De Camilli et al., 1984), demonstrates the origin of the amphiphysin II positive process from a small indentation at the basal pole of the cell (Fig. 6, A and B). Staining for glutamic acid decarboxylase, a marker of basket cell nerve terminals (Mugnaini and Oertel, 1985), shows that the amphiphysin IIpositive region of the axon coincides with its unmyelinated portion innervated by basket cells (Fig. 6, C and D). Labeling for myelin basic protein illustrates the sharp boundary between the amphiphysin II-positive portion of the axon and its myelinated portion (Fig. 6, E and F).
Figure 6.
Double immunofluorescence micrographs demonstrating the selective localization of amphiphysin II at axon initial segments. (A and B) Amphiphysin II-MAP2 immunostaining demonstrating the emergence of the amphiphysin II positive segment from the Purkinje cell body. (C and D) Amphiphysin II-GAD immunostaining demonstrating that the immunoreactive region of the axon corresponds to its region enclosed by the GABAnergic nerve terminals (arrows) of basket cells. (E and F) Amphiphysin II-myelin basic protein immunostaining demonstrating that amphiphysin II immunostaining terminates abruptly (arrowhead) at the site where the myelin sheath begins. Bar, 126 μm.
In addition to axon initial segments, amphiphysin II immunoreactivity was also observed in spots sparsely distributed in the gray matter (which contains axons as well as neuronal perikarya and dendrites) and more densely packed in the white matter (which contains axon tracts only). These spots are illustrated in Fig. 7, A and B, which show brain regions double stained for amphiphysin II and myelin basic protein. At high power, these immunoreactive structures appear as bright rings overlapping with axonal profiles (Fig. 7 C), very similar to the rings of amphiphysin II immunoreactivity visible at axon initial segments. The distribution of these structures and their fine morphology allows their identification as nodes of Ranvier (Peters et al., 1991).
Figure 7.
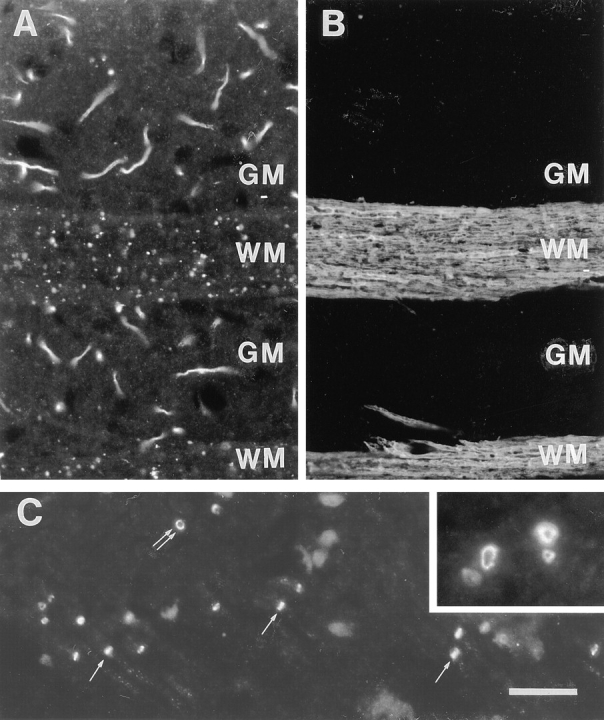
Localization of amphiphysin II at nodes of Ranvier. (A and B) Double immunofluorescence for amphiphysin II and myelin basic protein. Field shown is from the forebrain including two longitudinally sectioned white matter tracts. (A) Amphiphysin II positive axon initial segments are visible in the gray matter (GM). Small spots of amphiphysin II immunoreactivity visible on white matter tracts (WM) represent nodes of Ranvier. (C) White matter region in the brain stem demonstrating front (double arrows) and side (single arrows) views of nodes of Ranvier. The inset shows at high power a bundle of cross-sectioned axons demonstrating the localization of amphiphysin II in the cortical cytoplasm of nodes of Ranvier. Bar: (A and B) 27 μm; inset, 135 μm.
The presence of amphiphysin II in the cortical region of both axon initial segments and nodes of Ranvier is consistent with the functional and structural similarity of these two axonal portions (Waxman and Quick, 1978). An identical localization at both sites was previously reported for a neuron-specific isoform of ankyrin3 (ankryinG; Kapfhamer et al., 1995; Kordeli et al., 1995).
Localization of Amphiphysin II in Skeletal Muscle
The light microscopic localization of amphiphysin II in skeletal muscle is illustrated in Fig. 8. Amphiphysin II immunoreactivity appears as transverse striations along the muscle fiber (Fig. 8 A). These striations are within the I band, as shown by counterstaining of actin by fluorescent phalloidin (Fig. 8, B and C), and they flank the Z line, as shown by counterstaining for desmin (Fig. 8, D and E). The amphiphysin II stripes are similar to the stripes of ankyrin3 (ankyrinG) immunoreactivity (Fig. 8, F and G, as shown by double staining of ankyrin3 and actin). Ankyrin3 was previously shown to be concentrated along plasmalemmal T tubules (Flucher et al., 1990). Accordingly, the localization of amphiphysin II was also very similar to that of triadin, a marker of T tubules (Guo et al., 1994; data not shown). Immunoreactivity for clathrin heavy chain (monoclonal antibody X22), which was previously shown to be concentrated in muscle at I bands (Muñoz et al., 1995a ,b), formed stripes comprised between the amphiphysin II striations and the Z line, as shown by double staining of amphiphysin II and clathrin (Fig. 8, H and I). The glucose transporter, glut4, a protein that is internalized at least partially via clathrin coated vesicles (Garippa et al., 1996; Robinson et al., 1996), is also localized in proximity of T tubules (Muñoz et al., 1995a ). Glut4 immunoreactivity is centered around the M line, as shown by double labeling with anti-glut4 and anti-clathrin antibodies (Fig. 8, J and K).
Figure 8.
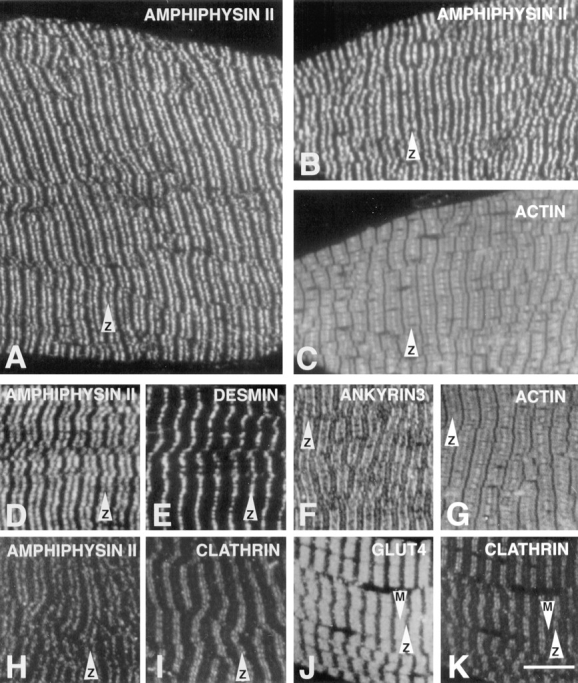
Immunofluorescence localization of amphiphysin II and other proteins of the sarcomere in skeletal muscle. Immunofluorescence of semithin frozen sections. Amphiphysin II immunoreactivity (A, B, D, and H) forms transverse bands that flank the Z line. B–K show pairs of double-fluorescence micrographs. (B and C) Amphiphysin II and actin (phalloidin staining); (D and E) amphiphysin II and desmin, a marker of the Z line; (F and G) ankyrin (ankyrin3) and actin; (H and I) amphiphysin II and clathrin heavy chain (antibody X22); (J and K) glut4 and clathrin. Arrows equal Z and M lines, as indicated. Bar, 7.9 μm.
The localization of amphiphysin II was further investigated by immunogold labeling of ultrathin frozen sections and found to be localized in correspondence with T tubules (Fig. 9, B and D). The specificity of this labeling was confirmed by labeling similar sections for desmin. As expected, desmin immunoreactivity was concentrated along the Z line and absent from the T system (Fig. 9, A and C).
Figure 9.
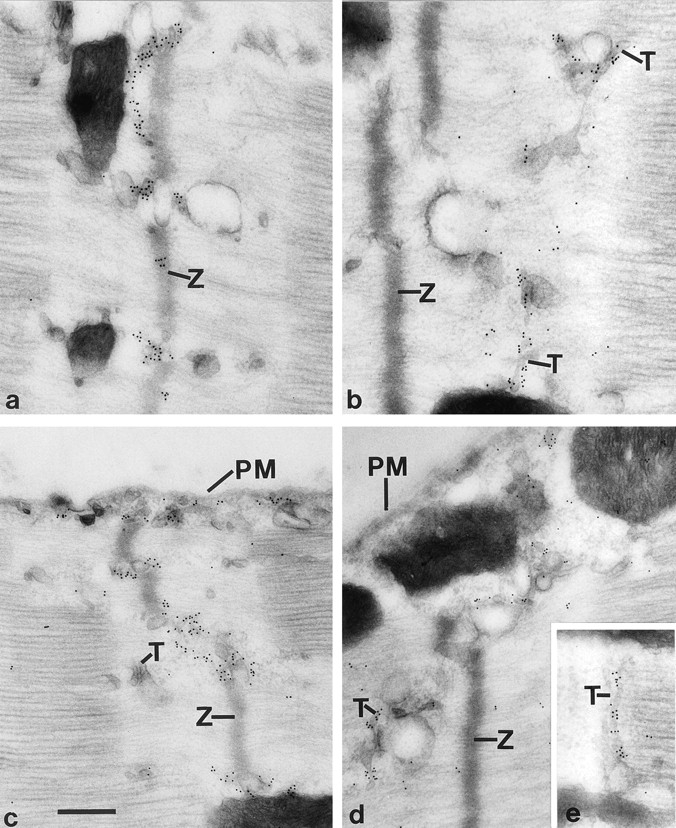
Comparison of the localizations of desmin and amphiphysin II in skeletal muscle by electron microscopy immunocytochemistry. Ultrathin frozen sections were labeled by immunogold for desmin (A and C) and amphiphysin II (B, D and E). Desmin immunoreactivity is localized on a network of filamentous structures that are in register with Z lines. Amphiphysin is selectively localized at the T system and is present on T tubules (E). Z, Z lines; T, T tubules; PM, plasmalemma. Bar: (A, B, and E) 300 nm; (C and D) 378 nm.
Cytoplasmic Localization of Amphiphysin II in Transfected Cells
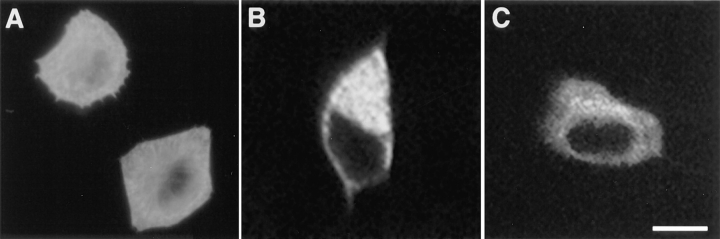
Neither in brain nor in muscle (Figs. 5–8) was amphiphysin II found to have a nuclear localization. This was in contrast to the nuclear localization of this protein (BIN1) reported by Sakamuro et al. (1996) in transfected HepG2 cells. We examined, therefore, the localization of amphiphysin IIa and IIb in transfected COS-7 cells as well as in HepG2 cells transfected with the same clone (17/12, and under the same experimental conditions) used by Sakamuro et al. (1996). In all cases the protein was primarily localized in the cytosol (Fig. 10 and data not shown).
Figure 10.
Immunofluorescence localization of amphiphysin II in transfected cells. (A) COS-7 cells transfected with clone 17-12 and examined by conventional epifluorescence light microscopy. (B and C) HepG2 cells transfected with clone 17/12 and examined by confocal microscopy. The slight fluorescence visible in A over the nuclei is out of the nuclei focal plane. Bar, (A) 12.6 μm; (B and C) 9.0 μm.
Discussion
We report here the characterization of an amphiphysin gene (amphiphysin II) that is primarily, but not exclusively, expressed in brain and skeletal muscle. The gene undergoes extensive alternative splicing. Amphiphysin II is substantially similar in amino acid sequence and domain structure to amphiphysin I. The strongest similarity is present in the A domain, which is predicted to form coiled coil structures (Sivadon et al., 1995), and in the D domain, which contains the SH3 domain (David et al., 1994). Domain A is characteristic of all proteins of the amphiphysin/ Rvs family identified so far, including the yeast protein Rvs161, which is composed of the A domain only (David et al., 1994; Sivadon et al., 1995). Due to these similarities, the two amphiphysins are likely to have homologous functions. However, their different cellular and subcellular localizations clearly indicate that their functions are not overlapping.
In brain, amphiphysin I is concentrated in the cortical cytoplasm of nerve terminals where it participates in synaptic vesicle endocytosis (David et al., 1996; Shupliakov et al., 1997). In contrast, amphiphysin II is concentrated in axon initial segments and nodes of Ranvier. The occurrence of clathrin coated pits and clathrin coated invaginations has been reported to occur more frequently at initial segments and nodes of Ranvier than at other locations along the axonal surface, with the exception of nerve terminals (Karlsson, 1967; Campos-Ortega et al., 1968; Conradi, 1969). Thus, an involvement of amphiphysin II in endocytosis is plausible. However, it is unlikely that the high and specific concentration of amphiphysin II present at these sites may be simply related to endocytosis.
A characteristic feature of the cortical cytoplasm at initial segments and nodes of Ranvier is the presence of a dense matrix underlying the plasmalemma (Palay et al., 1968; Conradi, 1969; Waxman and Quick, 1978). This submembranous cytoskeleton may participate in mediating the local enrichment of special adhesion molecules (members of the neurofascin/L1 family; Shiga and Oppenheim, 1991) and of proteins required for the generation and propagation of action potentials, such as Na+ channels (Srinivasan et al., 1988; Waxman and Ritchie, 1993), Na+/K+ ATPase (Nelson and Veshnock, 1987; Waxman and Ritchie, 1993), and Na+/Ca2+ exchangers (Waxman and Ritchie, 1993). The only unique component of this specialized cortical cytomatrix identified so far is a neuron-specific isoform of ankyrin3 (ankyrinG; Kordeli et al., 1995). Neuronal amphiphysin II may be a second component of this matrix. As in the case of amphiphysin II, the isoform of ankyrin expressed at axon initial segments and nodes of Ranvier is generated by alternative splicing of a gene (ankyrin3) that is widely expressed outside the nervous system, and at particularly high concentrations in skeletal muscle (Kordeli et al., 1995).
In skeletal muscle, amphiphysin II is concentrated around the plasmalemma of T tubules, and even here, it colocalizes with ankyrin3, previously shown to be a component of the submembranous cytoskeleton of T tubules (Flucher et al., 1990). Like axon initial segments and nodes of Ranvier, T tubules are enriched in proteins responsible for controlling ion permeability and transport (Lau et al., 1979; Flucher et al., 1990) and a specialized cytomatrix around the T tubules, including both amphiphysin II and ankyrin3, may help to define the composition and function of these plasmalemmal domains (Flucher et al., 1990). T tubules are not typically regarded as sites specialized for endocytosis. However, there is evidence that clathrin-mediated endocytosis may occur at this region. First, clathrin immunoreactivity (detected by monoclonal antibody X22) is present in proximity of T tubules as previously reported (Kaufman et al., 1990) and further confirmed by this study. This clathrin heavy chain is likely to correspond to the skeletal muscle specific clathrin recently described by several groups (Gong et al., 1996; Kedra et al., 1996; Lindsay et al., 1996; Sirotkin et al., 1996). Second, the glut4 transporter, which undergoes regulated surface exposure in response to insulin (Wang et al., 1996) and is internalized at least in part via clathrin coated vesicles (Garippa et al., 1996; Robinson et al., 1996), is concentrated along the T system and surrounding vesicles (Slot et al., 1991; Muñoz et al., 1995a ,b).
In yeast, mutations in either the Rvs161 and/or the RVS167 genes produce both endocytosis defects and defects in the function of the peripheral actin cytoskeleton, including abnormal polarity, uneven cell size and morphology, and delocalization of actin patches (Munn et al., 1995; Sivadon et al., 1995). The COOH-terminal region of Rvs167 was identified in a two hybrid screen for actin binding proteins (Amberg et al., 1995). More generally, yeast studies have demonstrated an important role of the actin cytoskeleton in endocytosis, thus raising the possibility that effects of RVS mutations on endocytosis and the peripheral cytoskeleton may be interrelated (Munn and Riezman, 1994; Amberg et al., 1995; Munn et al., 1995).
Our present demonstration that amphiphysin II is localized in the cortical cytomatrix of specialized regions of axons and muscle is consistent with the role of the RVS genes in actin function. A dual role in endocytosis and in the dynamics of the peripheral cytoskeleton may be a general characteristic of amphiphysin/Rvs family proteins. The strong implication of amphiphysin I in synaptic vesicle endocytosis may reflect the unique specialization of the presynaptic actin cytomatrix for this function.
An additional phenotype produced by mutations in the RVS genes is reduced viability upon starvation, a phenotype displayed by an inability of the cell to enter in stationary phase under these conditions (Crouzet et al., 1991; Bauer et al., 1993). Since amphiphysin I was shown to be an autoantigen in breast cancer (De Camilli et al., 1993; David et al., 1994, 1996), it was speculated (David et al., 1994) that proteins of the amphiphysin/Rvs family, like other proteins of the cortical cell cytomatrix that can act as tumor suppressors (Rubinfeld et al., 1993; Tsukita et al., 1993), may be directly implicated in cancer.
While this study was in progress, sequences of amphiphysin II isoforms were independently published in the context of two studies. A first study reported the identification of mouse muscle amphiphysin II during a search for novel SH3-containing proteins (Sparks et al., 1996). This protein (SH3P9) was not further characterized. A second study identified a fragment of murine amphiphysin II in a yeast two hybrid screen for MYC binding proteins (Sakamuro et al., 1996). The authors of this study went on to isolate a human amphiphysin II isoform, BIN1, which is identical to our clone 17/12, and to demonstrate that this protein is localized in the nucleus and has the properties of a tumor suppressor gene. These findings are consistent with the presence of a nuclear localization sequence in BIN1 (Sakamuro et al., 1996), which we show here to be encoded by splice fragment II (Fig. 1 B). In our study, however, we do not have any evidence for a nuclear localization of amphiphysin II in adult muscle or brain. Our results, therefore, argue for a primary function of amphiphysin II in the cytoplasm, although they clearly do not exclude that amphiphysin II may shuttle from the cytoplasm to the nucleus and that it may function in a signaling pathway from the cell periphery to the nucleus. It was shown previously that proteins of the submembranous cytoskeleton (e.g., the tight junction protein ZO1 [Gottardi et al., 1996]) have a nuclear localization under certain conditions. Thus, the possibility that amphiphysin II may participate in nuclear events and even be concentrated in the nucleus under certain functional states cannot be ruled out.
In conclusion, we suggest that amphiphysin/Rvs proteins may play a general role in the physiology of the peripheral cytoskeleton which underlies the plasmalemma. Different isoforms, generated either by distinct genes or by alternative splicing of the same genes, may serve to adapt this general role to specific functions of specialized cell surface domains. Given the central importance of the subplasmalemmal cytomatrix in a variety of cellular processes, including vesicular trafficking to and from the plasmalemma, generation of regional heterogeneity of plasmalemma, signal transduction, and regulation of cell–cell interaction, the components of this matrix are likely to have pleiotropic functions. Further studies of amphiphysin family members may not only reveal new aspects of the function of the peripheral cytoskeleton and endocytosis, but also help elucidate a novel signaling pathway from the cell surface to the nucleus. The reported connection between amphiphysin I autoimmunity and cancer (Folli et al., 1993; De Camilli et al., 1993) suggests that these studies may be of relevance to the biology of at least some forms of human cancer.
Acknowledgments
We thank Corinne Leprince for discussing unpublished data.
This study was supported by grants from the Donaghue Foundation, the Human Frontier Science Program Organization, and the National Institutes of Health (CA46128) to P. De Camilli. D. Grabs was a recipient of a Deutscher Akademischer Austauschdienst fellowship, C. David of a United States Army Medical Research and Development Command fellowship, and O. Cremona of Telethon and Human Frontier Science Program Organization long-term fellowships.
Footnotes
M.H. Butler and C. David contributed equally to this work.
Please address all correspondence to Pietro De Camilli, Department of Cell Biology and HHMI, Yale University School of Medicine, 295 Congress Avenue, New Haven, CT 06510. Tel.: (203) 737-4465; Fax: (203) 7371762; E-mail: pietro.decamilli@yale.edu
Detlev Grabs' current address is Institut für Anatomie and Spezielle Embryologie, Universität Fribourg, 1 Rue Gockel, CH-1700 Fribourg, Switzerland.
Ottavio Cremona's permanent address is Department of Medical Sciences, II Faculty of Medicine, University of Torino, Via Solaroli 17, 21100 Novara, Italy.
References
- Amberg DC, Basart E, Botstein D. Defining protein interactions with yeast actin in vivo. Nat Struct Biol. 1995;2:28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Bauer F, Urdaci M, Aigle M, Crouzet M. Alteration of a yeast SH3 protein leads to conditional viability with defects in cytoskeletal and budding patterns. Mol Cell Biol. 1993;13:5070–5084. doi: 10.1128/mcb.13.8.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue CW, Gross I, Vasavada H, Dynia DW, Wilson CM, Jacobs HD. Identification of Hox genes in newborn lung and effects of gestational age and retinoic acid on their expression. Am J Physiol. 1994;266:L448–454. doi: 10.1152/ajplung.1994.266.4.L448. [DOI] [PubMed] [Google Scholar]
- Brodsky FM. Clathrin structure characterized with monoclonal antibodies. I. Analysis of multiple antigenic sites. J Cell Biol. 1985;101:2047–2054. doi: 10.1083/jcb.101.6.2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega JA, Glees P, Neuhoff V. Ultrastructural analysis of individual layers in the lateral geniculate body of the monkey. Z Zellforsch Mikrosk Anat. 1968;87:82–100. doi: 10.1007/BF00326562. [DOI] [PubMed] [Google Scholar]
- Chang YC, Gottlieb DI. Characterization of the proteins purified with monoclonal antibodies to glutamic acid decarboxylase. J Neurosci. 1988;8:2123–2130. doi: 10.1523/JNEUROSCI.08-06-02123.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conradi S. Observations on the ultrastructure of the axon hillock and initial axon segment of lumbosacral motoneurons in the cat. Acta Physiol Scand Suppl. 1969;332:65–84. [PubMed] [Google Scholar]
- Crouzet M, Urdaci M, Dulau L, Aigle M. Yeast mutant affected for viability upon nutrient starvation: characterization and cloning of the RVS161 gene. Yeast. 1991;7:727–743. doi: 10.1002/yea.320070708. [DOI] [PubMed] [Google Scholar]
- David C, Solimena M, De Camilli P. Autoimmunity in stiff-Man syndrome with breast cancer is targeted to the C-terminal region of human amphiphysin, a protein similar to the yeast proteins, Rvs167 and Rvs161. FEBS (Fed Eur Biochem Soc) Letts. 1994;351:73–79. doi: 10.1016/0014-5793(94)00826-4. [DOI] [PubMed] [Google Scholar]
- David C, McPherson PS, Mundigl O, De Camilli P. A role of amphiphysin in synaptic vesicle endocytosis suggested by its binding to dynamin in nerve terminals. Proc Natl Acad Sci USA. 1996;93:331–335. doi: 10.1073/pnas.93.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P, Cameron R, Greengard P. Synapsin I (protein I), a nerve terminal-specific phosphoprotein. I. Its general distribution in synapses of the central and peripheral nervous system demonstrated by immunofluorescence in frozen and plastic sections. J Cell Biol. 1983;96:1337–1354. doi: 10.1083/jcb.96.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Camilli P, Miller PE, Navone F, Theurkauf WE, Vallee RB. Distribution of microtubule-associated protein 2 in the nervous system of the rat studied by immunofluorescence. Neuroscience. 1984;11:817–846. [PubMed] [Google Scholar]
- De Camilli P, Thomas A, Cofiell R, Folli F, Lichte B, Piccolo G, Meinck HM, Austoni M, Fassetta G, Bottazzo GF, et al. The synaptic vesicle-associated protein amphiphysin is the 128-kD autoantigen of StiffMan syndrome with breast cancer. J Exp Med. 1993;178:2219–2223. doi: 10.1084/jem.178.6.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarajan P, Stabach PR, Mann AS, Ardito T, Kashgarian M, Morrow JS. Identification of a small cytoplasmic ankyrin (AnkG119) in the kidney and muscle that binds β I σ spectrin and associates with the Golgi apparatus. J Cell Biol. 1996;133:819–830. doi: 10.1083/jcb.133.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flucher BE, Morton ME, Froehner SC, Daniels MP. Localization of the α1 and α2 subunits of the dihydropyridine receptor and ankyrin in skeletal muscle triads. Neuron. 1990;5:339–351. doi: 10.1016/0896-6273(90)90170-k. [DOI] [PubMed] [Google Scholar]
- Folli F, Solimena M, Cofiell R, Austoni M, Tallini G, Fassetta G, Bates D, Cartlidge N, Bottazzo GF, Piccolo G, et al. Autoantibodies to a 128kd synaptic protein in three women with the Stiff-Man syndrome and breast cancer. New Engl J Med. 1993;328:546–551. doi: 10.1056/NEJM199302253280805. [DOI] [PubMed] [Google Scholar]
- Garippa RJ, Johnson A, Park J, Petrush RL, McGraw TE. The carboxyl terminus of GLUT4 contains a serine-leucine-leucine sequence that functions as a potent internalization motif in Chinese hamster ovary cells. J Biol Chem. 1996;271:20660–20668. doi: 10.1074/jbc.271.34.20660. [DOI] [PubMed] [Google Scholar]
- Gong W, Emanuel BS, Collins J, Kim DH, Wang Z, Chen F, Zhang G, Roe B, Budarf ML. A transcription map of the DiGeorge and velocardio-facial syndrome minimal critical region on 22q11. Hum Mol Genet. 1996;5:789–800. doi: 10.1093/hmg/5.6.789. [DOI] [PubMed] [Google Scholar]
- Gottardi CJ, Arpin M, Fanning AS, Louvard D. The junction-associated protein, zonula occludens-1, localizes to the nucleus before the maturation and during the remodeling of cell-cell contacts. Proc Natl Acad Sci USA. 1996;93:10779–10784. doi: 10.1073/pnas.93.20.10779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabs D, Slepnev V, Songyang Z, David C, Lynch M, Cantley LC, De Camilli P. The SH3 domain of amphiphysin binds the proline rich domain of dynamin at a single site which defines a new SH3 binding consensus sequence. J Biol Chem. 1997;272:13419–13425. doi: 10.1074/jbc.272.20.13419. [DOI] [PubMed] [Google Scholar]
- Guo W, Jorgensen AO, Campbell KP. Characterization and ultrastructural localization of a novel 90-kDa protein unique to skeletal muscle junctional sarcoplasmic reticulum. J Biol Chem. 1994;269:28359–28365. [PubMed] [Google Scholar]
- Kapfhamer D, Miller DE, Lambert S, Bennett V, Glover TW, Burmeister M. Chromosomal localization of the ankyrinG gene (ANK3/ Ank3) to human 10q21 and mouse 10. Genomics. 1995;27:189–191. doi: 10.1006/geno.1995.1023. [DOI] [PubMed] [Google Scholar]
- Karlsson UL. Three-dimensional studies of neurons in the lateral geniculate nucleus of the rat. III. Specialized neuronal contacts in the neuropile. J Ultrastruct Res. 1967;17:137–157. doi: 10.1016/s0022-5320(67)80026-1. [DOI] [PubMed] [Google Scholar]
- Kaufman SJ, Bielser D, Foster RF. Localization of anti-clathrin antibody in the sarcomere and sensitivity of myofibril structure to chloroquine suggest a role for clathrin in myofibril assembly. Exp Cell Res. 1990;191:227–238. doi: 10.1016/0014-4827(90)90009-y. [DOI] [PubMed] [Google Scholar]
- Kedra C, Peyrard M, Fransson I, Collins JE, Dunham I, Roe BA, Dumanski JP. Characterization of a second human clathrin heavy chain polypeptide gene (CLH-22)from chromosome 22q11. Hum Mol Genet. 1996;5:625–631. doi: 10.1093/hmg/5.5.625. [DOI] [PubMed] [Google Scholar]
- Keller GA, Tokuyasu KT, Dutton AH, Singer SJ. An improved procedure for immunoelectron microscopy: ultrathin plastic embedding of immunolabeled ultrathin frozen sections. Proc Natl Acad Sci USA. 1984;81:5744–5747. doi: 10.1073/pnas.81.18.5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenig JH, Ikeda K. Disappearance and reformation of synaptic vesicle membrane upon transmitter release observed under reversible blockage of membrane retrieval. J Neurosci. 1989;9:3844–3860. doi: 10.1523/JNEUROSCI.09-11-03844.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordeli E, Lambert S, Bennett V. AnkyrinG. A new ankyrin gene with neural-specific isoforms localized at the axonal initial segment and node of Ranvier. J Biol Chem. 1995;270:2352–2359. doi: 10.1074/jbc.270.5.2352. [DOI] [PubMed] [Google Scholar]
- Kosaka T, Ikeda K. Possible temperature-dependent blockage of synaptic vesicle recycling induced by a single gene mutation in Drosophila. J Neurobiol. 1983;14:207–225. doi: 10.1002/neu.480140305. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lau HY, Caswell AH, Garcia H, Letelier L. Ouabain binding and coupling sodium, potassium and chloride transport in isolated transverse tubules from skeletal muscle. J Gen Physiol. 1979;74:335–349. doi: 10.1085/jgp.74.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichte B, Veh RW, Meyer HE, Kilimann MW. Amphiphysin, a novel protein associated with synaptic vesicles. EMBO (Eur Mol Biol Organ) J. 1992;11:2521–2530. doi: 10.1002/j.1460-2075.1992.tb05317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay EA, Rizzu P, Antonacci R, Jurecic V, Delmas-Mata J, Lee CC, Kim UJ, Scambler PJ, Baldini A. A transcription map in the CATCH22 critical region: identification, mapping, and ordering of four novel transcripts expressed in heart. Genomics. 1996;32:104–112. doi: 10.1006/geno.1996.0082. [DOI] [PubMed] [Google Scholar]
- Lupas A. Prediction and analysis of coiled-coil structures. Methods Enzymol. 1996;266:513–525. doi: 10.1016/s0076-6879(96)66032-7. [DOI] [PubMed] [Google Scholar]
- McPherson PS, Takei K, Schmid SL, De Camilli P. p145, a major Grb2-binding protein in brain, is co-localized with dynamin in nerve terminals where it undergoes activity-dependent dephosphorylation. J Biol Chem. 1994;269:30132–30139. [PubMed] [Google Scholar]
- McPherson PS, Garcia EP, Slepnev VI, David C, Zhang XM, Grabs D, Sossin WS, Bauerfeind R, Nemoto Y, De Camilli P. A presynaptic inositol-5-phosphatase. Nature (Lond) 1996;379:353–357. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- Mugnaini, E., and W.H. Oertel. 1985. An atlas of the distribution of GABAnergic neurons and terminals in the rat CNS as revealed by GAD immunocytochemistry. In Handbook of Chemical Neuroanatomy. Vol 4: GABA and Neuropeptides in the CNS. A. Bjorklund and T. Hokfelt, editors. Elsevier Science Publishers, Amsterdam, The Netherlands. 436–608.
- Munn AL, Riezman H. Endocytosis is required for the growth of vacuolar H(+)-ATPase-defective yeast: identification of six new END genes. J Cell Biol. 1994;127:373–386. doi: 10.1083/jcb.127.2.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn AL, Stevenson BJ, Geli MI, Riezman H. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1995;6:1721–1742. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz P, Rosemblatt M, Testar X, Palacin M, Thoidis G, Pilch PF, Zorzano A. The T-tubule is a cell-surface target for insulin-regulated recycling of membrane proteins in skeletal muscle. Biochem J. 1995a;307:393–400. doi: 10.1042/bj3120393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz P, Rosemblatt M, Testar X, Palacin M, Zorzano A. Isolation and characterization of distinct domains of sarcolemma and T-tubules from rat skeletal muscle. Biochem J. 1995b;307:273–280. doi: 10.1042/bj3070273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson WJ, Veshnock PJ. Ankyrin binding to (Na+ + K+)ATPase and implications for the organization of membrane domains in polarized cells. Nature (Lond) 1987;328:533–536. doi: 10.1038/328533a0. [DOI] [PubMed] [Google Scholar]
- Palay SL, Sotelo C, Peters A, Orkand PM. The axon hillock and the initial segment. J Cell Biol. 1968;38:193–201. doi: 10.1083/jcb.38.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters, A., S. Palay, and H. Webster. 1991. The axon. In The Fine Structure of the Nervous System. A. Peters, S.L. Palay, and H. Webster, editors. Oxford University Press, New York. 101–137.
- Robinson LJ, Pang S, Harris DS, Heuser J, James DE. Translocation of the glucose transporter (GLUT4) to the cell surface in permeabilized 3T3-L1 adipocytes: effects of ATP, insulin, and GTP gamma S and localization of GLUT4 to clathrin lattices. J Cell Biol. 1996;117:1181–1196. doi: 10.1083/jcb.117.6.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain SH, Masiarz FR, Munemitsu S, Polakis P. Association of the APC gene product with β-catenin. Science (Wash DC) 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- Sakamuro D, Elliott KJ, Wechsler-Reya R, Prendergast GC. BIN1 is a novel MYC-interacting protein with features of a tumour suppressor. Nat Gen. 1996;14:69–76. doi: 10.1038/ng0996-69. [DOI] [PubMed] [Google Scholar]
- Shiga T, Oppenheim MW. Immunolocalization studies of putative guidance molecules used by axons and growth cones of intersegemental interneurons in the chick embryo spinal cord. J Comp Neurol. 1991;310:234–252. doi: 10.1002/cne.903100208. [DOI] [PubMed] [Google Scholar]
- Shpetner HS, Vallee RB. Identification of dynamin, a novel mechanochemical enzyme that mediates interactions between microtubules. Cell. 1989;59:421–432. doi: 10.1016/0092-8674(89)90027-5. [DOI] [PubMed] [Google Scholar]
- Shupliakov O, Low P, Grabs D, Gad H, Chen H, David C, Takei K, De Camilli P, Brodin L. Synaptic vesicle endocytosis impaired by disruption of dynamin-SH3 domain interactions. Science (Wash DC) 1997;276:259–263. doi: 10.1126/science.276.5310.259. [DOI] [PubMed] [Google Scholar]
- Sirotkin H, Morrow B, DasGupta R, Goldberg R, Patanjali SR, Shi G, Cannizzaro L, Shprintzen R, Weissman S, Kucherlapati R. Isolation of a new clathrin heavy chain gene with muscle-specific expression from the region commonly deleted in the velo-cardio-facial syndrome. Human Mol Genet. 1996;5:617–624. doi: 10.1093/hmg/5.5.617. [DOI] [PubMed] [Google Scholar]
- Sivadon P, Bauer F, Aigle M, Crouzet M. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: The Rvs161 protein shares common domains with the brain protein amphiphysin. Mol Gen Genet. 1995;246:485–495. doi: 10.1007/BF00290452. [DOI] [PubMed] [Google Scholar]
- Slot JW, Geuze HJ, Gigengack S, James DE, Lienhard GE. Translocation of the glucose transporter GLUT4 in cardiac myocytes of the rat. Proc Natl Acad Sci USA. 1991;88:7815–7819. doi: 10.1073/pnas.88.17.7815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks AB, Hoffman NG, McConnell SJ, Fowlkes DM, Kay BK. Cloning of ligand targets: systemic isolation of SH3 domain-containing proteins. Nat Biotechnol. 1996;14:741–744. doi: 10.1038/nbt0696-741. [DOI] [PubMed] [Google Scholar]
- Srinivasan Y, Elmer L, Davis J, Bennett V, Angelides K. Ankyrin and spectrin associate with voltage-dependent sodium channels in brain. Nature (Lond) 1988;333:177–180. doi: 10.1038/333177a0. [DOI] [PubMed] [Google Scholar]
- Takei K, McPherson PS, Schmid SL, De Camilli P. Tubular membrane invaginations coated by dynamin rings are induced by GTP-γ S in nerve terminals. Nature (Lond) 1995;374:186–190. doi: 10.1038/374186a0. [DOI] [PubMed] [Google Scholar]
- Tokuyasu KT. Use of poly(vinylpyrrolidone) and poly(vinyl alcohol) for cryoultramicrotomy. Histochem J. 1989;21:163–171. doi: 10.1007/BF01007491. [DOI] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Itoh M, Nagafuchi A, Yonemura S, Tsukita S. Submembranous junctional plaque proteins include potential tumor suppressor molecules. J Cell Biol. 1993;123:1049–1053. doi: 10.1083/jcb.123.5.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Südhof TC, Anderson RGW. The appendage domain of α-adaptin is a high affinity binding site for dynamin. J Biol Chem. 1995;270:10079–10083. doi: 10.1074/jbc.270.17.10079. [DOI] [PubMed] [Google Scholar]
- Wang W, Hansen PA, Marshall BA, Holloszy JO, Mueckler M. Insulin unmasks a COOH-terminal glut4 epitope and increases glucose transport across T-tubules in skeletal muscle. J Cell Biol. 1996;135:415–430. doi: 10.1083/jcb.135.2.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waxman SG, Quick DC. Intra-axonal ferric ion-ferrocyanaide staining of nodes of Ranvier and initial segments in central myelinated fibers. Brain Res. 1978;144:1–10. doi: 10.1016/0006-8993(78)90430-4. [DOI] [PubMed] [Google Scholar]
- Waxman SG, Ritchie JM. Molecular dissection of the myelinated axon. Ann Neurol. 1993;33:121–136. doi: 10.1002/ana.410330202. [DOI] [PubMed] [Google Scholar]