Abstract
Pex mutants of the yeast Yarrowia lipolytica are defective in peroxisome assembly. The mutant strain pex16-1 lacks morphologically recognizable peroxisomes. Most peroxisomal proteins are mislocalized to a subcellular fraction enriched for cytosol in pex16 strains, but a subset of peroxisomal proteins is localized at, or near, wild-type levels to a fraction typically enriched for peroxisomes. The PEX16 gene was isolated by functional complementation of the pex16-1 strain and encodes a protein, Pex16p, of 391 amino acids (44,479 D). Pex16p has no known homologues. Pex16p is a peripheral protein located at the matrix face of the peroxisomal membrane. Substitution of the carboxylterminal tripeptide Ser-Thr-Leu, which is similar to the consensus sequence of peroxisomal targeting signal 1, does not affect targeting of Pex16p to peroxisomes. Pex16p is synthesized in wild-type cells grown in glucose-containing media, and its levels are modestly increased by growth of cells in oleic acid–containing medium. Overexpression of the PEX16 gene in oleic acid– grown Y. lipolytica leads to the appearance of a small number of enlarged peroxisomes, which contain the normal complement of peroxisomal proteins at levels approaching those of wild-type peroxisomes.
Peroxisomes, along with the glyoxysomes of plants and the glycosomes of trypanosomes, comprise the microbody family of organelles. Peroxisomes compartmentalize a number of essential biochemical functions, including the β-oxidation of fatty acids (Lazarow and de Duve, 1976) and the decomposition of H2O2 by catalase (de Duve and Baudhuin, 1966). Peroxisomes are indispensable for normal human development and physiology, as shown by the lethality of genetic disorders such as Zellweger syndrome in which peroxisomes fail to assemble normally (Lazarow and Moser, 1994).
The question of how peroxisomes assemble has received a great deal of attention in recent years. Peroxisomal proteins are synthesized on free polysomes in the cytosol. Most soluble proteins of the matrix are targeted by one of two types of peroxisomal targeting signals (PTS)1. PTS1 is a carboxyl-terminal tripeptide with the consensus structure (Ser/Ala/Cys)(Lys/Arg/His)(Leu/Met). PTS2 is a sometimes cleaved amino-terminal signal with the consensus structure (Arg/Lys)(Leu/Val/Ile)(X)5(His/Gln)(Leu/ Ala) (for reviews see Subramani, 1993, 1996; Rachubinski and Subramani, 1995). The PTSs of peroxisomal membrane proteins remain less well defined, although Dyer et al. (1996) have recently shown that the membrane protein PMP47 of the yeast Candida boidinii is targeted to peroxisomes by a hydrophilic loop of 20 amino acids located between two membrane-spanning domains. Peroxisomes, and the related glycosomes, are different from mitochondria and chloroplasts in that they can translocate folded and oligomeric proteins (Glover et al., 1994; McNew and Goodman, 1994, 1996; Walton et al., 1995; Häusler et al., 1996).
Complementation of peroxisome assembly mutants, collectively known as pex mutants (Distel et al., 1996), in both yeast and mammalian species has proven invaluable in identifying proteins called peroxins required for peroxisome assembly. Peroxins have been identified that act as receptors for PTS1 or PTS2 motifs (for review see McNew and Goodman, 1996). However, while the analysis of pex mutants has identified an ever-increasing number of peroxins required for peroxisomal protein targeting and import, much less is known about the mechanisms controlling peroxisome size and number. New peroxisomes are thought to arise by fission of preexisting peroxisomes (Lazarow and Fujiki, 1985), and, therefore, peroxisomal templates must be present under all conditions. A peroxisome proliferative response can be elicited in various yeast species by switching from a fermentable carbon source such as glucose to a carbon source that can be metabolized solely by peroxisomes, most often oleic acid or methanol. Two models for the temporal order of events of peroxisome proliferation have been proposed. In Hansenula polymorpha, peroxisomes first grow, and then smaller peroxisomes bud off mature peroxisomes (Veenhuis et al., 1979). In contrast, peroxisome proliferation and fission precede protein import in C. boidinii (Veenhuis and Goodman, 1990) and most other yeast species (for review see Subramani, 1993).
A few proteins have been implicated in directly regulating peroxisome number and size rather than having indirect effects on these processes through general defects in peroxisomal protein import. Oversynthesis of the peroxisomal integral membrane proteins Pex11p of Saccharomyces cerevisiae (Marshall et al., 1995) and C. boidinii (Sakai et al., 1995) and Pex3p and Pex10p of H. polymorpha (Baerends et al., 1996; Tan et al., 1995) leads to proliferation of normal peroxisomes, while cells lacking Pex11p of S. cerevisiae (Erdmann and Blobel, 1995; Marshall et al., 1995) and C. boidinii (Sakai et al., 1995) have fewer but larger peroxisomes than usual. Here we report the isolation and characterization of the gene PEX16 of the yeast Yarrowia lipolytica encoding the peroxin Pex16p. Pex16p is peripherally associated with the matrix face of the peroxisomal membrane. Interestingly, overexpression of the PEX16 gene in oleic acid–grown Y. lipolytica results in the appearance of enlarged peroxisomes that contain the normal complement of proteins at levels approaching those of wild-type peroxisomes.
Materials and Methods
Strains, Culture Conditions, and Microbial Techniques
The Y. lipolytica strains used in this study are listed in Table I. Growth was at 30°C. Strains containing plasmids were initially grown in YND (0.67% yeast nitrogen base without amino acids, 2% glucose) medium to an OD600 of ∼1.5 and then shifted to YNO (0.67% yeast nitrogen base without amino acids, 0.05% [wt/vol] Tween 40, 0.1% [wt/vol] oleic acid) medium to proliferate peroxisomes. Media were supplemented with uracil, leucine, lysine, and histidine each at 50 μg/ml, as required. Strains not containing plasmids were grown as above in YPD (1% yeast extract, 2% peptone, 2% glucose) medium and then shifted to YPBO (0.3% yeast extract, 0.5% peptone, 0.5% K2HPO4, 0.5% KH2PO4, 1% Brij-35, 1% [wt/ vol] oleic acid) medium to proliferate peroxisomes. DNA manipulation and growth of Escherichia coli were performed as previously described (Ausubel et al., 1989).
Table I.
Yarrowia lipolytica Strains Used in This Study
| Strain | Genotype | |
|---|---|---|
| E122 * | MatA, ura3-302, leu2-270, lys8-11 | |
| 22301-3 * | MatB, ura3-302, leu2-270, his1 | |
| pex16-1 ‡ | MatA, ura3-302, leu2-270, lys8-11, pex16-1 | |
| P16TR ‡ | MatA, ura3-302, leu2-270, lys8-11, p16HC2(LEU2) | |
| P16KO-8A ‡ | MatA, ura3-302, leu2-270, lys8-11, pex16::LEU2 | |
| P16KO-8B ‡ | MatB, ura3-302, leu2-270, his1, pex16::LEU2 | |
| D16-E1x8B ‡ | MatA/MatB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, +/pex16::LEU2 | |
| D16-8Ax22 ‡ | MatA/MatB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, pex16::LEU2/+ | |
| D16-P16x8B ‡ | MatA/MatB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, pex16-1/pex16::LEU2 | |
| D16-P16x22 ‡ | MatA/MatB, ura3-302/ura3-302, leu2-270/leu2-270, lys8-11/+, +/his1, pex16-1/+ | |
| pex16-HA ‡ | MatA, ura3-302, leu2-270, lys8-11, pex16-1:: PEX16HA-URA3 | |
| pex16-TH ‡ | MatA, ura3-302::PEX16TH-URA3, leu2-270, lys8-11, pex16-1 |
C. Gaillardin (Institut National de la Recherche Agronomique-CNRS) ThivervalGrignon.
This study.
Cloning, Sequencing, and Integrative Disruption of the PEX16 Gene
The pex16-1 mutant strain was isolated from randomly mutagenized Y. lipolytica strain E122, as previously described (Nuttley et al., 1993). The PEX16 gene was isolated by functional complementation of the pex16-1 strain using a Y. lipolytica genomic DNA library (Nuttley et al., 1993). Leu+ transformants were replica plated onto selective YNO agar plates and screened for their ability to utilize oleic acid as a sole carbon source. Total DNA was isolated from colonies that recovered growth and used to transform Escherichia coli for plasmid recovery. Restriction fragments prepared from genomic inserts were subcloned and tested for their ability to functionally complement the pex16-1 strain. The smallest genomic DNA fragment capable of complementation was sequenced in both directions.
Targeted integrative disruption of the PEX16 gene was performed with the LEU2 gene of Y. lipolytica. A 2.2-kbp SphI fragment containing the LEU2 gene was ligated into the PEX16 gene cleaved with SphI, replacing a 0.9-kbp fragment. This construct was cleaved with ClaI/HindIII to liberate the LEU2 gene flanked by PEX16 gene sequences. This linear construct was used to transform Y. lipolytica to leucine prototrophy. Leu+ transformants were replica plated onto YNO- agar to screen for the ole− phenotype. Correct integration was confirmed by Southern blot analysis. Disruption strains were crossed with wild-type and pex16-1 mutant strains, and the resultant diploids were checked for growth on YNO-agar plates.
Cell Fractionation, Protein Isolation, and Protease Protection
Total cellular protein was isolated by glass bead disruption of cells in 25 mM Tris-HCl (pH 7.5), 100 mM KCl, 1 mM EDTA, 10% (vol/vol) glycerol, 0.1 mM DTT plus protease inhibitors (0.5 mM PMSF, and pepstatin, chymostatin, antipain, and leupeptin each at 1 μg/ml). Cells were subjected to subcellular fractionation to yield a postnuclear supernatant (PNS). The PNS was subjected to centrifugation at 20,000 g max to yield a pellet (20KgP) enriched for peroxisomes and mitochondria and a supernatant (20KgS), as previously described (Aitchison et al., 1991). Peroxisomes were purified from the 20KgP by isopycnic centrifugation on a discontinuous sucrose gradient (Titorenko et al., 1996). Peroxisomal subfractions were prepared by extraction at 4°C for 30 min with one of Ti8 buffer (10 mM Tris-HCl, pH 8.5, 5 mM EDTA, 0.5 mM PMSF), TS buffer (Ti8 buffer containing 0.5 M KCl; Erdmann and Blobel, 1995), or carbonate buffer (0.1 M Na2CO3, pH 11.5, 0.5 mM PMSF), followed by centrifugation at 200,000 g max at 4°C for 30 min in a rotor (TLA 120.2; Beckman Instruments, Fullerton, CA).
Protease protection experiments were performed on 20KgP fractions isolated in the absence of protease inhibitors. 0, 2, 5, 10, or 25 μg of trypsin was combined with 200 μg of protein, with or without 0.5% (vol/vol) Triton X-100, in a final volume of 100 μl. Reactions were incubated at 4°C for 30 min and terminated by addition of hot SDS-PAGE sample buffer (Laemmli, 1970) and immediate boiling. Samples were subjected to SDSPAGE, followed by immunoblotting.
Preparation of Antibodies
Antibodies to Pex16p were raised in guinea pig and rabbit against a maltose-binding protein–Pex16p fusion. The open reading frame (ORF) of the PEX16 gene was amplified from the plasmid p16N1 by PCR, using primers 249 (5′) and 224 (3′) (Table II). The product was digested with EcoRI and HindIII and ligated into the vector pMAL-c2 (New England Biolabs, Beverly, MA) in-frame and downstream of the gene encoding maltose-binding protein.
Table II.
Oligonucleotides
| Oligonucleotide | Sequence | |
|---|---|---|
| 222 | 5′-TCA TAA GCT TTG AGA ACC CCG AAG A-3′ | |
| 224 | 5′-GCT TAA GCT TCC AAT CAT CAA TCG CTT AGA-3′ | |
| 249 | 5′-TAC GAA TTC ATG ACG GAC AAG CTG GTC AA-3′ | |
| 261 | 5′-TTA AGC TTA AAG ATC TGC GGT GAA GTA GTA CCG AT-3′ | |
| THpr5′ | 5′-CAG ATC TAA CCT ACC GG-3′ | |
| THpr3′ | 5′-TGA ATT CGG TCC AAA GTG-3′ | |
| THtr5′ | 5′-GAG TGA ATT CAC ATA CAA G-3′ | |
| THtr3′ | 5′-GAG ATC TAC GAC CTG G-3′ | |
| 7-HA | 5′-GAT CCG CTA GCC ATG TAC CCA TAC GAC GTC CCA GAC | |
| GC GAT CGG TAG ATG GGT ATG CTG CAG GGT CTG | ||
| D P L A M Y P Y D V P D | ||
| TAC GCT GCC ATG TAC CCA TAC GAC GTC CCA GAC TAC | ||
| ATG CGA CGG TAC ATG GGT ATG CTG CAG GGT CTG ATG | ||
| Y A A M Y P Y D V P D Y | ||
| GCT GCC ATG GGT AAG GGT GAA TAG AAG AGG AAG ATC T-3′ | ||
| CGA CGG TAC CCA TTC CCA CTT ATC TTC TCC TTC | ||
| A A M G K G E – |
Antibodies to acyl-CoA oxidase, isocitrate lyase, thiolase, malate synthase, and Pex2p (formerly Pay5p [Eitzen et al., 1996]) were prepared as previously described (Eitzen et al., 1996). Antibodies to Pex5p (formerly Pay32p [Szilard et al., 1995]) and the carboxyl-terminal SKL-tripeptide motif were prepared as described (Szilard et al., 1995). 12CA5 monoclonal antibodies against the influenza virus hemagglutinin (HA) antigen were purchased from the Berkeley Antibody Company (Berkeley, CA).
Immunoblotting and detection of antigen–antibody complexes by enhanced chemiluminescence (Amersham Life Sciences, Mississauga, Ontario, Canada) were performed as described (Szilard et al., 1995).
Microscopical Analysis
Y. lipolytica cells were harvested 8 h after shifting from glucose-containing medium to oleic acid–containing medium. Whole cells and subcellular fractions were prepared for electron microscopy by fixation in 1.5% KMnO4 for 20 min, followed by embedding in TAAB 812 resin (Marivac, Halifax, Nova Scotia, Canada). For immunoelectronmicroscopy, subcellular fractions were fixed in 3% (wt/vol) paraformaldehyde/0.2% (wt/vol) glutaraldehyde for 1 h at 4°C and embedded in LR Gold resin (Electron Microscopy Sciences, Fort Washington, PA). Immunodetection was performed with rabbit anti-SKL primary antibodies and goat anti–rabbit IgG secondary antibodies conjugated to 10 nm colloidal gold (Sigma Chemical Co., Mississauga, Ontario, Canada). Immunofluorescence microscopy was performed as described (Aitchison et al., 1992; Szilard et al., 1995).
Epitope Tagging of Pex16p and Overexpression of the PEX16 Gene
Various PEX16 expression plasmids were constructed as follows. The PEX16 gene, along with 965 and 16 bp of genomic DNA 5′ and 3′, respectively, to the ORF, was amplified by PCR of p16N1 (Fig. 2 A) using primers 222 (5′) and 224 (3′) (Table II) and inserted as a blunt-ended fragment into the SmaI site of pGEM7Zf(+) (Promega Corp., Madison, WI). The insert was excised with HindIII and ligated into a Y. lipolytica shuttle vector to yield p16CN, which expresses mRNA for the full-length Pex16p. The PEX16 gene was also amplified by PCR of p16N1 using primers 222 (5′) and 261 (3′) (Table II) and inserted into pGEM7Zf(+), as above. The insert was excised and cloned into a shuttle vector, as above, to give p16Δ, which contains a modified PEX16 gene encoding Pex16p lacking the carboxyl-terminal amino acids Ser-Thr-Leu. p16Δ was cleaved with BglII, which cuts immediately before the stop codon, and the double-stranded oligonucleotide 7-HA (Table II) encoding the HA-epitope (Wilson et al., 1984) was ligated into this site, creating p16HA, which expresses mRNA for a Pex16p fused near its carboxyl terminus to two copies of the HAepitope (Pex16p-HA). p16CN and p16HA were used to transform the pex16-1 and P16KO-8A mutant strains.
Figure 2.

Cloning and analysis of the PEX16 gene. (A) Complementing activity of inserts, restriction map analysis, and targeted gene disruption strategy for the PEX16 gene. (Solid lines) Y. lipolytica genomic DNA; (open boxes) vector DNA. The ORFs of the PEX16 and LEU2 genes are indicated by the wide arrows. The (+) symbol denotes the ability and the (−) symbol the inability of an insert to confer growth on oleic acid to pex16-1. B, BamHI; C, ClaI; H, HindIII; S, SalI; Sp, SphI. (B) Nucleotide sequence of the PEX16 gene and deduced amino acid sequence of Pex16p. (Underlined residues) Predicted membrane-associated helices; (doubly underlined residues) predicted transmembrane α-helix. These sequence data are available from EMBL/GenBank/DDBJ under accession number U75433. (C) Hydropathy profile of Pex16p calculated according to Kyte and Doolittle (1982) with a window size of 15 amino acids. Four hydrophobic domains predicted to interact with membranes are underlined.
For PEX16 overexpression studies, the promoter and terminator regions of the Y. lipolytica thiolase gene were amplified by PCR of plasmid pS106 (Berninger et al., 1993) using the oligonucleotide pairs THpr5′/ THpr3′ and THtr5′/THtr3′, respectively (Table II). Amplified products were cloned into the plasmid pGEM7Zf(+) to create the expression cassette vector pTEC with a unique EcoRI cloning site between the promoter and terminator regions. The ORF of the PEX16 gene was amplified by PCR of p16N1 using oligonucleotides 249 (5′) and 224 (3′) (Table II) and cloned into pGEM7Zf(+) as a blunt-ended fragment. The insert was excised by digestion with EcoRI and ligated into the EcoRI site of pTEC. The PEX16 ORF flanked by the thiolase gene promoter and terminator regions was excised by digestion with BglII and cloned into a shuttle vector to make the plasmid p16TH.
PEX16 constructs encoding Pex16p-HA or flanked by the thiolase promoter/terminator regions were integrated into the Y. lipolytica genome by gene conversion (Rothstein, 1991) to control for gene copy number and to ensure stability of recombinant gene expression. The insert of p16HA or p16TH, along with the URA3 gene, was cloned into the plasmid pSP73 (Promega). The HA-tagged version of the PEX16 gene was integrated into the pex16-1 site by cutting upstream of the PEX16 gene. The thiolase gene promoter/terminator version of the PEX16 gene was integrated into the endogenous ura3-302 site by cutting within the URA3 gene of the integration construct. Linearized DNA was introduced into the pex16-1 mutant by electroporation, and Ura+ transformants were selected. Correct integration was determined by Southern blot analysis.
Analytical Procedures
Enzymatic activities of the peroxisomal marker catalase (Luck, 1963) and the mitochondrial markers cytochrome c oxidase (Douma et al., 1985) and fumarase (Tolbert, 1974) were measured by established procedures. Protein concentration was measured as described by Bradford (1976) using ovalbumin as a standard. Total nucleic acid was isolated by glass bead lysis and phenol extraction, as previously described (Eitzen et al., 1995). Northern blot and Southern blot analyses were performed as described by Ausubel et al. (1989). Densitometry was performed using a laser densitometer (Ultrascan XL; LKB Instruments, Bromma, Sweden) (Szilard et al., 1995).
Results
Isolation of the PEX16 Gene
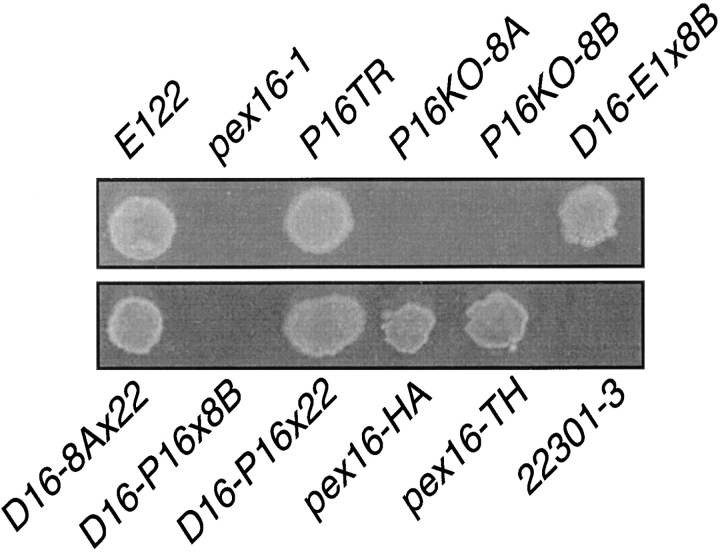
The pex16-1 mutant was isolated from randomly mutagenized Y. lipolytica cells by screening for the inability to use oleic acid as a sole carbon source (Fig. 1). Several biochemical and morphological criteria (data presented below) were used to determine that this strain was affected in peroxisome assembly (pex, or formerly pay, phenotype) (Nuttley et al., 1993). The PEX16 gene was isolated from a library of Y. lipolytica genomic DNA by functional complementation of the pex16-1 strain. Approximately 106 leucine prototrophy (Leu+) transformants were screened. Two strains recovered growth on oleic acid (ole+). Total DNA was isolated from these strains, and the complementing plasmids were recovered by transformation of E. coli. The recovered plasmids, p16N1 and p16C2, were mapped by restriction endonuclease digestion and were found to share a 5.5-kbp region (Fig. 2 A). Subfragments of this region were cloned, and the minimum complementing subfragment was localized to a 2.1-kbp HindIII/ClaI restriction fragment contained within plasmid p16HC2 (Fig. 2 A) in the transformed strain P16TR (Fig. 1).
Figure 1.
Growth of various Y. lipolytica strains on oleic acid– containing medium. The strains listed in Table I were grown for 3 d on YNO agar. The appearance of the original pex16-1 mutant is compared to that of the wild-type strain E122; the complemented strain P16TR; the gene disruption strains P16KO-8A and P16KO-8B; the diploid strains D16-E1x8B, D16-8Ax22, D16P16x8B, and D16-P16x22; the strain pex16-HA synthesizing Pex16p-HA; and the strain pex16-TH expressing the PEX16 gene from the thiolase promoter. The wild-type strain 22301-3 was not supplemented for its auxotrophic requirements. Growth on YNO requires at least one copy of the intact PEX16 gene.
Sequencing of the 2.1-kbp insert of p16HC2 revealed an ORF encoding a protein of 391 amino acids, Pex16p, with a predicted molecular weight of 44,479 (Fig. 2 B). Hydropathy analysis predicted the presence of several hydrophobic segments capable of membrane interaction (Fig. 2 C). Pex16p was predicted to contain one membrane-spanning α-helix and three membrane-associated helices (Fig. 2 B). A search of protein data bases using the GENINFO(R) BLAST Network Service (Blaster) of the National Center for Biotechnology Information revealed no significant homology between Pex16p and any other known protein.
The putative PEX16 gene was disrupted by targeted integration of the Y. lipolytica LEU2 gene to make the strains P16KO-8A and P16KO-8B in the A and B mating types, respectively (Table I). The PEX16 gene was disrupted in such a manner as to lack its initiating methionine codon. Disruption strains P16KO-8A and P16KO-8B could not grow on oleic acid (Fig. 1) and had the morphological and biochemical characteristics of the original pex16-1 strain (see below). The diploid strains D16-8Ax22 and D16-P16x22 from the mating of strains P16KO-8A and pex16-1, respectively, to wild-type strain 22301-3, and the diploid strain D16-E1x8B from the mating of wild-type strain E122 to strain P16KO-8B (Table I), could grow on oleic acid–containing medium (Fig. 1), showing the recessive nature of the pex16-1 mutation. The diploid strain D16-P16x8B made by mating the original pex16-1 mutant strain to strain P16KO-8B (Table I) could not grow on oleic acid-containing medium (Fig. 1). Accordingly, the ability to utilize oleic acid as the sole carbon source required at least one intact copy of the PEX16 gene, thereby confirming that the authentic PEX16 gene had been cloned.
pex16 Cells Lack Normal Peroxisomes but Show Evidence of Peroxisomal Structures
Normal peroxisomes of Y. lipolytica appear as round vesicular structures, 0.2–0.5 μm in diameter, with a granular electron-dense core and a single unit membrane (Fig. 3 A). The original mutant strain pex16-1 (Fig. 3 B) and the disruption strain P16KO-8A (Fig. 3 D) grown in oleic acid– containing medium lacked normal peroxisomes. However, these strains did show clusters of small (40–50 nm diam) vesicles (Fig. 3, B and D, arrows, and B, inset) and occasionally larger (100–150 nm diam) vesicular structures (Fig. 3, B and D, arrowheads, and D, inset). Morphologically similar structures could be seen in the wild-type strain (compare Fig. 3 A); however, extensive clustering of the smaller vesicles was rarely, if ever, observed in these cells. The transformed strain P16TR had the appearance of the wildtype strain and showed normal peroxisome morphology (Fig. 3 C).
Figure 3.
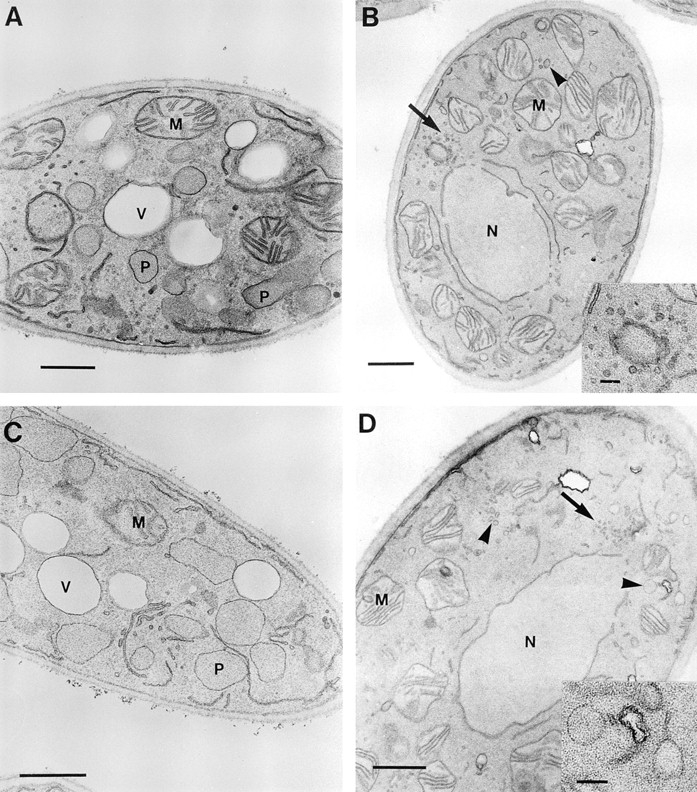
Ultrastructure of wild-type and pex16 mutant strains. The E122 (A), pex16-1 (B), P16TR (C), and P16KO-8A (D) strains were grown in YEPD medium to an OD600 of ∼1.5, transferred to YPBO medium at a dilution of 1:4, and grown for an additional 8 h in YPBO. Cells were fixed in KMnO4 and processed for electron microscopy. P, peroxisome; M, mitochondrion; N, nucleus; V, vacuole. (Arrows) Vesicular structures of 40–50 nm diam. (Arrowheads) Vesicular structures 100–150 nm diam. Inset in B shows a cluster of vesicles of 40–50 nm diam. Inset in D shows vesicles of 100–150 nm. Bars: (A– D) 0.5 μm; (inset) 0.1 μm.
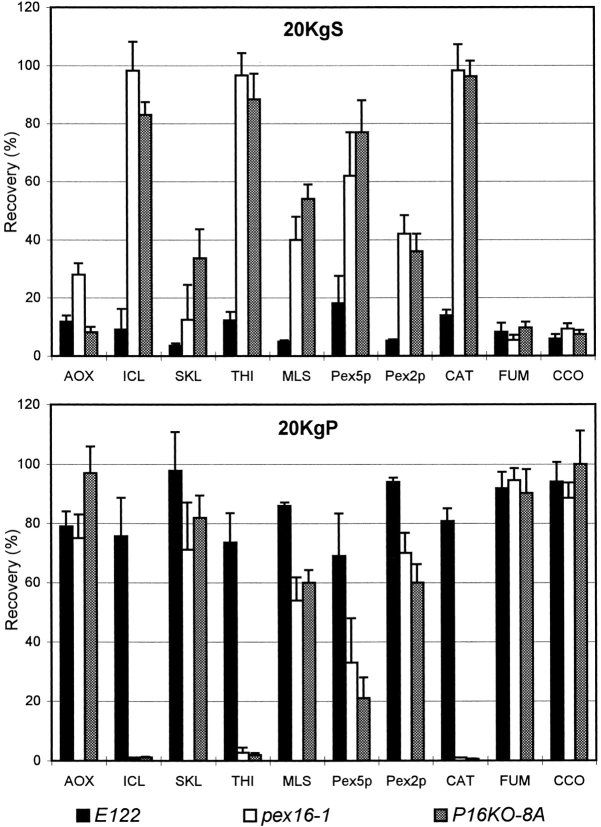
Although pex16 mutants lacked normal peroxisomes, subcellular fractionation provided evidence of peroxisomal structures in these strains. Wild-type and mutant strains were grown in oleic acid–containing medium to proliferate peroxisomes and then were fractionated into a 20KgP fraction enriched for peroxisomes and mitochondria and a 20KgS fraction, as described in Materials and Methods. In the wild-type strain E122, >75% of each peroxisomal protein was localized to the 20KgP fraction (Fig. 4). In contrast, in the original mutant strain pex16-1 and in the disruption strain P16KO-8A, several peroxisomal proteins were almost completely mislocalized to the 20KgS fraction, while others were only partially mislocalized (Fig. 4). Isocitrate lyase, thiolase, and catalase were found almost exclusively (85.5–99.4%) in the 20KgS fraction of the mutant strains. Malate synthase and the peroxisomal membrane proteins Pex2p (Eitzen et al., 1996) and Pex5p (Szilard et al., 1995) were partially mislocalized (38–79%) to the 20KgS, while acyl-CoA oxidase and a 62-kD anti-SKL reactive polypeptide were localized to the 20KgP fraction at levels comparable to those for the wild-type strain. The mitochondrial markers cytochrome c oxidase and fumarase were preferentially localized to the 20KgP fraction in both the wild-type and mutant strains (Fig. 4). Protease protection experiments showed that peroxisomal matrix proteins localized to the 20KgP fraction of the pex16-1 mutant were resistant to digestion by external protease in the absence, but not in the presence, of detergent (Fig. 5), consistent with their localization within membrane-enclosed structures.
Figure 4.
Peroxisomal proteins are mislocalized to the 20KgS fraction to varying extents in pex16 mutants. Wildtype strain E122 (solid bar) and mutant strains pex16-1 (open bar) and P16KO-8A (stippled bar) were grown as described in the legend to Fig. 3. Cells were subjected to subcellular fractionation to yield 20KgS and 20KgP fractions. The distributions of the enzymatic activities of catalase (CAT), fumarase (FUM), and cytochrome c oxidase (CCO) and of the immunosignals of acyl-CoA oxidase (AOX), isocitrate lyase (ICL), a 62-kD antiSKL reactive polypeptide (SKL), thiolase (THI), malate synthase (MLS), Pex5p, and Pex2p to the 20KgS and 20KgP fractions are given as the percent recovery of the respective total enzymatic activity or immunosignal in the PNS fraction. Immunoblots were quantitated by densitometry. Values reported are the means ± SD of four independent experiments, each analyzed in duplicate.
Figure 5.
Peroxisomal matrix proteins localized to the 20KgP fraction of the pex16-1 mutant are resistant to the action of external protease. The 20KgP fraction of mutant strain pex16-1 was isolated in the absence of protease inhibitors. 200 μg of protein of this fraction was incubated with 0, 2, 5, 10, and 25 μg of trypsin in the absence (−) or presence (+) of 0.5% (vol/vol) Triton X-100 on ice for 30 min. Reactions were terminated by addition of hot SDS-PAGE sample buffer and immediate boiling. Samples were analyzed by immunoblotting with anti–acyl-CoA oxidase (AOX), anti-SKL (SKL), and anti-thiolase (THI) antibodies.
Immunofluorescence analysis of oleic acid–grown wildtype cells probed with anti-SKL antibodies showed a punctate pattern of staining characteristic of peroxisomes (Fig. 6 A). In contast, pex16-1 cells (Fig. 6 B) probed with the same antibodies showed either a more generalized pattern of staining or a punctate pattern suggestive of peroxisomal structures that were smaller and less fluorescent than wild-type peroxisomes (Fig. 6 A). In an attempt to gain some initial insight into the nature of these structures, the 20KgP fractions of the wild-type and mutant strains were subfractionated by isopycnic centrifugation on discontinuous sucrose density gradients (see Materials and Methods). Fractions were analyzed for density, protein content, catalase and fumarase activities (Fig. 7 A), and by immunoblotting with antibodies to peroxisomal matrix (anti-SKL reactive polypeptides, acyl-CoA oxidase, and thiolase) and membrane (Pex2p) proteins (Fig. 7 B). In the wild-type strain E122, peroxisomal proteins were localized primarily to fractions 3–5, peaking in fraction 4 at a density of 1.21 g/cm3, while mitochondria were well separated from peroxisomes in fractions 9–11, peaking at 1.18 g/cm3. In the pex16-1 and P16KO-8A mutant strains, two peaks of peroxisomal proteins were observed, a lesser peak at a density of 1.21 g/cm3 and a greater peak at a density of 1.16 g/cm3 (fractions 9–12). It is noteworthy that the more slowly migrating anti-SKL reactive polypeptide of 64 kD was not present in the immunoblots of the fractions of the pex16 mutant strains (Fig. 7 B). This 64-kD polypeptide corresponds to isocitrate lyase (Eitzen, G.A., and R.A. Rachubinski, unpublished results), and as presented in Fig. 4, most isocitrate lyase was mislocalized to the 20KgS in the pex16 strains.
Figure 6.

Immuno-microscopic analysis of whole cells and subcellular fractions of the wild-type and pex16-1 mutant strains. The wild-type strain E122 (A) and the mutant strain pex16-1 (B) were grown for 8 h in YPBO, as described in the legend to Fig. 3. Cells were processed for immunofluorescence microscopy with rabbit antiSKL antibodies and FITCconjugated goat anti-rabbit IgG antibodies. (C–F) Subcellular fractions were isolated by isopycnic gradient analysis of 20KgP fractions of the wild-type E122 and the mutant pex16-1 strains as described in Materials and Methods and the legend to Fig. 7. Gradient fractions 4 and 11 of the wild-type strain (C and D, respectively) and of the pex16-1 strain (E and F, respectively) were fixed with paraformaldehyde/glutaraldehyde and processed for immuno-electronmicroscopy with anti-SKL antibodies. Peroxisomes in fraction 4 of the wild-type strain (C) and vesicular structures in both fraction 4 (E) and fraction 11 (F) of the pex16-1 strain are decorated with immunogold. Mitochondria are essentially undecorated. Bars: (A and B) 5 μm; (C, D, and F) 0.5 μm; (E) 0.25 μm.
Figure 7.
Isopycnic density gradient analysis of 20KgP fractions of the wild-type and pex16 strains. Strains were grown as described in the legend to Fig. 3 and subjected to subcellular fractionation. The 20KgP from each strain was subjected to centrifugation on a discontinuous sucrose gradient, as described in Materials and Methods. Each gradient was collected in 18 equivolume fractions. (A) Distributions of catalase, fumarase, and protein are shown for the strains E122 (-○-), pex16-1 (-x-), and P16KO-8A (-▪-). The stippled line in the panel at the top shows the density profile (g/cm3) of the gradient. Results are the means of two independent experiments, each analyzed in duplicate. (B) Equal volumes of gradient fractions were analyzed by immunoblotting. Abbreviations are as in Fig. 4.
Gradient fractions were also analyzed by immuno-electronmicroscopy using anti-SKL antibodies (Fig. 6). Fraction 4 (1.21 g/cm3, the normal density of peroxisomes) from the wild-type strain showed mainly large, round electron-dense profiles characteristic of peroxisomes (Fig. 6 C). These profiles were decorated with gold particles. The occasional mitochondria present in this fraction were largely undecorated by gold particles (Fig. 6 C). Fraction 4 of the pex16-1 mutant showed predominantly round, electron-dense profiles decorated by gold particles (Fig. 6 E). These structures were smaller in diameter than wild-type peroxisomes. Wild-type fraction 11 (1.16 g/cm3) contained predominantly undecorated mitochondria (Fig. 6 D). Fraction 11 from the pex16-1 mutant strain contained mitochondria and, in addition, small electron-dense structures decorated with gold particles (Fig. 6 F).
Pex16p Is a Peripheral Protein Preferentially Associated with the Matrix Face of the Peroxisomal Membrane
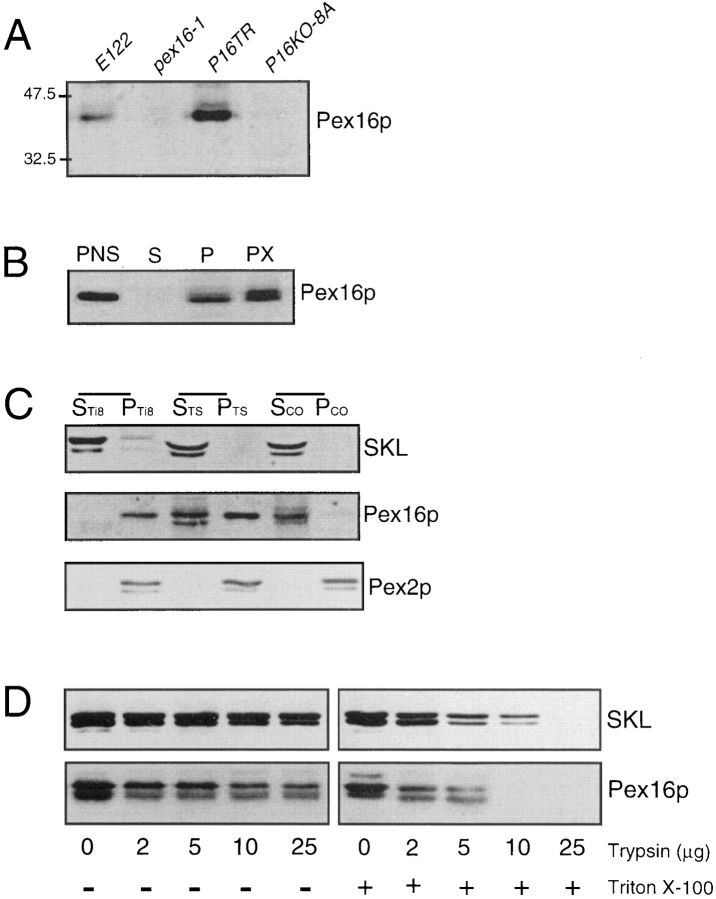
Antibodies raised against a maltose-binding protein–Pex16p fusion protein recognized a polypeptide of ∼43 kD in extracts of oleic acid-grown E122 cells but not of pex16-1 cells (Fig. 8 A). The molecular mass of this polypeptide is close to the predicted molecular mass of Pex16p, 44,479 D. No immunoreactive 43-kD polypeptide was seen in the lysate of the disruption strain P16KO-8A, but it was present in the lysate of the transformed strain P16TR (Fig. 8 A). Therefore, the antibodies specifically recognize Pex16p.
Figure 8.
Pex16p is a peripheral protein preferentially associated with the matrix face of the peroxisomal membrane. (A) Immunoblot analysis of whole cell extracts (50 μg of protein) of strains grown for 8 h in YPBO medium, as described in the legend to Fig. 3. Anti-Pex16p antibodies recognize a polypeptide of ∼43 kD in lysates of the wild-type strain E122 and the transformed strain P16TR but not in lysates of the original mutant strain pex16-1 or the disruption strain P16KO-8A. The numbers at left indicate the migrations of molecular mass standards (in kilodaltons). (B) Immunoblot analysis of subcellular fractions PNS, 20KgS (S), and 20KgP (P), and of whole peroxisomes (PX, 30 μg of protein) of the wild-type strain grown as in A and probed with anti-Pex16p antibodies. Equal fractions (0.1% of the total volume) of the PNS, 20KgS, and 20KgP were loaded. (C) Immunoblot analysis of peroxisomes separated into pellet (P) and supernatant (S) fractions by treatment with Ti8, TS, or sodium carbonate buffer. The upper blot was probed with anti-SKL antibodies to detect peroxisomal matrix proteins. The middle blot was probed with anti-Pex16p antibodies. The lower blot was probed with antibodies to the peroxisomal integral membrane protein Pex2p. (D) Protease protection analysis of anti-SKL reactive polypeptides and Pex16p. The 20KgP fraction (200 μg of protein) isolated in the absence of protease inhibitors from the wildtype strain E122 was incubated with the protease trypsin in the absence (−) or presence (+) of 0.5% (vol/vol) Triton X-100.
Immunoblot analysis of subcellular fractions and peroxisomes purified from oleic acid–grown wild-type cells, with anti-Pex16p antibodies, showed Pex16p to be localized to peroxisomes (Fig. 8 B, lane PX). Pex16p was absent from the 20KgS fraction (Fig. 8 B, lane S). Lysis of peroxisomes with Ti8 buffer, followed by high speed centrifugation, showed Pex16p to be localized exclusively to the pellet containing membrane proteins (Fig. 8 C, middle and bottom, lanes PTi8) and not to the supernatant containing soluble matrix proteins (Fig. 8 C, top, lane STi8). Increasing the stringency of the extraction buffer by the addition of 0.5 M KCl to the Ti8 buffer (TS buffer) led to partial extraction of Pex16p (Fig. 8 C, middle, compare lane PT S to lane STS) but not of the integral membrane protein Pex2p (Fig. 8 C, bottom, compare lane PTS to lane STS) from the membrane pellet. The more quickly migrating species of Pex16p in lane STS was probably due to nonspecific proteolysis. Treatment of peroxisomes with 0.1 M Na2CO3, pH 11.5, led to the complete extraction of Pex16p but not of Pex2p from the membrane pellet (Fig. 8 C, middle and bottom, respectively; compare lanes PCO to lanes SCO). Therefore, Pex16p shows the characteristics of a protein that is peripherally associated with the peroxisomal membrane.
To examine the association of Pex16p with the peroxisomal membrane, the 20KgP fraction isolated from the wildtype strain grown in oleic acid–containing medium was subjected to protease protection analysis (Fig. 8 D). Pex16p was protected from the action of external protease in the absence, but not in the presence, of the detergent Triton X-100. Peroxisomal matrix proteins recognized by anti-SKL antibodies behaved similarly to Pex16p in this analysis (Fig. 8 D). These results indicate that Pex16p is associated preferentially with the matrix face of the peroxisomal membrane.
Carboxyl Terminus of Pex16p Is Not Essential for Its Function or Targeting
Pex16p has at its carboxyl terminus the tripeptide sequence, Ser-Thr-Leu (Fig. 2 B). We investigated whether this tripeptide was a variant of the prototypical PTS1 sequence Ser-Lys-Leu and was necessary for targeting Pex16p to peroxisomes. The plasmid p16HA encodes a modified Pex16p (Pex16p-HA) that has two copies of the HA epitope tag appended to its carboxyl terminus and ends in the tripeptide, Lys-Gly-Glu, which does not resemble the PTS1 consensus motif. Transformation of strains pex16-1 and P16KO-8A with p16HA reestablished growth on oleic acid medium (data not shown). Therefore, Pex16p-HA was capable of functional complementation of the pex16 strains.
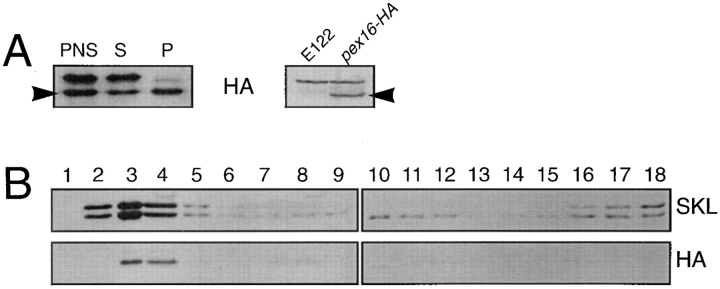
To ensure that functional complementation of the pex16 strains by the plasmid p16HA was not due to overexpression of the modified PEX16 gene, we constructed the strain pex16-HA (Table I), in the pex16-1 background, that harbors a single copy of the modified PEX16 gene under the control of the PEX16 promoter and coding for Pex16p-HA. This strain recovered growth on oleic acid medium (Fig. 1). Immunoblot analysis of subcellular fractions prepared from the pex16-HA strain grown in oleic acid–containing medium showed localization of Pex16pHA preferentially to the 20KgP fraction (Fig. 9 A, arrowhead, lane P) and to purified peroxisomes (Fig. 9 B). A polypeptide with reduced electrophoretic mobility relative to Pex16p-HA was nonspecifically recognized by 12CA5 antibodies, as shown by its detection in wild-type E122 cells not synthesizing Pex16p-HA (Fig. 9 A, right). Our results show that the amino acids Ser-Thr-Leu at the carboxyl terminus of Pex16p are not essential for its function or targeting to peroxisomes.
Figure 9.
Pex16p-HA is targeted to peroxisomes. (A) Immunoblot analysis of strain pex16-HA grown 8 h in YPBO medium, as described in the legend to Fig. 3, and then subjected to subcellular fractionation to yield PNS, 20KgS (S), and 20KgP (P) fractions. Equal portions of the fractions (0.1% of the total volume) were probed with 12CA5 antibodies to localize Pex16p-HA (arrowhead). (B) Immunoblot analysis of the 20KgP of the pex16HA strain fractionated by isopycnic density gradient centrifugation. Equal volumes of the fractions were probed with anti-SKL and 12CA5 antibodies. Anti-SKL reactive polypeptides and Pex16p-HA colocalize to fractions with the characteristic density of peroxisomes.
Oversynthesis of Pex16p Results in Fewer but Enlarged Peroxisomes
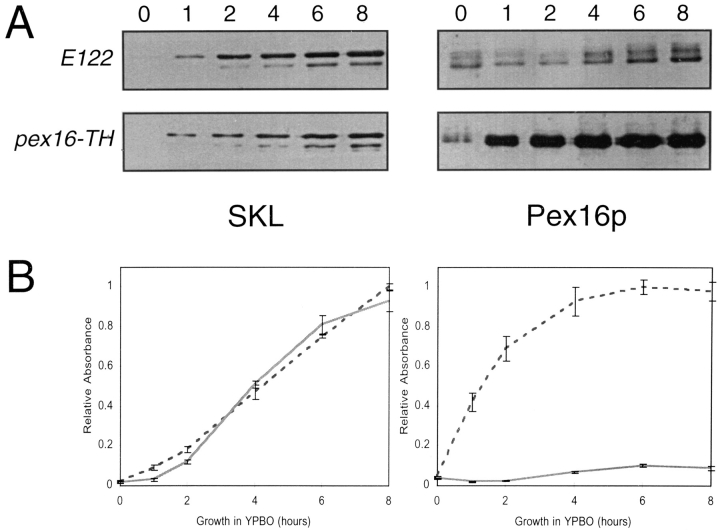
Immunofluorescence analysis of oleic acid–grown cells probed with antibodies directed against peroxisomal matrix proteins showed a punctate pattern characteristic of peroxisomes in wild-type cells (Fig. 10 A, and compare Fig. 6 A) but not in pex16 cells (compare Fig. 6 B). Expression of PEX16 in the transformed strain P16TR restored the characteristic punctate pattern of staining of peroxisomes (Fig. 10 B). To examine the effects of oversynthesis of Pex16p, the strain pex16-TH (Table I) expressing the PEX16 gene from the thiolase gene promoter was grown in oleic acid–containing medium. Immunoblot analysis showed that Pex16p synthesis was increased 10-fold after 8 h of growth in oleic acid–containing medium in the strain pex16-TH as compared with the wild-type strain E122 (Fig. 11, Pex16p), while the levels of expression of peroxisomal matrix proteins in the two strains remained unchanged (Fig. 11, SKL). It is interesting to note that Pex16p was synthesized by wild-type cells grown in glucose-containing medium (time = 0 h) and that the level of Pex16p only modestly increased during growth in oleic acid-containing medium (Fig. 11).
Figure 10.
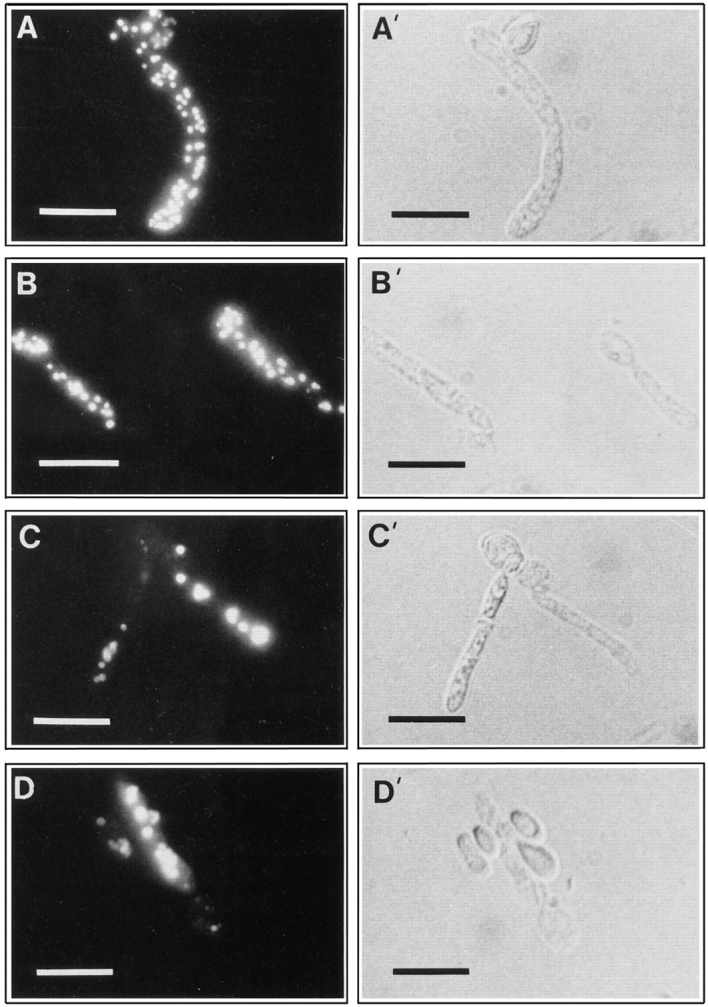
Oversynthesis of Pex16p results in fewer but enlarged peroxisomes. The strains E122 (A), P16TR (B), and pex16-TH (C and D) were grown for 8 h in YPBO medium, as described in the legend to Fig. 3. Cells were processed for immunofluorescence microscopy with rabbit anti-thiolase and FITCconjugated goat anti–rabbit IgG antibodies. (A′–D′), Nomarski images of (A–D). Unpermeabilized cells do not exhibit an immunofluorescent signal. Bars, 5 μm.
Figure 11.
Pex16p is oversynthesized in the strain pex16-TH. (A) Immunoblot analysis of cell lysates of the wild-type strain E122 and the strain pex16-TH. Strains were grown to an OD600 of ∼1.5 in YEPD medium (time = 0 h) and transferred to YPBO medium. Samples were removed at various times after transfer to YPBO medium. Lysates (50 μg of protein) of cells sampled at times 0, 1, 2, 4, 6, and 8 h (numbers at top) after transfer to YPBO medium were analyzed by immunoblotting with anti-SKL and anti-Pex16p antibodies. (B) Quantitation of immunoblots as in A. Values reported are the means ± SD for three independent experiments. Solid line, E122; dashed line, pex16-TH.
Immunofluorescence microscopy of the pex16-TH strain showed the effects of oversynthesis of Pex16p. After 8 h of growth in oleic acid-containing medium, cells of the pex16TH strain contained 1–5 enlarged peroxisomes (Fig. 10, C and D) instead of the 25–50 peroxisomes normally seen in wild-type cells grown under the same conditions (Figs. 6 A and 10 A). Since the ability to grow in oleic acid–containing medium was restored in the strain pex16-TH (Fig. 1), these enlarged peroxisomes are functional.
Electron microscopic analysis showed that pex16-TH cells contained peroxisomes with an average diameter of 0.87 ± 0.14 μm, while wild-type cells contained peroxisomes with an average diameter of 0.36 ± 0.10 μm. The increased size of peroxisomes in the pex16-TH strain apparently does not affect their inheritance, since cell sections routinely showed large peroxisomes in both halves of a dividing cell (Fig. 12 A).
Figure 12.
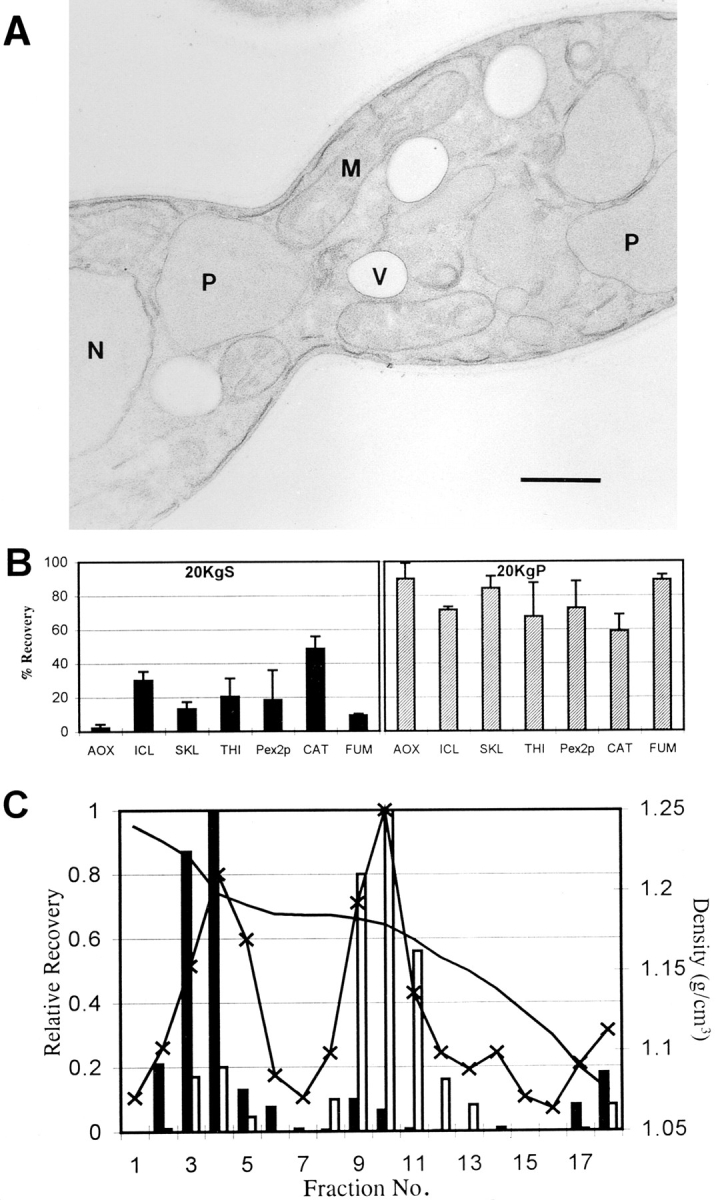
Characterization of the enlarged peroxisomes of the pex16TH strain. (A) Electron micrograph of a dividing cell of strain pex16-TH. The cell appears normal except for enlarged peroxisomes. Enlarged peroxisomes are present in both halves of the dividing cell. (B) The PEX16 overexpression strain, pex16-TH, was grown for 8 h in YPBO medium and subjected to subcellular fractionation to yield 20KgS and 20KgP fractions. The distribution of the enzymatic activities of catalase (CAT) and fumarase (FUM) and of the immunosignals of acyl-CoA oxidase (AOX), isocitrate lyase (ICL), a 62-kD antiSKL reactive polypeptide (SKL), thiolase (THI), and Pex2p to the 20KgS and 20KgP fractions are reported as the percent recovery of the respective total enzymatic activity or immunosignal in the PNS fraction. Immunoblots were quantitated by densitometry. Values reported are the means ±SD of two independent experiments, each analyzed in duplicate. (C) Isopycnic density gradient analysis of the 20KgP from the strain pex16-TH grown for 8 h in YPBO medium. Distribution of protein (–x–), catalase (solid bars), fumarase (open bars), and density (solid line). Values reported are the means of two independent experiments, each analyzed in duplicate. P, peroxisome; M, mitochondrion, N, nucleus. Bar, 0.5 μm.
Peroxisomal proteins were preferentially localized to the 20KgP isolated from the pex16-TH strain at levels approaching those of the 20KgP from the wild-type strain (Fig. 12 B). The reduction in levels of peroxisomal proteins in the 20KgP fraction from the pex16-TH strain may be due to an increased fragility of the enlarged peroxisomes vis-à-vis wild-type peroxisomes during subcellular fractionation and/or to a reduced overall number of protein import sites in the enlarged peroxisomes. The enlarged peroxisomes of the pex16-TH strain could be isolated by isopycnic centrifugation of the 20KgP on a discontinuous sucrose gradient (Fig. 12 C). These enlarged peroxisomes peaked at a density of 1.21 g/cm3, the same as wild-type peroxisomes (compare Fig. 7 A), and contained catalase (Fig. 12 C) and all other peroxisomal matrix and membrane proteins tested (data not shown). Mitochondria from the pex16-TH strain peaked at a density of 1.18 g/cm3, like those of the wild-type strain (compare Fig. 7 A).
Discussion
Here we report the following: the isolation and characterization of Y. lipolytica pex16 mutant strains, the cloning and sequencing of the PEX16 gene, the identification and characterization of the peroxin Pex16p, and an analysis of the effects of overexpression of the PEX16 gene.
Absence of Functional Peroxisomes but Evidence of Peroxisomal Structures in pex16 Mutant Strains
pex16 mutant strains cannot assemble functional peroxisomes. They are unable to grow using oleic acid as the sole carbon source, and under conditions of peroxisome induction, they fail to proliferate morphologically normal peroxisomes. However, subcellular fractionation showed that pex16 strains import a subset of peroxisomal proteins at, or near, wild-type levels, suggesting that they are not generalized mutants of peroxisomal protein import. While isocitrate lyase, thiolase, and catalase are completely mislocalized to a 20,000 g max supernatant fraction (20KgS) in pex16 mutants, other peroxisomal matrix (acyl-CoA oxidase, a 62-kD anti-SKL reactive polypeptide, malate synthase) and membrane (Pex2p and Pex5p) proteins are localized to an organellar fraction pelletable at 20,000 g max (20KgP).
Peroxisomal proteins pelletable to the 20KgP are contained in elements that equilibrate at densities of 1.21 and 1.16 g/cm3. Preliminary immunocytochemical analysis suggests that both of these fractions contain vesicular structures of varying diameters that were labeled by anti-SKL antibodies and immunogold (compare Fig. 6, E and F). These or similar vesicular structures were visible in electron micrographs of whole cells of the pex16 mutant strains and, to a lesser extent, of the wild-type strain (compare Fig. 6, B and A, respectively). In contrast, immunoelectronmicrographs of fractions from wild-type cells showed a preponderance of decorated profiles typical of peroxisomes in the fraction equilibrating at a density of 1.21 g/ cm3 (compare Fig. 6 C), while the fraction equilibrating at a density of 1.16 g/cm3 showed primarily mitochondrial profiles that were largely undecorated (compare Fig. 6 D). We do not know the origin or fate of the structures containing peroxisomal proteins that are found in pex16 cells. Perhaps they represent intermediates along a pathway of peroxisome biogenesis or distinct individual peroxisomes somehow compromised in their normal biogenetic progression. Whether either of these scenarios or a completely different scenario is true awaits further investigation.
Carboxyl-terminal Tripeptide Ser-Thr-Leu of Pex16p Is Not Essential for Its Targeting to Peroxisomes
Pex16p has at its carboxyl terminus the tripeptide Ser-ThrLeu. This sequence is very similar to the consensus sequence of PTS1 motifs, (Ser/Ala/Cys)(Lys/Arg/His)(Leu/ Met). We therefore investigated whether this tripeptide motif is required for the targeting of Pex16p to peroxisomes. Elimination of the tripeptide by addition of the HA epitope to the carboxyl terminus of Pex16p does not prevent its targeting to peroxisomes (compare Fig. 9). Therefore, the carboxyl-terminal tripeptide of Pex16p is not required for its targeting to peroxisomes.
Peroxisomes are able to import oligomeric and folded proteins (Glover et al., 1994; McNew and Goodman, 1994, 1996; Walton et al., 1995). The subunit composition of Pex16p is unknown. However, the gene encoding Pex16pHA was expressed in the P16KO-8A background, in which the ORF of the nuclear PEX16 gene is deleted. Accordingly, targeting of Pex16p-HA to peroxisomes cannot be the result of multimerization with a wild-type Pex16p having its carboxyl-terminal tripeptide intact. However, we cannot rule out the possibility that Pex16p-HA could gain access to the peroxisome by heterodimerizing with a different peroxisomal protein. Nevertheless, such a scenario would still be consistent with the carboxyl-terminal tripeptide of Pex16p not being necessary for its targeting to peroxisomes.
What Could be the Function of Pex16p?
What role might Pex16p play in peroxisome biogenesis? Our results indicate that Pex16p is a component of the peroxisome assembly machinery and suggest that Pex16p may in fact have more than one function.
Pex16p may facilitate the import of some peroxisomal proteins, notably of catalase, thiolase, and isocitrate lyase. These enzymes are not localized to peroxisomal structures that pellet to the 20KgP but are mislocalized to the 20KgS in pex16 cells (compare Fig. 4). However, the levels of these enzymes in the enlarged peroxisomes of the PEX16 overexpression strain approach the levels found in wildtype peroxisomes (compare Figs. 10 and 12). Whether facilitation of import of this subset of peroxisomal proteins can be attributed directly to the functioning of Pex16p cannot be answered at this time.
Our results also suggest that Pex16p may be involved in peroxisome proliferation. Interestingly, overexpression of the PEX16 gene in oleic acid–grown Y. lipolytica results in a reduced number of enlarged peroxisomes as compared to wild-type cells. Other peroxins have been implicated in peroxisomal proliferation or fission, although their actions contrast with those of Pex16p. Overexpression of Pex3p and Pex10p in H. polymorpha (Baerends et al., 1996; Tan et al., 1995) and Pex11p in S. cerevisiae and C. boidinii (Marshall et al., 1995; Sakai et al., 1995) leads to a proliferation of normal peroxisomes, while elimination of Pex11p in these yeast species leads to a small number of enlarged peroxisomes (Erdmann and Blobel, 1995; Marshall et al., 1995; Sakai et al., 1995). Whether Pex16p and these peroxins act themselves as positive and negative factors of peroxisome proliferation cannot be answered at this time. It will be interesting to determine whether Pex16p interacts with these peroxins and to identify novel partners of Pex16p so as to provide insight into the molecular machinery controlling peroxisome structure, growth, and proliferation.
Acknowledgments
We thank Honey Chan for assistance with electron microscopy.
Abbreviations used in this paper
- HA
hemagglutinin
- ORF
open reading frame
- PNS
postnuclear supernatant
- PTS
peroxisomal targeting signal
Footnotes
G.A. Eitzen is the recipient of a Studentship from the Alberta Heritage Foundation for Medical Research. R.K. Szilard is the recipient of a Studentship from the Medical Research Council (MRC) of Canada. R.A. Rachubinski is an MRC Scientist and an International Research Scholar of the Howard Hughes Medical Institute. This work was supported by a grant from the MRC to R.A. Rachubinski.
Please address all correspondence to Richard A. Rachubinski, Department of Cell Biology and Anatomy, University of Alberta, Medical Sciences Building 5-14, Edmonton, Alberta T6G 2H7, Canada. Tel.: (403) 492-9868; Fax: (403) 492-9278; E-mail: rrachubi@anat.med.ualberta.ca
References
- Aitchison JD, Murray WW, Rachubinski RA. The carboxyl-terminal tripeptide Ala-Lys-Ile is essential for targeting Candida tropicalistrifunctional enzyme to yeast peroxisomes. J Biol Chem. 1991;266:23197–23203. [PubMed] [Google Scholar]
- Aitchison JD, Szilard RK, Nuttley WM, Rachubinski RA. Antibodies directed against a yeast carboxyl-terminal peroxisomal targeting signal specifically recognize peroxisomal proteins from various yeasts. Yeast. 1992;8:721–734. doi: 10.1002/yea.320080905. [DOI] [PubMed] [Google Scholar]
- Ausubel, F.J., R. Brent, R.E. Kingston, D.D. Moore, J.G. Seidman, J.A. Smith, and K. Struhl. 1989. In Current Protocols in Molecular Biology. Green Publishing Associates, New York. 3.1–3.17, 13.11–13.12.
- Baerends RJS, Rasmussen SW, Hilbrands RE, van der Heide M, Faber KN, Reuvekamp PTW, Kiel JAKW, Cregg JM, van der Klei IJ, Veenhuis M. The Hansenula polymorpha PER9gene encodes a peroxisomal membrane protein essential for peroxisome assembly and integrity. J Biol Chem. 1996;271:8887–8894. doi: 10.1074/jbc.271.15.8887. [DOI] [PubMed] [Google Scholar]
- Berninger G, Schmidtchen R, Casel G, Knorr A, Rautenstrauss K, Kunau W-H, Schweizer E. Structure and metabolic control of the Yarrowia lipolyticaperoxisomal 3-oxoacyl-CoA thiolase gene. Eur J Biochem. 1993;216:607–613. doi: 10.1111/j.1432-1033.1993.tb18180.x. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- De Duve C, Baudhuin P. Peroxisomes (microbodies and related particles) Physiol Rev. 1966;46:323–357. doi: 10.1152/physrev.1966.46.2.323. [DOI] [PubMed] [Google Scholar]
- Distel B, Erdmann R, Gould SJ, Blobel G, Crane DI, Cregg JM, Dodt G, Fujiki Y, Goodman JM, Just WW, et al. A unified nomenclature for peroxisome biogenesis. J Cell Biol. 1996;135:1–3. doi: 10.1083/jcb.135.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douma AC, Veenhuis M, de Koning W, Evers M, Harder W. Dihydroxyacetone synthase is localized in the peroxisomal matrix of methanolgrown Hansenula polymorpha. . Arch Microbiol. 1985;143:237–243. [Google Scholar]
- Dyer JM, McNew JA, Goodman JM. The sorting sequence of the peroxisomal integral membrane protein PMP47 is contained within a short hydrophilic loop. J Cell Biol. 1996;133:269–280. doi: 10.1083/jcb.133.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitzen GA, Aitchison JD, Szilard RK, Veenhuis M, Nuttley WM, Rachubinski RA. The Yarrowia lipolytica gene PAY2encodes a 42kDa peroxisomal integral membrane protein essential for matrix protein import and peroxisome enlargement but not for peroxisome membrane proliferation. J Biol Chem. 1995;270:1429–1436. doi: 10.1074/jbc.270.3.1429. [DOI] [PubMed] [Google Scholar]
- Eitzen GA, Titorenko VI, Smith JJ, Veenhuis M, Szilard RK, Rachubinski RA. The Yarrowia lipolytica gene PAY5encodes a peroxisomal integral membrane protein homologous to the mammalian peroxisome assembly factor PAF-1. J Biol Chem. 1996;271:20300–20306. doi: 10.1074/jbc.271.34.20300. [DOI] [PubMed] [Google Scholar]
- Erdmann R, Blobel G. Giant peroxisomes in oleic acid-induced Saccharomyces cerevisiaelacking the peroxisomal membrane protein Pmp27p. J Cell Biol. 1995;128:509–523. doi: 10.1083/jcb.128.4.509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover JR, Andrews DW, Rachubinski RA. Saccharomyces cerevisiaeperoxisomal thiolase is imported as a dimer. Proc Natl Acad Sci USA. 1994;91:10541–10545. doi: 10.1073/pnas.91.22.10541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Häusler T, Stierhof Y-D, Wirtz E, Clayton C. Import of a DHFR hybrid protein into glycosomes in vivo is not inhibited by the folate analogue aminopterin. J Cell Biol. 1996;132:311–324. doi: 10.1083/jcb.132.3.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lazarow PB, de Duve C. A fatty acyl-CoA oxidizing system in rat liver peroxisomes: enhanced by clofibrate, a hypolipidemic drug. Proc Natl Acad Sci USA. 1976;73:2043–2046. doi: 10.1073/pnas.73.6.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- Lazarow, P.B., and H.W. Moser. 1994. Disorders of peroxisome biogenesis. In The Metabolic Basis of Inherited Disease. 7th Ed. C.R. Scriver, A.L. Beaudet, W.S. Sly, and A.D. Valle, editors. McGraw-Hill, New York. 2287–2324.
- Luck, H. 1963. Catalase. In Methods of Enzymatic Analysis. H.-U. Bergmeyer, editor. Academic Press, New York. 885–888.
- Marshall PA, Krimkevich YI, Lark RH, Dyer JM, Veenhuis M, Goodman JM. Pmp27 promotes peroxisomal proliferation. J Cell Biol. 1995;129:345–355. doi: 10.1083/jcb.129.2.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew JA, Goodman JM. An oligomeric protein is imported into peroxisomes in vivo. J Cell Biol. 1994;127:1245–1257. doi: 10.1083/jcb.127.5.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNew JA, Goodman JM. The targeting and assembly of peroxisomal proteins: some old rules do not apply. Trends Biochem Sci. 1996;21:54–58. [PubMed] [Google Scholar]
- Nuttley WM, Brade AM, Gaillardin C, Eitzen GA, Glover JR, Aitchison JD, Rachubinski RA. Rapid identification and characterization of peroxisomal assembly mutants in Yarrowia lipolytica. . Yeast. 1993;9:507–517. [Google Scholar]
- Rachubinski RA, Subramani S. How proteins penetrate peroxisomes. Cell. 1995;83:525–528. doi: 10.1016/0092-8674(95)90091-8. [DOI] [PubMed] [Google Scholar]
- Rothstein R. Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol. 1991;194:281–301. doi: 10.1016/0076-6879(91)94022-5. [DOI] [PubMed] [Google Scholar]
- Sakai Y, Marshall PA, Saiganji A, Takabe K, Saiki H, Kato N, Goodman JM. The Candida boidinii peroxisomal membrane protein Pmp30 has a role in peroxisomal proliferation and is functionally homologous to Pmp27 from Saccharomyces cerevisiae. . J Bacteriol. 1995;177:6773–6781. doi: 10.1128/jb.177.23.6773-6781.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramani S. Protein import into peroxisomes and biogenesis of the organelle. Annu Rev Cell Biol. 1993;9:445–478. doi: 10.1146/annurev.cb.09.110193.002305. [DOI] [PubMed] [Google Scholar]
- Subramani S. Protein translocation into peroxisomes. J Biol Chem. 1996;271:32483–32486. doi: 10.1074/jbc.271.51.32483. [DOI] [PubMed] [Google Scholar]
- Szilard RK, Titorenko VI, Veenhuis M, Rachubinski RA. Pay32p of the yeast Yarrowia lipolyticais an intraperoxisomal component of the matrix protein translocation machinery. J Cell Biol. 1995;131:1453–1469. doi: 10.1083/jcb.131.6.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan X, Waterham HR, Veenhuis M, Cregg JM. The Hansenula polymorpha PER8gene encodes a novel peroxisomal integral membrane protein involved in proliferation. J Cell Biol. 1995;128:307–319. doi: 10.1083/jcb.128.3.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titorenko VI, Eitzen GA, Rachubinski RA. Mutations in the PAY5 gene of the yeast Yarrowia lipolyticacause the accumulation of multiple subpopulations of peroxisomes. J Biol Chem. 1996;271:20307–20314. doi: 10.1074/jbc.271.34.20307. [DOI] [PubMed] [Google Scholar]
- Tolbert NE. Isolation of subcellular organelles of metabolism on isopycnic sucrose gradients. Methods Enzymol. 1974;31:734–746. doi: 10.1016/0076-6879(74)31077-4. [DOI] [PubMed] [Google Scholar]
- Veenhuis M, Goodman JM. Peroxisome assembly: membrane proliferation precedes the induction of the abundant matrix proteins in the methylotrophic yeast Candida boidinii. . J Cell Sci. 1990;96:583–590. doi: 10.1242/jcs.96.4.583. [DOI] [PubMed] [Google Scholar]
- Veenhuis M, Keizer I, Harder W. Characterization of peroxisomes in glucose-grown Hansenula polymorphaand their development after the transfer of cells into methanol-containing media. Arch Microbiol. 1979;120:167–175. [Google Scholar]
- Walton PA, Hill PE, Subramani S. Import of stably folded proteins into peroxisomes. Mol Biol Cell. 1995;6:675–683. doi: 10.1091/mbc.6.6.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]