Abstract
We cloned the myo2 gene of Schizosaccharomyces pombe, which encodes a type II myosin heavy chain, by virtue of its ability to promote diploidization in fission yeast cells. The myo2 gene encodes 1,526 amino acids in a single open reading frame. Myo2p shows homology to the head domains and the coiledcoil tail of the conventional type II myosin heavy chain and carries putative binding sites for ATP and actin. It also carries the IQ motif, which is a presumed binding site for the myosin light chain. However, Myo2p apparently carries only one IQ motif, while its counterparts in other species have two. There are nine proline residues, which should break α-helix, in the COOH-terminal coiled-coil region of Myo2p. Thus, Myo2p is rather unusual as a type II myosin heavy chain. Disruption of myo2 inhibited cell proliferation. myo2Δ cells showed normal punctate distribution of interphase actin, but they produced irregular actin rings and septa and were impaired in cell separation. Overproduction of Myo2p was also lethal, apparently blocking actin relocation. Nuclear division proceeded without actin ring formation and cytokinesis in cells overexpressing Myo2p, giving rise to multinucleated cells with dumbbell morphology. Analysis using tagged Myo2p revealed that Myo2p colocalizes with actin in the contractile ring, suggesting that Myo2p is a component of the ring and responsible for its contraction. Furthermore, genetic evidence suggested that the acto–myosin system may interact with the Ras pathway, which regulates mating and the maintenance of cell morphology in S. pombe.
Cell division is an essential event for cell proliferation. Many eukaryotic cells divide at their medial region by virtue of an acto–myosin system constituting a contractile ring. The involvement of actin and type II myosin in the contractile ring, as well as some additional proteins, has been inferred in various organisms (37). However, little is known about how this ring is framed at a specific period of the cell cycle and what regulates its timely constriction (for review see reference 12).
Fission yeast Schizosaccharomyces pombe is a unicellular eukaryotic organism genetically tractable, which has provided an excellent model system to study cell cycle regulation (18). In recent years, this microbe has been also used conveniently in the study of cellular morphogenesis and cytokinesis (for reviews see references 10, 32, 39). Fission yeast has a rodlike morphology and divides by medial cleavage using an actin-based contractile ring, like most animal cells (24). During interphase, actin patches are seen at both ends of S. pombe cells, where the cell volume is increased. As S. pombe cells enter mitosis, they stop elongation, and actin patches disappear from the cell tips. In early M-phase, actin relocates to the plasma membrane at the cell equator surrounding a nucleus, forming a contractile ring (24). After nuclear division, the contractile ring constricts, and a septum starts to grow centripetally after the constriction. The actin ring gradually loses its visibility as the septum grows, and many actin dots appear in the vicinity of the septum, which are probably involved in the supply of septal materials. When the septum has been assembled and the cytoplasm separated, it undergoes digestion so that two daughter cells can be split.
Previous studies have identified genes and gene products that are pertinent to cytokinesis in S. pombe. The cdc3 and cdc8 genes encode profilin and tropomyosin, respectively (1, 2). These two proteins are required for general actin organization, and a mutant strain defective in either of them exhibits abnormal actin distribution in all phases of the cell cycle (1, 2, 6). In contrast, certain temperaturesensitive mutants, as typically analyzed by Chang et al. (6), show normal actin patches during interphase but produce either aberrant actin rings or no ring at all during M-phase. One of the genes causing this phenotype is cdc4, which encodes a putative myosin light chain (27). Cdc4p is a 141– amino acid polypeptide that is similar to EF-hand proteins and it apparently localizes to the contractile ring during cytokinesis (27). This implies that S. pombe also uses type II myosin to execute cell division.
Mammalian type II myosin is a hexamer, composed of two copies each of the heavy chain, the essential light chain, and the regulatory light chain (for review see reference 21). While Cdc4p has been assigned to be a putative light chain of myosin II, so far there is no report on the characterization of a myosin heavy chain in S. pombe. S. pombe is favorably accepted as a model organism to study cytokinesis, but one may note that its cytokinesis does not appear to be perfectly analogous to that of animal cells: for example, actin rings emerge at an earlier stage of M-phase in fission yeast than in animal cells, and cytokinesis is associated with septum formation in fission yeast, which never happens in animals. Thus, it will be necessary to clarify the molecular architecture of the contractile ring of this microbe and compare it with those of other organisms to assess the extent to which S. pombe can serve as a model organism in the study of cytokinesis. An obvious deficit in this regard is the lack of knowledge about its type II myosin heavy chain.
During a screening for high–copy number suppressors of the mating deficiency in S. pombe ras1–related mutants, we fortuitously obtained cDNA clones that elevated the frequency of diploidization in haploid host cells. These clones turned out to derive from a single gene, named myo2, which appeared to encode a type II myosin heavy chain. Here we show that the myo2 gene product, Myo2p, is a component of the contractile ring. Its association to a shrinking contractile ring has been live-recorded. Loss of myo2 function did not affect interphase actin distribution and nuclear division, but cells defective in myo2 generated irregular actin rings and abnormal septa. They were impaired in cell separation and could not proliferate properly. Excessive overexpression of myo2 was also lethal. It blocked cytokinesis and enhanced multinucleation. Relocation of actin to the contractile ring was apparently impaired in cells overexpressing myo2. In addition, unusual structural features of Myo2p as a type II myosin heavy chain will be discussed.
Materials and Methods
Strains, Media, and Transformation of S. pombe
S. pombe strains relevant to this study are listed in Table I. Media used have been described previously (9, 16, 30, 38). Transformation of S. pombe cells was done either by electroporation (35) or according to the lithium method developed for Saccharomyces cerevisiae (14).
Table I.
Strains Used in This Study
| Strain | Genotype | |
|---|---|---|
| JY450 | h 90 ade6-M216 leu1 | |
| JZ281 | h 90 ade6-M210 leu1 mam2 | |
| JZ489 | h 90/h 90 ade6-M210/ade6-M216 leu1/leu1 ura4-D18/ura4-D18 | |
| JX124 | h 90 ade6-M216 leu1 ura4-D18 ral1::ura4 + | |
| JX273 | h 90 ade6-M210 leu1 ura4-D18 ral3::ura4 + | |
| JX572 | h 90/h 90 ade6-M210/ade6-M216 leu1/leu1 ura4-D18/ura4-D18 | |
| myo2::ura4 + /myo2 + | ||
| JX573 | h 90 ade6-M216 leu1 ura4-D18 myo2::ura4 + |
JX573 was kept as a transformant.
Isolation of High-Copy Suppressors of ral3Δ
To screen for high–copy number plasmids that might suppress conjugation deficiency caused by loss of ral3 function, a homothallic haploid ral3Δ strain JX273 was transformed to Leu+ with an S. pombe cDNA library based on the expression vector pREP3 (26). Transformants were plated on synthetic sporulation medium (SSA)1 and incubated at 30°C for 5–6 d. Formed colonies were exposed to iodine vapor, which stains sporulated cells dark brown (16). We picked up brownish colonies. Because diploid ral3Δ cells are sporulation proficient, we reexamined these transformants to exclude fortuitous fusants generated during the transformation process. We eventually selected 25 transformants, whose sporulation proficiency was inheritable, from about 100,000 original Leu+ transformants. Plasmids were recovered from them.
Cloning and Sequencing of the Genomic myo2 DNA
To isolate the complete myo2 gene, genomic DNA of wild-type S. pombe was digested with BamHI, separated by electrophoresis in a 1.0% agarose gel, and blotted to a nylon filter (Genescreen Plus; Dupont/NEN, Wilmington, DE). The filter was then probed by a 32P-labeled NotI-SaII fragment carrying the myo2 cDNA. This probe hybridized to a band corresponding to 2.8-kb in size. BamHI fragments of S. pombe genomic DNA sizing between 2.5 and 3.0 kb were pooled and inserted into pBlueScriptSK+ (Stratagene, La Jolla, CA). Colonies of Escherichia coli transformed with this library were screened by the cDNA probe, and plasmids were recovered from positive colonies. All plasmids tested were found to carry the same BamHI fragment. Nucleotide sequencing indicated that the insert encompassed the cDNA sequence but was likely to lack the initiation codon. We therefore repeated a similar cloning procedure with HindIII digests of S. pombe genomic DNA and isolated a 4.2-kb-long HindIII fragment that overlapped with the NH2-terminal region of the BamHI fragment. The putative initiation codon was found on this HindIII fragment. By combining the cloned HindIII and BamHI fragments, we reconstructed a 5.5-kb-long genomic fragment carrying the entire myo2 gene.
For the determination of nucleotide sequences, DNA fragments to be examined were unidirectionally deleted by exonuclease III and S1 nuclease (Takara Shuzo, Kyoto, Japan), and each deletion product was sequenced by an automated DNA sequencer (model 4000L; LI-Cor Inc., Lincoln, NE). The entire myo2 gene was sequenced in both directions at least once. Regions showing unusual features of a myosin heavy chain were sequenced repeatedly.
Gene Disruption
A 4.0-kb HincII fragment within the myo2 open reading frame (ORF) was replaced by a ura4 + cassette (15). A ura4 − diploid strain JZ489 was transformed with a DNA fragment carrying the myo2::ura4 + allele, and Ura+ transformants were selected. Southern analysis confirmed that they were heterozygous for the myo2 locus, i.e., myo2::ura4 +/myo2 +. One transformant (JX572) was allowed to sporulate on malt-extract medium. Tetrads in each ascus displayed segregation of two viable and two nonviable spores. All spores that gave rise to colonies were Ura−. We similarly constructed another derivative of JZ489, which carried only a short deletion in the COOH-terminal region of the myo2 ORF. A 0.2-kb HindIII fragment was replaced by the ura4 + cassette in this case. Tetrad analysis of this diploid strain gave essentially the same results as JX572.
Plasmids
pREP1, pREP3, and pREP81 carry the nmt1 promoter, which is repressed by thiamine (3, 25, 26). The former two plasmids carry the authentic nmt1 promoter, whereas the promoter on pREP81 is mutated and weaker than the authentic one by about 100-fold. To express the entire myo2 gene from the nmt1 promoter, nucleotides −3 to 3 of the gene (CCTATG) were replaced by the NdeI target sequence (CATATG) according to a standard protocol for site-directed mutagenesis (23), and a 5.0-kb fragment downstream of this NdeI site was cloned into pREP81. For the construction of Myo2p fused to the jellyfish green fluorescent protein (GFP), we used a plasmid pGFT81, which carries an enhanced version of GFP (S65T) in the vector pREP81. Nucleotides −7 to −2 of the myo2 gene (TAGCCC) were replaced by the BglII target sequence (AGATCT), and a 5.0-kb fragment downstream of this BglII site was connected in frame to the 3′ end of the GFP ORF on pGFT81. To express GFP-fused Myo2p in a myo2Δ haploid strain, a diploid strain JX572 (myo2Δ/myo2 +) was transformed with the resultant plasmid (pGFT81-myo2), and sporulation was induced in the transformant. We could recover Ura+ Leu+ haploid progeny, and one of them was kept as a myo2Δ haploid strain JX573 carrying pGFT81-myo2.
Fluorescence Microscopy
Staining of cells for fluorescence microscopy was performed as previously described (34). Observation was done by using an Axioscope (Carl Zeiss, inc., Thornwood, NY) with appropriate sets of filters for GFP, TRITC, and Calcofluor and 4′,6-diamidino-2-phenylindole (DAPI). Images were recorded with a cooled charge-coupled device camera and analyzed by the Argus software (Hamamatsu Photonics, Hamamatsu, Japan). A microscope (model BX40; Olympus, Tokyo, Japan) was also used and images were photographed (Tri-X Pan 400 ASA films; Eastman Kodak Co., Rochester, NY) in this case. Time lapse recording of GFP-Myo2p localization was performed on a computer (model 9500/100 Power Macintosh; Apple Computer Inc., Cupertino, CA), using the public domain NIH Image program developed at the U.S. National Institutes of Health (Bethesda, MD) and available from zippy.nimh.nih.gov.
Results
Isolation of a Gene That Converts ral3Δ Haploid Cells to Sporulation Proficiency by Enhancing Diploidization
The ral3/scd2 gene is known to function in the Ras signaling pathway in fission yeast, and loss of its function causes deficiency in conjugation (5, 13). We screened for high– copy number suppressors of the conjugation deficiency of ral3Δ, and as described in Materials and Methods, we obtained 25 candidates. We analyzed these plasmids by Southern hybridization and excluded ones carrying ral3 cDNA. Only three plasmids turned out not to carry ral3. These plasmids could transform a homothallic haploid ral3Δ strain JX273 to a sporulation-proficient line, relatively weakly but reproducibly.
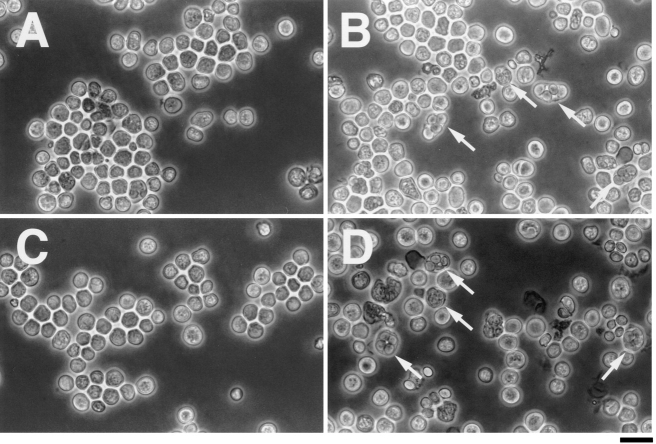
Restriction mapping of the three plasmids suggested that two of them were identical, and partial sequence analysis revealed that the third clone also carried an insert overlapping with them (data not shown). We named the former two identical clones as pCHE1 and analyzed it further. JX273 cells transformed with pCHE1 sporulated at a frequency of about 5%, and most of the asci contained four spores. These spores could germinate and grow efficiently, suggesting that diploid cells gave rise to them. However, all of the asci appeared to be azygotic in morphology (Fig. 1 B), implying that they were unlikely to have undergone mating before sporulation.
Figure 1.
Induction of sporulation by pCHE1 in ral3Δ and ral1Δ haploid cells. JX273 (ral3Δ) and JX124 (ral1Δ) were transformed with either pCHE1 or the vector pREP3. Transformants were streaked in patches on SSA medium and incubated at 30°C for 4 d to allow them to sporulate after saturation of the growth. Each sample was examined under the phase-contrast microscope: (A) JX273 (ral3Δ) transformed with pREP3; (B) JX273 with pCHE1; (C) JX124 (ral1Δ) with pREP3; and (D) JX124 with pCHE1. Arrows indicate sporulated cells. Bar, 10 μm.
The ral1/scd1 gene functions in the same Ras signaling pathway as ral3, and the ral1Δ and the ral3Δ strains exhibit similar phenotypes to each other (5, 13). We transformed a haploid ral1Δ strain JX124 with pCHE1. The transformant could sporulate on SSA at a low frequency (about 5%), indicating that the effect of pCHE1 is not confined to ral3Δ cells. Again, the asci did not appear to have undergone mating (Fig. 1 D). pCHE1 could elevate sporulation in homothallic haploid ral1Δ or ral3Δ cells, but it was unable to promote mating in heterothallic haploid cells (data not shown). These observations suggest that the pCHE1transformed cells acquired diploidy through some irregularity at mitosis rather than through conjugation.
To investigate the effect of pCHE1 on diploidization in the ral1 + ral3 + background, we transformed a homothallic haploid mam2 mutant JZ281 with pCHE1. The mam2 gene encodes the receptor for the mating pheromone P-factor (20), and the mam2 mutant has been used conveniently to detect diploidization (4, 6) because it does not mate at all but can sporulate once diploidized. JZ281 transformed with pCHE1 sporulated at a very low but significant frequency (about 0.5%). This supports the idea that pCHE1 can elevate the frequency of spontaneous diploidization in any kind of S. pombe cells.
The Cloned Gene Encodes a Myosin Heavy Chain
pCHE1 carried a 2.1-kb-long cDNA insert. Nucleotide sequence analysis revealed that it encodes a polypeptide that has homology to coiled-coil proteins. However, this cDNA carried no stop codon before the first methionine codon and appeared not to cover the complete ORF. This view was supported by the observation that the cDNA hybridized to a transcript of about 4.5 kb in length (data not shown). We therefore isolated a 5.5-kb-long genomic DNA segment encompassing the cDNA, as described in Materials and Methods. This segment contained an ORF encoding 1,526 amino acids, which did not appear to have introns and matched the estimated size of the transcript (Fig. 2, A and B). The molecular mass of the deduced gene product was 176.3 kD. We looked for proteins that show similarity to the gene product by using the FASTA homology search algorithm (33). The NH2-terminal part of the gene product was similar to the head domains of the myosin heavy chain (Fig. 2 C), and the COOH-terminal part was similar to a number of coiled-coil proteins including myosin heavy chains (Fig. 2 A). Furthermore, the gene product apparently carried an ATP-binding site at residues 150–188, an actin-binding site at residues 634–655, and an IQ motif, which is thought to be a binding site for the myosin light chain, at residues 765–775 (Fig. 2 B). These results strongly suggest that the cloned gene encodes a heavy chain of fission yeast type II myosin, and hence we call it myo2 hereafter. Curiously, the coiled-coil region of S. pombe Myo2p carries nine proline residues, at positions 1089, 1245, 1251, 1375, 1383, 1390, 1396, 1445, and 1447 (Fig. 2, B and C). Proline is a potent α-helix breaker and is seen only rarely in the tail region of type II myosin of other organisms.
Figure 2.
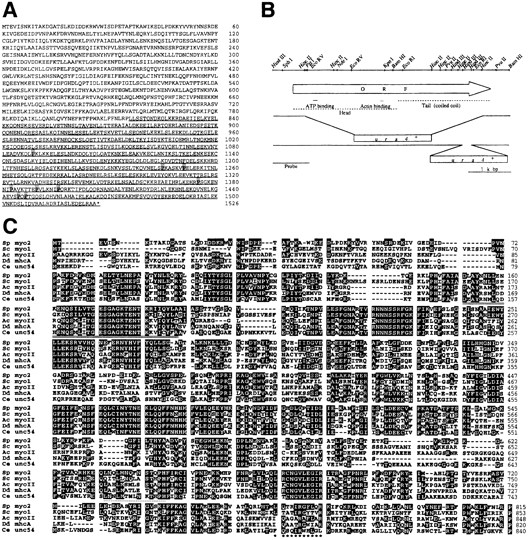
Structural features of the myo2 gene product. (A) The deduced amino acid sequence of Myo2p. Numbering of the amino acid residues is given on the right side. The putative α-helical coiled-coil region is underlined. Proline residues scattered in this region are shadowed. (B) A restriction map of the genomic myo2 locus. The open arrow indicates the extent and direction of the myo2 ORF, which encodes 1,526 amino acids. Areas corresponding to the head and tail regions of the myosin heavy chain are shown by broken lines. The putative ATP- and actin-binding sites are indicated by solid lines. Two kinds of disruption constructs used to replace the chromosomal myo2 gene are schematically shown below the arrow. Proper disruption of the chromosomal gene was confirmed by the probe shown at the bottom in Southern blot analysis. (C) Alignment of the head region of Myo2p with type II myosin heavy chains of other species. Numbering of the amino acid residues for each protein is given on the right side. Five proteins are compared here. From top to bottom: the S. pombe myo2 gene product, the Saccharomyces cerevisiae MYO1 gene product (43, 45), Acanthamoeba castellanii myosin II (17), the Dictyostelium discoideum mhcA gene product (8, 44), and the Caenorhabditis elegans unc54 gene product (19, 28). Amino acid residues identical to those of S. pombe Myo2p are shown in white against black. The putative actin-binding site of Myo2p is underscored with a dotted line, and the putative ATP-binding site is doubly underlined. The putative IQ motif is marked with asterisks underneath. The sequence data of myo2 are available from GenBank/EMBL/DDBJ under accession number AB000544.
Phenotype of the myo2-disrupted Strain
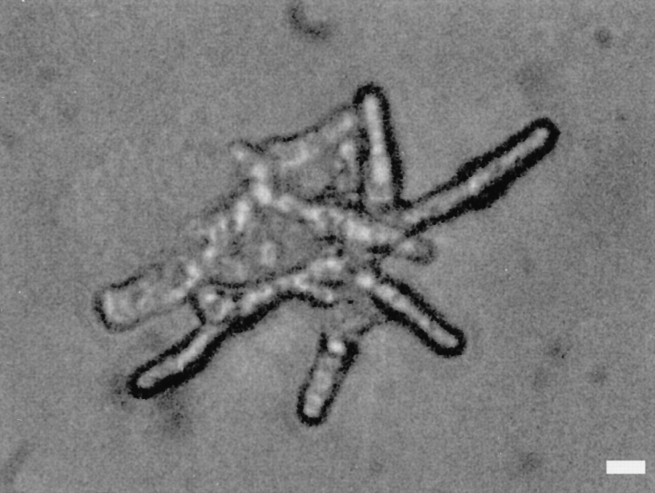
Disruption of the myo2 gene by insertion of an S. pombe ura4 + cassette was carried out as described in Materials and Methods. Sporulation was induced in a resultant diploid strain JX572, which is heterozygous for myo2 (myo2:: ura4 +/myo2 +). Tetrad dissection suggested that two of the ascospores were viable and able to form colonies, but the other two were not. The viable progeny were all Ura−. Essentially the same results were obtained in the analyses of two types of disruption constructs, in which either a 4.0kb-long (JX572) or a 0.2-kb-long fragment was deleted from the myo2 ORF (Fig. 2 B). Thus, we conclude that disruption of myo2 is lethal. However, microscopic observation of deduced myo2-disrupted spores revealed that they could germinate and grow up to some extent. Under the phase-contrast microscope, the disruptants appeared to be impaired in cell separation after septation (Fig. 3). They exhibited elongated and sometimes branched cell morphology as a terminal phenotype.
Figure 3.
Terminal morphology of a deduced myo2-disrupted spore. Tetrad dissection of asci produced by an myo2::ura4 +/ myo2 + diploid strain JX572 was done, and dissected spores were grown on rich medium YPD at 30°C for 3 d. A phase-contrast micrograph of a germinated myo2Δ spore is shown. Bar, 10 μm.
To confirm that the observed lethality of myo2Δ cells was indeed due to loss of Myo2p, the myo2::ura4 +/myo2 + diploid strain JX572, which carried the 4.0-kb deletion that removed 7/8 of the entire myo2 ORF, was transformed with pREP81-myo2, which could express the entire myo2 ORF from the weak nmt1 promoter. Sporulation was induced in the transformant, and progeny spores were analyzed. Ura+ offspring could be recovered in this case, indicating that the myo2::ura4 + defect was complemented by the expression of myo2 carried on pREP81. One of the myo2::ura4 + haploid strains thus obtained was named JX573 and used in the subsequent analyses. JX573 carrying pREP81-myo2 halted growth if the nmt1 promoter was shut off by the addition of thiamine. These observations reinforce that loss of Myo2p is lethal. In addition, this shut-off system allowed us to investigate the effects of loss of myo2 expression precisely, as described below.
Terminal Phenotypes Caused by Loss of myo2 Expression
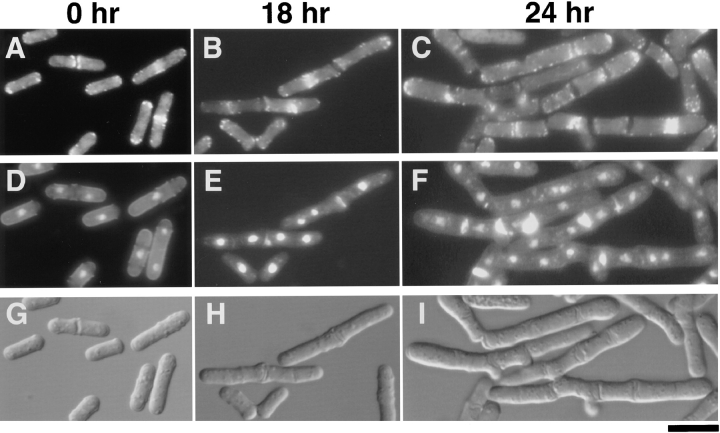
Cells of JX573 (myo2Δ) carrying pREP81-myo2 were grown in the absence of thiamine to the exponential phase, and thiamine was added to the medium. Cells were then observed chronologically under the microscope (Fig. 4). They began to cease growth ∼18 h after the addition of thiamine, displaying elongated cell shape. DAPI staining revealed that many elongated cells were undergoing a second round of nuclear division, although cell separation after the first division was not completed (Fig. 4 E). Staining with phalloidin-TRITC indicated that cell cycle–dependent redistribution of actin was nearly normal in these cells, and actin rings could be observed over the nuclei undergoing a second round of mitosis (Fig. 4 B). However, the rings looked somewhat irregular. They were sometimes thinner, sometimes broader, and sometimes biased. At 24 h after the addition of thiamine, elongated cells with multiple septa appeared (Fig. 4 I). The diameter of these cells was considerably larger than that of normal S. pombe cells. Calcofluor staining showed that some septa were abnormally thickened, whereas some were not clearly stained by the dye although visible in Nomarski (Fig. 4 F). The septal material did not appear to cross the entire cell in some cases. Each compartment partitioned by the septa carried one nucleus (Fig. 4, F and I). Even branched cells were seen at this stage (Fig. 4 I), but localization of actin appeared generally normal, except that the rings were ill shaped in one way or another (Fig. 4 C). All these observations imply that the function of myo2 may not be necessary for cell growth per se, but that a failure of proper cell separation may gradually disturb and eventually block cell proliferation in the myo2 disruptant (see Discussion).
Figure 4.
Morphological changes caused by depletion of Myo2p. Cells of an myo2Δ strain (JX573) carrying pREP81-myo2 were grown in liquid MM supplemented with adenine (50 μg/ml). When the cell concentration reached 5 × 106/ml, thiamine was added to the medium at the final concentration of 6 μg/ml to repress myo2 expression from the nmt1 promoter. Cells were sampled chronologically, fixed, and stained with phalloidin-TRITC, DAPI, and Calcofluor. Micrographs showing either actin localization (A–C), nuclei and septa (D–F), or Nomarski views (G–I) are displayed for the same cell population. Time after the addition of thiamine is indicated on the top. Bar, 10 μm.
Overproduction of Myo2p Blocks Cytokinesis
Wild-type S. pombe cells (JY450) were transformed with either pREP1-myo2 or the vector pREP1. The nmt1 promoter carried on pREP1 is about 100 times stronger than that on pREP81. These two kinds of transformants grew at similar rates in the presence of thiamine. However, cells carrying pREP1-myo2 hardly grew up in the absence of thiamine, while cells carrying pREP1 grew normally under the same conditions, indicating that overproduction of Myo2p is inhibitory to cell growth (data not shown).
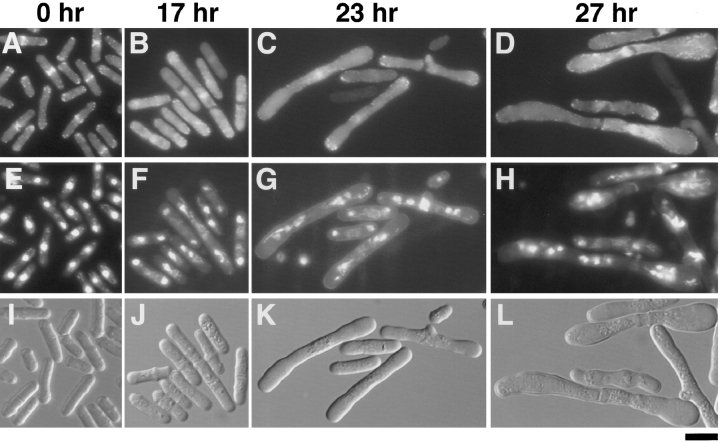
To analyze the deleterious effects of Myo2p overproduction on cellular activities, JY450 cells carrying pREP1myo2 were incubated in liquid minimal medium (MM) containing no thiamine (Fig. 5). The nmt1 promoter becomes active usually ∼14 h after the removal of thiamine from the medium (25). The pREP1-myo2 transformant began to generate elongated cells 17 h after the removal of thiamine, suggesting a failure in cell division (Fig. 5 J). Nearly all cells were elongated and their diameter enlarged after 23 h (Fig. 5 K). DAPI staining revealed that cells at 23 h carried multiple nuclei. Double staining of the cells with Calcofluor and DAPI indicated that a second round of nuclear division had occurred in large cells in the absence of preceding cell division (Fig. 5 G). Some cells carried a single septum in the central region, which looked somewhat irregular. Some others showed accumulation of actin, but no apparent septum formation, at the middle of the cell. Actin dots were observed at both tips of many cells, but we could hardly detect actin rings surrounding nuclei in myo2-overexpressing cells (Fig. 5 C). Remarkably, new rounds of nuclear division appeared to proceed in the absence of actin ring formation in these cells (Fig. 5, C and G). At 27 h after the removal of thiamine, most cells became swollen at both ends of the cell, looking like dumbbells (Fig. 5 L). Each cell displayed a number of nuclei (Fig. 5 H). A septum was occasionally seen at the middle of the cell, but not at any other position. Abnormal accumulation of actin was observed here and there, sometimes shaping cables. Blobs stained by Calcofluor were also seen (Fig. 5, D and H).
Figure 5.
Morphological changes caused by myo2 overexpression. JY450 cells transformed with pREP1-myo2 were grown in liquid MM supplemented with adenine and thiamine, and overexpression of myo2 was induced by shifting the cells to the same medium with no thiamine. Cells were sampled chronologically after the removal of thiamine and analyzed as in Fig. 4. Micrographs showing actin localization (A–D), nuclei and septa (E–H), and Nomarski views (I–L) are displayed for the same cell population. Time after the removal of thiamine is indicated on the top. Bar, 10 μm.
The above results suggest that nuclear division takes place without proper cytokinesis in cells overproducing Myo2p, resulting in the formation of multinucleated cells. The Myo2p overproducer shows two obvious defects in cytokinesis. One is a failure in actin ring formation, and the other is the absence of septation and subsequent cell separation, except for the occasional formation of a single septum at the middle, which is likely to have embarked before the cell starts to accumulate Myo2p. Probably the latter defect is a consequence of the former.
Localization of the Myo2 Protein
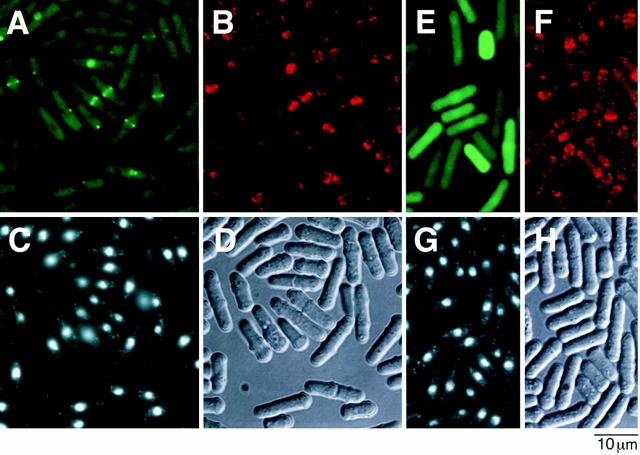
To examine cellular localization of Myo2p, we fused Myo2p to the COOH terminus of an enhanced version of GFP by gene engineering, and expressed the fusion protein from the weak nmt1 promoter in S. pombe cells. Expression of GFP-Myo2p could rescue the lethality of the myo2Δ strain, indicating that the fusion protein can provide the Myo2p function. Cells of JX573 (myo2Δ) expressing GFPMyo2p, constructed as described in Materials and Methods, were harvested in the exponential phase, fixed, and then doubly stained with DAPI and phalloidin-TRITC to detect DNA and F-actin, respectively (Fig. 6). Soon after the initiation of mitosis, as judged by DAPI staining, emission of GFP-Myo2p was detected as a band over the nucleus at the medial region of the cell (Fig. 6, A and C). This band apparently colocalized with actin stained by phalloidin-TRITC (Fig. 6, A and B). By examining microscopic views at several focal planes, it was concluded that GFPMyo2p and actin were both in a ring structure. The GFPMyo2p ring apparently decreased its diameter when the two chromatin regions were separated at a later stage of mitosis. A live recording of cytokinesis of cells expressing GFP-Myo2p clearly demonstrated that GFP-Myo2p was on the contractile ring (Fig. 7). GFP-Myo2p became a bright fluorescent dot at the final stage of cytokinesis. A septum became clearly visible concomitantly with the disappearance of this dot, and two daughter cells separated subsequently (Fig. 7). Fluorescence of GFP, not fused to Myo2p, was detected rather uniformly over the whole cell (Fig. 6 E), suggesting that the observed localization of GFP-Myo2p represents that of the genuine Myo2 protein. This was confirmed by the following immunological analysis: One copy of hemagglutinin (HA) tag was fused to the NH2 terminus of Myo2p, and HA-Myo2p was expressed in myo2Δ cells. Growing cells were fixed and stained with anti-HA antibodies. HA-Myo2p was detected in a ring structure, like GFP-Myo2p (data not shown).
Figure 6.
Cellular localization of Myo2p fused to GFP. Cells of JX573 carrying pGFT81-myo2, grown in liquid MM supplemented with adenine, were harvested at the concentration of 5 × 106/ml. They were fixed, then stained with phalloidin-TRITC and DAPI, and observed under the microscope (A–D). Cells of JY450 carrying pGFT81, which expressed GFP alone, were examined as a control (E–H). Micrographs show GFP fluorescence (A and E), phalloidin-TRITC–stained actin (B and F), DAPI-stained nuclei (C and G), and Nomarski views (D and H) for the same cell population.
Figure 7.
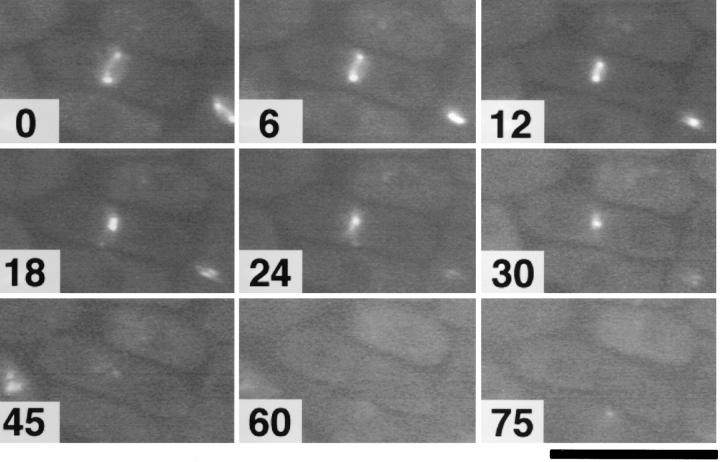
Live recording of the cytokinetic process in JX573 carrying pGFT81-myo2. Cells growing at room temperature in liquid MM supplemented with adenine were observed without fixation under the fluorescence microscope, and images were recorded at 3-min intervals. A typical example, displaying contraction of the Myo2p ring, is shown here. For the first 30 min, images taken every 6 min are shown, and for the next 45 min, images taken every 15 min are shown. The Myo2p dot disappeared around 54 min, the septum became clearly visible at 57 min, and the daughter cells began to separate around 69 min in this analysis.
The observed localization of GFP-myo2 strongly suggests that Myo2p is a component of the contractile ring and participates in its contraction by interacting with actin. However, while phalloidin-TRITC staining revealed the presence of punctate F-actin at the growing ends of interphase cells, no emission of GFP-myo2 was detected at these areas. This may suggest that Myo2p is essential for the formation of a functional contractile ring but not for other aspects of the F-actin functions.
Finally, we note that emission of GFP-Myo2p could be detected as a single dot in a considerable number of interphase cells, which were likely to be in the G2 phase. Most of these dots appeared to associate to one side of the nucleus (Fig. 6 A), and actin was unlikely to be included in them (Fig. 6 B). However, a small number of cells showed a dot at the tip of the cell, and actin appeared to associate to such a dot (Fig. 6, A and B). The significance of these dots remains as an interesting question (see Discussion).
Discussion
The myo2 gene product identified in this study showed similarity to the type II myosin heavy chain in a number of structural features. In addition, Myo2p colocalized with actin overlying mitotic nuclei and could be seen on a shrinking contractile ring. These results strongly suggest that Myo2p composes S. pombe type II myosin, which interacts with actin in the contractile ring and generates force for cytokinesis.
Type II myosin heavy chains known to date carry two IQ motifs, agreeing with the hexameric structure of myosin II. However, we found only one perfect IQ motif (IQXXXRGXXXR) at residues 765–775 in S. pombe Myo2p. A similar sequence to the IQ motif (LQANLQVYNEFR) exists at residues 791–802, but this sequence does not have the central arginine, which is believed to be critical for the binding to the myosin light chain. Thus, we suspect that the type II myosin heavy chain of S. pombe may interact with only one myosin light chain. Interestingly, McCollum et al. (27) observed in their immunoprecipitation analysis that the putative myosin light chain encoded by S. pombe cdc4 coprecipitated with a 200-kD protein, which they suspected to be a myosin heavy chain, but with no obvious second light chain. Their observation may reinforce the possibility that S. pombe myosin II is rather eccentric and consists of two heavy chains and two light chains.
Another unusual feature of S. pombe type II myosin heavy chain is that it carries nine proline residues in the COOH-terminal region. This region is important for the coiled-coil structure of myosin II, and there is usually no proline, as it breaks α-helix. However, S. cerevisiae myosin heavy chain has six proline residues in this region (43), and Acanthamoeba myosin heavy chain has one (17). Electronmicroscopic analysis revealed a hinge in the COOH-terminal rod of Acanthamoeba myosin II (17). The proline residues in S. pombe Myo2p cluster roughly in four locations, and hence the Myo2p rod is likely to be bent at four positions. Physiological significance of these hinges remains unknown.
This study has shown that type II myosin heavy chain is essential for cell proliferation in S. pombe. However, the S. cerevisiae MYO1 gene, encoding a type II myosin heavy chain, is apparently dispensable for growth (36, 45). Although MYO1-disrupted cells grow slower, they are essentially viable. These cells carry out incomplete cell division and form a continuous chain during the growth. One report describes that the MYO1-disruptant also shows a defect in nuclear migration, which often results in the generation of cells that have several nuclei or cells that have none (45). However, another report describes that the MYO1-disruptant has no strong defect in nuclear migration (36). Our observation suggested that the myo2 disruptant of S. pombe does not have a defect in nuclear migration.
Cells lacking Myo2p can form actin rings over dividing nuclei, although the rings do not appear to be organized properly. These cells are sectioned subsequently by septa, which are again mostly irregular. However, no cell separation follows. Each compartment sectioned by septa carries one nucleus, indicating that nuclear migration is not hampered in these cells. These results also indicate that initial disposition of the actin ring in the middle of the cell can take place in the absence of Myo2p, but subsequent assembly of the ring cannot proceed properly without it.
One apparent phenotype shown by the myo2Δ strain is the deficiency in cell separation, like S. cerevisiae myo1Δ, but another is the ultimate arrest of cell proliferation, unlike S. cerevisiae myo1Δ. Several mutants defective in cell separation have been described in S. pombe, and sep1-1 is a typical example (40). Although the identity of the sep1 gene product is unknown, this mutant apparently shows very similar morphology to the myo2 disruptant, forming septated cells with multiple nuclei. However, sep1-1 cells are viable, displaying mycelial cell growth (40), as are S. cerevisiae myo1Δ cells. This implies that the lethality of the myo2 disruptant may not be a simple outcome of the lack of cell separation.
Overexpression of myo2 also arrests cell proliferation, giving rise to enlarged and multinucleated cells. This indicates that an excessive amount of Myo2p inhibits cytokinesis. An increase in the copy number of the S. pombe actin gene is lethal (29). It appears that the components of cytoskeletal structures must be produced stoichiometrically to ensure proper cell proliferation.
Cdc4p is the only myosin light chain so far identified in S. pombe, which is apparently involved in the regulation of cell division (6, 27, 31). Hence, it would be reasonable to speculate that Cdc4p and Myo2p form a complex and cooperate in function. However, disruptions of cdc4 and of myo2 do not result in the same phenotype. Loss of cdc4 function blocks actin ring formation, but loss of myo2 function does not. Contrarily, overproduction of Myo2p blocks actin relocation and formation of rings. These observations lead us to speculate that the myosin heavy chain Myo2p may inhibit proper relocation of actin if it is not associated by the light chain Cdc4p. Intriguingly, certain cdc4 temperature-sensitive mutants show very similar phenotypes to myo2Δ at the restrictive temperature. They accumulate multiple nuclei and multiple improper actin rings and septa but fail to complete cytokinesis (27). Thus, one possibility is that these mutant Cdc4 proteins block the function of Myo2p at the restrictive temperature and cause phenotypes similar to those of myo2Δ.
A curious observation we encountered was that Myo2p forms a single dot in interphase cells, which apparently associates to one side of the nucleus. Although this may simply reflect a trivial artifact brought by overexpression of Myo2p, it will certainly be interesting if this dot plays any role in mediating a nuclear signal for the organization of the contractile ring. Recent analyses have shown that the mid1/dmf1 gene product is an important element in assuring correct placement of the division septum in fission yeast (6, 41). Mid1p is nuclear in interphase and relocates to form a medial ring at the cell cortex before the actin ring formation (41). Thus, it is tempting to speculate that the dot may have relevance to the function of Mid1p, although more analysis is obviously required to reach any firm conclusion. A small number of cells showed a dot of Myo2p at the tip of the cell. We suspect that this dot is likely to represent a remnant of the contractile ring that failed to be digested completely, although this is also to be confirmed.
The original myo2 clone pCHE1 carried a truncated myo2 ORF, which could produce only the COOH-terminal one-third of the complete gene product. pCHE1 could not complement the myo2Δ strain, indicating that the truncated gene product is not fully functional (our unpublished observation). We moderately overexpressed the complete myo2 gene and found that it could promote diploidization, although it slowed the growth of the host cell at the same time (our unpublished observation). A logical inference from these observations will be that the diploidization does not require the total function of Myo2p, but may result from sequestration of a factor that interacts with the COOH-terminal region of Myo2p. The fact that excessive overexpression of the entire myo2 gene is cytotoxic may explain why we could isolate only truncated myo2 cDNAs in our original screening, in which we used the strongest nmt1 promoter.
pCHE1 could diploidize both ral − and ral + cells, but the frequency of sporulation induced by pCHE1 was clearly higher in ral1Δ or ral3Δ cells than in ral + cells. This may suggest the presence of a functional interaction between Myo2p and the Ras pathway involving Ral1p and Ral3p. In this connection, we note that alterations in the S. pombe byr4 gene, characterized by Song et al. (42), can cause phenotypes similar to those of myo2 mutants. The null mutation in byr4 causes cell cycle arrest in late mitosis with multiple rounds of septation. As byr4Δ cells typically form multiple septa between two nuclei, their phenotype is not exactly the same as that of myo2Δ cells. However, like overexpression of myo2, overexpression of byr4 inhibits cytokinesis without affecting karyokinesis and generates multinucleate cells. In both cases, actin relocation during the cell cycle appears to be hampered. Furthermore, the inhibition of cytokinesis by byr4 overexpression is exacerbated by the null allele of ral1, and moderate expression of byr4 promotes diploidization in ral1Δ cells, suggesting a link between Byr4p and Ral1p (42). Thus, although the molecular function of Byr4p remains unclear, Myo2p and Byr4p may be functionally related to each other, and furthermore, they may sequester and block the same essential factor for cytokinesis when they are excessively overproduced. Further analysis of the relationship between the Ras pathway and the contractile ring in fission yeast will be important to elucidate the regulation of cytokinesis by signal transduction via small GTPases, as is an emerging topic also in S. cerevisiae and Drosophila (7, 11, 22).
Acknowledgments
We thank R. Tsuruta, Y. Iino, and Y. Watanabe for their kind assistance in the construction of plasmids used in this study. We have been informed by J. Hyams that his group cloned and analyzed the myo2 gene independently. Their preliminary report (abstract) can be seen in May, K.M., F.Z. Watts, N. Jones, and J.S. Hyams. Mol. Biol. Cell. 1996. 7:196a. We appreciate his communication of the results before publication.
This work was supported by Grants-in-Aid from the Ministry of Education, Science, Sports, and Culture of Japan.
Footnotes
1. Abbreviations used in this paper: DAPI, 4′,6-diamidino-2-phenylindole; GFP, green fluorescent protein; HA, hemagglutinin; MM, minimal medium; ORF, open reading frame; SSA, synthetic sporulation medium.
Address all correspondence to Dr. Masayuki Yamamoto, Department of Biophysics and Biochemistry, Graduate School of Science, University of Tokyo, P.O. Hongo, Tokyo 113, Japan. Tel.: 81-3-3814-9620. Fax: 81-35802-2042. E-mail: myamamot@ims.u-tokyo.ac.jp
References
- 1.Balasubramanian MK, Helfman DM, Hemmingsen SM. A new tropomyosin essential for cytokinesis in the fission yeast S. pombe. . Nature (Lond) 1992;360:84–87. doi: 10.1038/360084a0. [DOI] [PubMed] [Google Scholar]
- 2.Balasubramanian MK, Hirani BR, Burke JD, Gould KL. The Schizosaccharomyces pombe cdc3 +gene encodes a profilin essential for cytokinesis. J Cell Biol. 1994;125:1289–1301. doi: 10.1083/jcb.125.6.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- 4.Broek D, Bartlett R, Crawford K, Nurse P. Involvement of p34cdc2in establishing the dependency of S phase on mitosis. Nature (Lond) 1991;349:388–393. doi: 10.1038/349388a0. [DOI] [PubMed] [Google Scholar]
- 5.Chang EC, Barr M, Wang Y, Jung V, Xu HP, Wigler MH. Cooperative interaction of S. pombeproteins required for mating and morphogenesis. Cell. 1994;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 6.Chang F, Woollard A, Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- 7.Cvrckova F, De Virgilio C, Manser E, Pringle JR, Nasmyth K. Ste20-like protein kinases are required for normal localization of cell growth and for cytokinesis in budding yeast. Genes Dev. 1995;9:1817–1830. doi: 10.1101/gad.9.15.1817. [DOI] [PubMed] [Google Scholar]
- 8.DeLozanne A, Lewis M, Spudich JA, Leinwand LA. Cloning and characterization of a nonmuscle myosin heavy chain cDNA. Proc Natl Acad Sci USA. 1985;82:6807–6810. doi: 10.1073/pnas.82.20.6807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egel R, Egel-Mitani M. Premeiotic DNA synthesis in fission yeast. Exp Cell Res. 1974;88:127–134. doi: 10.1016/0014-4827(74)90626-0. [DOI] [PubMed] [Google Scholar]
- 10.Fankhauser C, Simanis V. Cold fission: splitting the pombe cell at room temperature. Trends Cell Biol. 1994;4:96–101. doi: 10.1016/0962-8924(94)90182-1. [DOI] [PubMed] [Google Scholar]
- 11.Field CM, al-Awar O, Rosenblatt J, Wong ML, Alberts B, Mitchison TJ. A purified Drosophilaseptin complex forms filaments and exhibits GTPase activity. J Cell Biol. 1996;133:605–616. doi: 10.1083/jcb.133.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fishkind DJ, Wang YL. New horizons for cytokinesis. Curr Opin Cell Biol. 1995;7:23–31. doi: 10.1016/0955-0674(95)80041-7. [DOI] [PubMed] [Google Scholar]
- 13.Fukui, Y., and M. Yamamoto. 1988. Isolation and characterization of Schizosaccharomyces pombe mutants phenotypically similar to ras1−. Mol. Gen. Genet. 215:26–31. [DOI] [PubMed]
- 14.Gietz RD, Schiestl RH. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 15.Grimm C, Kohli J, Murray J, Maundrell K. Genetic engineering of Schizosaccharomyces pombe: a system for gene disruption and replacement using the ura4gene as a selectable marker. Mol Gen Genet. 1988;215:81–86. doi: 10.1007/BF00331307. [DOI] [PubMed] [Google Scholar]
- 16.Gutz, H., H. Heslot, U. Leupold, and N. Loprieno. 1974. Schizosaccharomyces pombe. In Handbook of Genetics. Vol. 1. R.D. King, editor. Plenum Publishing Corp., New York. 395–446.
- 17.Hammer JA, Bowers B, Paterson BM, Korn ED. Complete nucleotide sequence and deduced polypeptide sequence of a nonmuscle myosin heavy chain gene from Acanthamoeba: evidence of a hinge in the rodlike tail. J Cell Biol. 1987;105:913–925. doi: 10.1083/jcb.105.2.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayles J, Nurse P. A review of mitosis in the fission yeast Schizosaccharomyces pombe. . Exp Cell Res. 1989;184:273–286. doi: 10.1016/0014-4827(89)90327-3. [DOI] [PubMed] [Google Scholar]
- 19.Karn J, Brenner S, Barnett L. Protein structural domains in the Caenorhabditis elegans unc-54myosin heavy chain gene are not separated by introns. Proc Natl Acad Sci USA. 1983;80:4253–4257. doi: 10.1073/pnas.80.14.4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kitamura K, Shimoda C. The Schizosaccharomyces pombe mam2 gene encodes a putative pheromone receptor which has a significant homology with the Saccharomyces cerevisiaeSte2 protein. EMBO (Eur Mol Biol Organ) J. 1991;12:3743–3751. doi: 10.1002/j.1460-2075.1991.tb04943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knight, A., T. Rowe, and J. Kendrick–Jones. 1994. Myosins. In The Encyclopaedia of Molecular Biology. J. Kendrew, editor. Blackwell Scientific, Cambridge, MA. 699–703.
- 22.Kohno H, Tanaka K, Mino A, Umikawa M, Imamura H, Fujiwara T, Qadota H, Watanabe T, Ohya Y, Takai Y. Bni1p implicated in cytoskeletal control is a putative target of Rho1p small GTP binding protein in Saccharomyces cerevisiae. . EMBO (Eur Mol Biol Organ) J. 1996;15:6060–6068. [PMC free article] [PubMed] [Google Scholar]
- 23.Kunkel TA. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marks J, Hyams J. Localization of F-actin through the cell division cycle of Schizosaccharomyces pombe. . Eur J Cell Biol. 1985;39:27–32. [Google Scholar]
- 25.Maundrell K. nmt1of fission yeast. A highly transcribed gene completely repressed by thiamine. J Biol Chem. 1990;265:10857–10864. [PubMed] [Google Scholar]
- 26.Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- 27.McCollum D, Balasubramanian MK, Pelcher LE, Hemmingsen SM, Gould KL. Schizosaccharomyces pombe cdc4 +gene encodes a novel EF-hand protein essential for cytokinesis. J Cell Biol. 1995;130:651–660. doi: 10.1083/jcb.130.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McLachlan AD, Karn J. Periodic charge distributions in the myosin rod amino acid sequence match cross-bridge spacings in muscle. Nature (Lond) 1982;299:226–231. doi: 10.1038/299226a0. [DOI] [PubMed] [Google Scholar]
- 29.Mertins P, Gallwitz D. A single intronless action gene in the fission yeast Schizosaccharomyces pombe: nucleotide sequence and transcripts formed in homologous and heterologous yeast. Nucleic Acids Res. 1987;15:7369–7379. doi: 10.1093/nar/15.18.7369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. . Methods Enzymol. 1990;194:795–826. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 31.Nurse P. Mutants of the fission yeast Schizosaccharomyces pombewhich alter the shift between cell proliferation and sporulation. Mol Gen Genet. 1985;198:497–502. [Google Scholar]
- 32.Nurse P. Fission yeast morphogenesis-posing the problems. Mol Biol Cell. 1994;5:613–616. doi: 10.1091/mbc.5.6.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pichova A, Streiblova E. Features of the cell periphery in the deformed ras1 negative cells of Schizosaccharomyces pombe. . Exp Mycology. 1992;16:178–187. [Google Scholar]
- 35.Prentice HL. High efficiency transformation of Schizosaccharomyces pombeby electroporation. Nucleic Acids Res. 1992;20:621. doi: 10.1093/nar/20.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodriguez JR, Paterson BM. Yeast myosin heavy chain mutant: maintenance of the cell type specific budding pattern and the normal deposition of chitin and cell wall components requires an intact myosin heavy chain gene. Cell Motil Cytoskel. 1990;17:301–308. doi: 10.1002/cm.970170405. [DOI] [PubMed] [Google Scholar]
- 37.Satterwhite LL, Pollard TD. Cytokinesis. Curr Opin Cell Biol. 1992;4:43–52. doi: 10.1016/0955-0674(92)90057-j. [DOI] [PubMed] [Google Scholar]
- 38.Sherman, F., G. Fink, and J. Hicks. 1986. Methods in Yeast Genetics: Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. 163–167.
- 39.Simanis V. The control of septum formation and cytokinesis in fission yeast. Semin Cell Biol. 1995;6:79–87. doi: 10.1016/1043-4682(95)90004-7. [DOI] [PubMed] [Google Scholar]
- 40.Sipiczki M, Grallert B, Miklos I. Mycelial and syncytial growth in Schizosaccharomyces pombeinduced by novel septation mutations. J Cell Sci. 1993;104:485–493. doi: 10.1242/jcs.104.2.485. [DOI] [PubMed] [Google Scholar]
- 41.Sohrmann M, Fankhauser C, Simanis V. The dmf1/mid1gene is essential for correct positioning of division septum in fission yeast. Genes Dev. 1996;10:2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- 42.Song K, Mach KE, Chen CY, Reynolds T, Albright CF. A novel suppressor of ras1 in fission yeast, byr4, is a dosage-dependent inhibitor of cytokinesis. J Cell Biol. 1996;133:1307–1319. doi: 10.1083/jcb.133.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sweeney FP, Pocklington MJ, Orr E. The yeast type II myosin heavy chain: analysis of its predicted polypeptide sequence. J Muscle Res Cell Motil. 1991;12:61–68. doi: 10.1007/BF01781175. [DOI] [PubMed] [Google Scholar]
- 44.Warrick HM, De Lozanne A, Leinwand LA, Spudich JA. Conserved protein domains in a myosin heavy chain gene from Dictyostelium discoideum. . Proc Natl Acad Sci USA. 1986;83:9433–9437. doi: 10.1073/pnas.83.24.9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watts FZ, Shiels G, Orr E. The yeast MYO1gene encoding a myosin-like protein required for cell division. EMBO (Eur Mol Biol Organ) J. 1987;6:3499–3505. doi: 10.1002/j.1460-2075.1987.tb02675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]