Abstract
The chloroplastic inner envelope protein of 110 kD (IEP110) is part of the protein import machinery in the pea. Different hybrid proteins were constructed to assess the import and sorting pathway of IEP110. The IEP110 precursor (pIEP110) uses the general import pathway into chloroplasts, as shown by the mutual exchange of presequences with the precursor of the small subunit of ribulose-1,5-bisphosphate carboxylase (pSSU). Sorting information to the chloroplastic inner envelope is contained in an NH2-proximal part of mature IEP110 (110N). The NH2-terminus serves to anchor the protein into the membrane. Large COOH-terminal portions of this protein (80–90 kD) are exposed to the intermembrane space in situ. Successful sorting and integration of IEP110 and the derived constructs into the inner envelope are demonstrated by the inaccessability of processed mature protein to the protease thermolysin but accessibility to trypsin, i.e., the imported protein is exposed to the intermembrane space. A hybrid protein consisting of the transit sequence of SSU, the NH2-proximal part of mature IEP110, and mature SSU (tpSSU-110N-mSSU) is completely imported into the chloroplast stroma, from which it can be recovered as soluble, terminally processed 110NmSSU. The soluble 110N-mSSU then enters a reexport pathway, which results not only in the insertion of 110N-mSSU into the inner envelope membrane, but also in the extrusion of large portions of the protein into the intermembrane space. We conclude that chloroplasts possess a protein reexport machinery for IEPs in which soluble stromal components interact with a membrane-localized translocation machinery.
A central question in chloroplast biogenesis deals with the mechanisms of intraorganellar sorting of nuclear-encoded plastid preproteins to the different subcompartments. Chloroplasts are highly structured, and six suborganellar locations can be distinguished; i.e., the outer and inner envelope membranes (IEPs),1 the thylakoid membrane network, the intermembrane space between the two envelope membranes, the stroma, and the thylakoid lumen. Nuclear-coded precursor proteins exist for all these compartments, which use the general import machineries that are present in both envelope membranes (Tranel et al., 1995; Brink et al., 1995; Knight and Gray, 1995; Hageman et al., 1990; Grossmann et al., 1980). An envelope transfer stroma-targeting sequence as a cleavable NH2-terminal extension is the common denominator for all of the preproteins (for review see Cline and Henry, 1996; Lübeck et al., 1997). Further transport into and across the thylakoid membranes is accomplished by four or five distinct pathways that involve (a) a chloroplast Sec A homologue (Knott and Robinson, 1994; Nohara et al., 1995; Berghöfer et al., 1995; Robinson et al., 1994; Yuan et al., 1994) and probably Sec Y and Sec E to complement the system (Laidler et al., 1995); (b) a unique, ΔpH-dependent pathway whose components have not yet been identified (Cline et al., 1993; Robinson et al., 1994); (c) a SRP54 homologue–dependent integration of thylakoid proteins (Franklin and Hoffman, 1993; Hoffman and Franklin, 1994; Yuan et al., 1993); (d) a so-called spontaneous thylakoid insertion pathway (Michl et al., 1994); and (e) a routing pathway that could involve a chloroplast homologue of the NEM-sensitive factor (Huguency et al., 1995; Malhotra et al., 1988).
The nuclear-coded chloroplastic IEPs identified to date are synthesized in the cytosol with a stroma-targeting presequence (Flügge et al., 1989; Brink et al., 1995; Knight and Gray 1995; for a review see Lübeck et al., 1997). The data indicate that the presequence of the inner envelope– localized phosphate-triose-phosphate translocator is cleaved off by the general stromal-processing peptidase (Brink et al., 1995; Knight and Gray, 1995). The topogenic signal for the integration into the inner envelope, however, seems to be contained in the mature part of the protein. Since it was not possible to separate the translocation process of the phosphate-triose-phosphate translocator from the inner envelope insertion process, there remained the question of how the insertion pathway took place, e.g., via a membrane-associated form or via a soluble stromal intermediate followed by a putative anchestral reexport pathway.
We have recently identified the IEP of 110 kD (IEP110) as a constituent of the chloroplastic protein import machinery (Kessler and Blobel, 1996; Lübeck et al., 1996). IEP110 is anchored into the membrane by an NH2-terminal hydrophobic stretch of amino acids with the potential to form an α helix. The topology of IEP110 can be assessed by treating intact chloroplasts with the protease trypsin. Trypsin penetrates the chloroplastic outer envelope and degrades proteins in the intermembrane space, as well as those exposed on the outer surface of the inner envelope (Marshall et al., 1990; Lübeck et al., 1996), while leaving the inner envelope membrane and the chloroplast integrity intact. IEP110 is sensitive to trypsin, demonstrating that the majority of the protein, ∼80–90 kD, is exposed to the intermembrane space between the two envelopes (Lübeck et al., 1996). This asymmetrical arrangement of IEP110 might result in distinguishable import stages in contrast to the phosphate-triose-phosphate translocator that is deeply buried in the membrane (Flügge et al., 1989). The data presented in this paper reveal the existence of a reexport pathway that can direct proteins in a posttranslocational manner from the stroma into the chloroplastic inner envelope independently of a cleavable NH2-terminal presequence. Furthermore, the export machinery is capable of translocating polypeptide domains across the inner envelope into the intermembrane space.
Materials and Methods
Chloroplast Protein Import Assays
Chloroplasts were isolated from the pea leaves of 10–12-d-old plants and purified on silica sol gradients (Waegemann and Soll, 1991). Chlorophyll (chl) was determined as described (Arnon, 1949). A standard import assay contained chloroplasts equivalent to 30 μg chl in 100 μl import mix (330 mM sorbitol, 50 mM Hepes-KOH, pH 7.6, 3 mM MgSO4, 10 mM methionine, 10 mM cysteine, 20 mM potassium gluconate, 10 mM NaHCO3, 2% (wt/vol) BSA, 3 mM ATP, and 1–10% of reticulocyte lysate in vitro–synthesized 35S-labeled precursor proteins. Import was carried out for 20 min at 25°C. Chloroplasts were reisolated from the import assay by centrifugation at 4°C through a Percoll cushion (40% [vol/vol] Percoll in 300 mM sorbitol, 50 mM Hepes-KOH, pH 7.6), washed once in Hepes-sorbitol (Waegemann and Soll, 1991), and used for further treatments. Prebinding of precursor proteins to chloroplasts was done under conditions identical to those described above, except that all steps were carried out at 0°C.
Chloroplasts were treated with the protease thermolysin in 300 mM sorbitol, 50 mM Hepes-KOH, pH 7.6, 0.5 mM CaCl2 for 20 min at 4°C at 200 μg protease/mg chl. The reaction was terminated by the addition of 10 mM EDTA, and chloroplasts were recovered by centrifugation through a 40% Percoll cushion and washed once (Joyard et al., 1983; Cline et al., 1984). Chloroplasts (equivalent to 200 μg chl) were treated with 200 μg trypsin (10700 Nα-benzoyl-l-arginine ethyl estes U/mg from bovine pancrease) in 300 mM sorbitol, 50 mM Hepes-KOH, pH 8.0, and 0.1 mM CaCl2 for 1 h at 20°C in a final volume of 200 μl. The reaction was stopped by the addition of 1 mM PMSF and a fivefold excess of soybean trypsin inhibitor (Marshall et al., 1990; Lübeck et al., 1996). Chloroplasts were recovered and washed as described above. Protease inhibitors were present at all subsequent steps. Chlorophyll was determined (Arnon, 1949), and equal amounts of organelles were loaded onto the SDS-PAGE on a chl basis.
After hypotonic lysis in 10 mM Hepes-KOH, pH 7.6, chloroplasts were separated into a soluble stromal and a total membrane fraction by centrifugation for 10 min at 165,000 g. Soluble proteins were precipitated with TCA. Import products were analyzed by SDS-PAGE (Laemmli, 1970) followed by fluorography (Bonner and Laskey, 1974). Separation of envelope and thylakoid membranes was achieved after hypotonic lysis of chloroplasts (see above) by centrifugation for 30 s at 2,000 g. The resulting pellet contained thylakoid membranes. The supernatant was once again centrifuged for 10 min at 165,000 g. Soluble proteins were precipitated with TCA and treated as described above. The pellet fraction, containing a mixture of envelope and remaining thylakoid membranes, was resuspended in 100 μl Hepes-KOH, pH 7.6, and layered over a 300-μl sucrose cushion (1 M sucrose, 10 mM Hepes-KOH, pH 7.6). After centrifugation for 10 min at 165,000 g, thylakoid membranes had pelleted through the cushion and were pooled with the above thylakoid fraction. The supernatant, including the sucrose cushion, was diluted 1:5 with 10 mM HepesKOH, pH 7.6, and envelope membranes were recovered by centrifugation for 10 min at 165,000 g.
Construction of cDNAs Coding for IEP110 Hybrid Proteins and Synthesis of Labeled Proteins
cDNAs coding for the different hybrid proteins were constructed by recombinant PCR (Higuchi, 1990). Products were cloned into a vector suitable for in vitro transcription and controlled by DNA sequencing (Sanger et al., 1977). The original cDNAs coding for pIEP110 and pSSU have been described (Lübeck et al., 1996; Klein and Salvucci, 1992). In vitro transcription was done using either T7- or SP6-RNA polymerase, as outlined before (Salomon et al., 1990). Proteins were synthesized in reticulocyte lysate in the presence of 35S-labeled methionine and cysteine (1175Ci/ mmol) for 1.5 h at 30°C (Salomon et al., 1990). Overexpression of pSSU was done in Escherichia coli BL21 (DE3) cells using the pET vector system (Novagen, Madison, WI) (Waegemann and Soll, 1995). pSSU was isolated from inclusion bodies and solubilized in 8 M urea. The final urea concentration in the assays never exceeded 80 mM.
Results
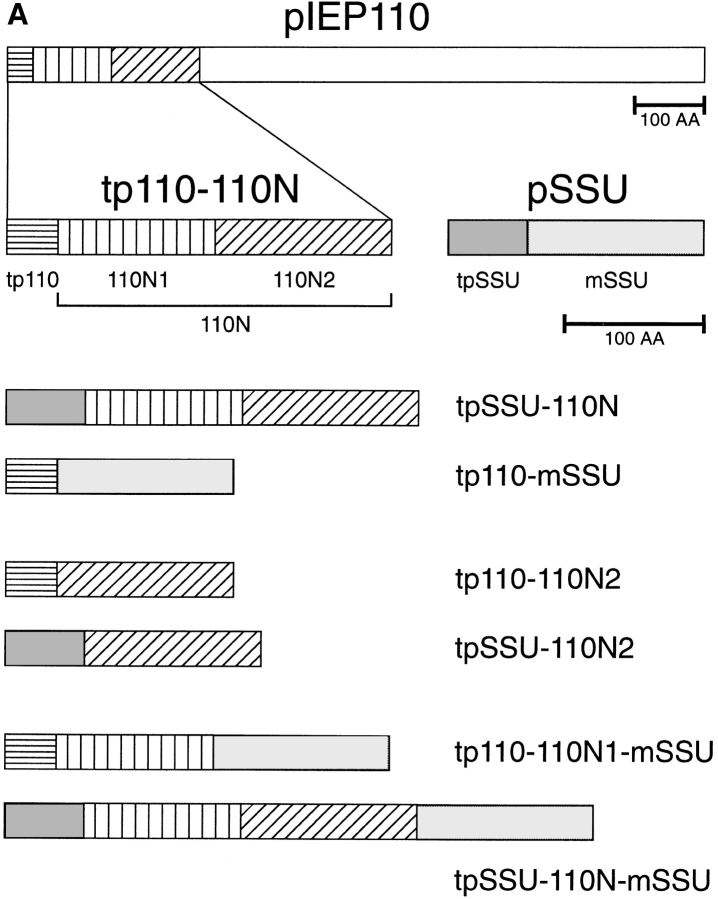
The role of IEP110 in protein import and its specific topology prompted us to study its import in some detail. The protein sequence of IEP110 shows the potential to form a hydrophobic α helix in the NH2 terminus (amino acids 100–140, Fig. 1 B), which seems to anchor the protein into the inner envelope membrane (Lübeck et al., 1996; Kessler et al., 1996). Initial results had indicated that pIEP110 uses the general import pathway into chloroplasts; however, the translation and import efficiencies of full-length precursor of IEP110 (pIEP110) were rather low, so we decided to use a truncated version of pIEP110, namely the transit peptide of 110 and the NH2-proximal part of mature IEP110 (tp110-110N; Fig. 1 A), which comprises amino acids 1–269, to study its import characteristics.
Figure 1.
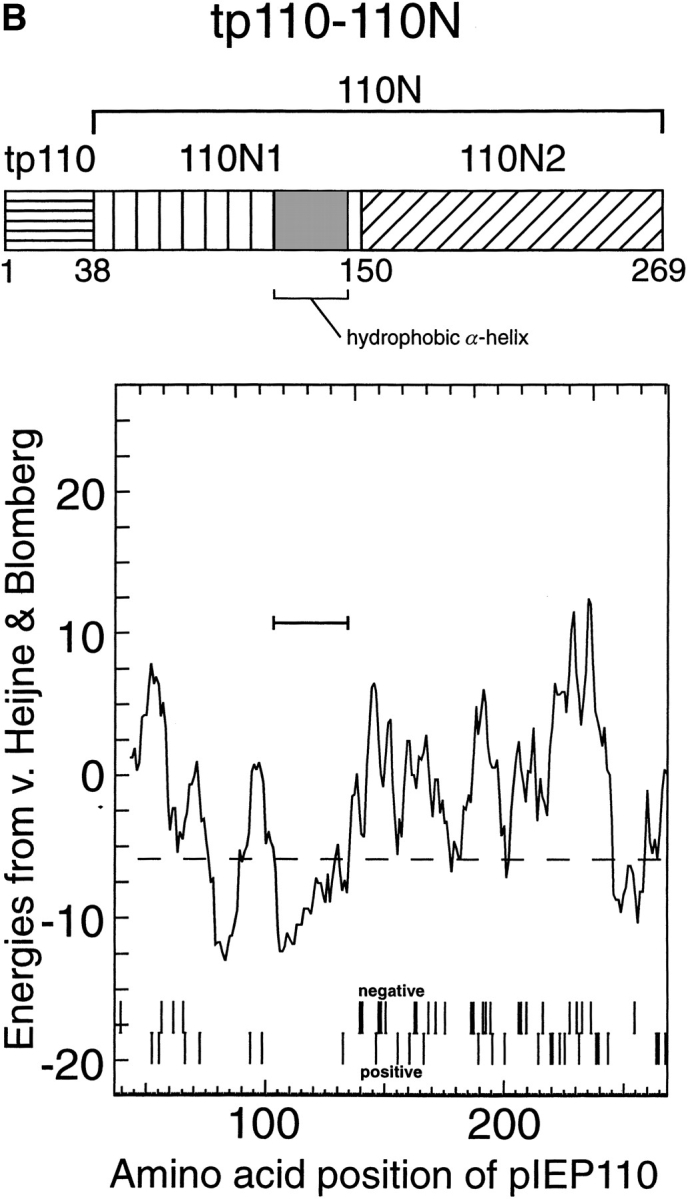
(A) Peptide domain combination of various hybrid precursor proteins used in this study and their nomenclature. (AA, amino acids; tp, transit peptide; 110N, amino acids 39–269 of pIEP110; 110N1, amino acids 39–149 of IEP110; 110N2, amino acids 150–269 of pIEP110). (B) Detailed charge distribution and hydropathy analysis of tp110-110N.
After import of the tp110-110N translation product into intact pea chloroplasts under standard import conditions, the terminally processed form was recovered almost exclusively in the envelope membrane fraction (Fig. 2 A). Processed 110N was resistant to extraction at pH 11.5 (0.1 M Na2CO3), indicating a stable insertion into the membrane similar to in situ (Lübeck et al., 1996). No 110N was detectable in the soluble protein fraction of chloroplasts, and only very little was detectable in the thylakoid membrane fraction (Fig. 2 A, lane 5). Under thylakoid isolation procedures such as those used here, these were shown to be contaminated with envelope membranes (Keegstra and Youssif, 1986; Joyard et al., 1991). Therefore, we conclude that the low amount of 110N found in the thylakoid fraction results from contamination with envelope membranes.
Figure 2.
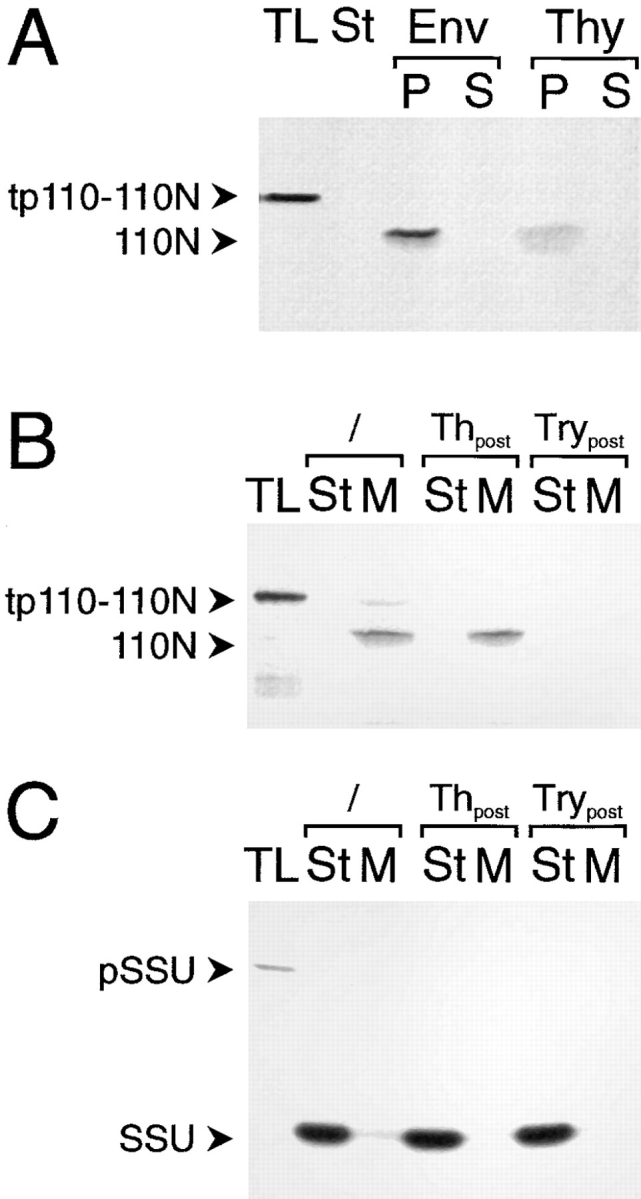
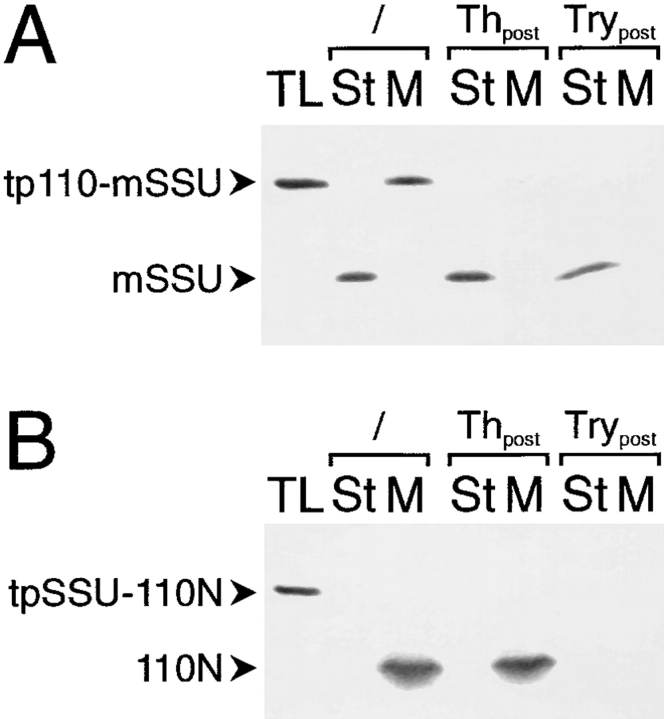
Inner envelope– targeting and insertion information is contained in the NH2 terminus of IEP110. A truncated IEP110 precursor, namely tp110-110N, comprising amino acids 1–269, was used in standard import experiments with purified pea chloroplasts (equivalent to 30 μg chl; A and B). (A) Chloroplasts were separated into different fractions containing either envelope membranes (Env), soluble stroma (St), and thylakoid membranes (Thy). Membranes were subsequently extracted with Na2CO3 at pH 11.5 and further separated into a pellet (P) and soluble fraction (S). (B) Inserted, terminally processed 110N is sensitive to the protease trypsin but not to thermolysin. After completion of a standard import reaction, chloroplasts were either not treated (lanes 2 and 3) or treated with protease (thermolysin [Th], lanes 4 and 5; trypsin [Try], lanes 6 and 7). Organelles were reisolated through a 40% (vol/vol) Percoll cushion washed once. Chlorophyll concentration was determined, and equal amounts of organelles were used for further analysis. Organelles were separated into a soluble stroma (St) and membrane (M) fraction. (C) pSSU is imported into the chloroplast stroma, where the processed mature form is resistant to both proteases, thermolysin and trypsin. All experimental conditions are as decribed in B. 35S-labeled translation product (TL) is shown as an internal standard, 10% of which was added to a standard import reaction. Fluorograms are shown.
The COOH terminus of IEP110 that is exposed to the intermembrane space in intact pea chloroplasts is inaccessible to the noninvasive protease thermolysin, but is degraded by trypsin, which proteolytically penetrates through the outer envelope (Lübeck et al., 1996). Imported mature 110N was recovered in a thermolysin-inaccessible but trypsin-accessible manner (Fig. 2 B), giving further evidence that tp110-110N is correctly translocated, processed, and inserted into the inner envelope membrane. Control import experiments (Fig. 2 C) using the precursor of the small subunit of ribulose-1,5-bisphosphate carboxylase (pSSU), a protein localized in the stroma, show that imported mature SSU is inaccessible to trypsin. These results indicate that the chloroplastic inner envelope was not penetrated by trypsin under these experimental conditions, corroborating earlier results that used the latency of the Hill reaction to measure chloroplast intactness after trypsin treatment (Lübeck et al., 1996). We conclude that tp110-110N can be used as a bona fide substrate to study the import of pIEP110 into pea chloroplasts, since both full-length and truncated precursors are targeted to the inner envelope membrane and exhibit identical behavior to extraction at pH 11.5 and sensitivity to trypsin.
The IEP110 Transit Peptide Contains Stromal Targeting Information
Next, we wanted to know if the targeting information present in the presequence of IEP110 is essential for translocation into the organelle and insertion into the inner envelope. Therefore, the presequences between 110N and SSU were mutually exchanged, resulting in the constructs tpSSU-110N and tp110-mSSU (see Fig. 1). Import experiments into pea chloroplasts clearly show that the transit peptide of IEP110 directs the SSU passenger protein into the soluble stroma phase, where it is processed and resistant to thermolysin and to trypsin treatment (Fig. 3 A, lanes 2–7). The import efficiency of tp110-mSSU, however, is less than 50% of the authentic pSSU. The transit peptide of SSU also directed the 110N domain of IEP110 to chloroplasts in the reverse construct (Fig. 3 B, lanes 2 and 3). The processed 110N was recovered in a thermolysin-insensitive but trypsin-sensitive manner, indicating its correct insertion and orientation in the inner envelope membrane. We conclude that tp110 contains only stromaltargeting envelope transfer information, and that the topogenic signal for envelope targeting to the inner envelope membrane and correct insertion is present in the 110N domain of IEP110.
Figure 3.
The presequence of IEP110 contains only stroma-targeting information. The presequence of IEP110 and SSU were mutually exchanged, and the resulting hybrids, tp110-mSSU and tpSSU-110N, were used under standard chloroplast import conditions. (A) Chloroplasts were separated into a soluble stroma phase and a total membrane fraction either before or after treatment with thermolysin or trypsin. (B) The stroma-directing presequence of SSU (tpSSU) cannot deviate 110N to the stroma. Import and subfractions of chloroplasts are as decribed in A. All further experimental conditions and abbreviations are as decribed in Fig. 2.
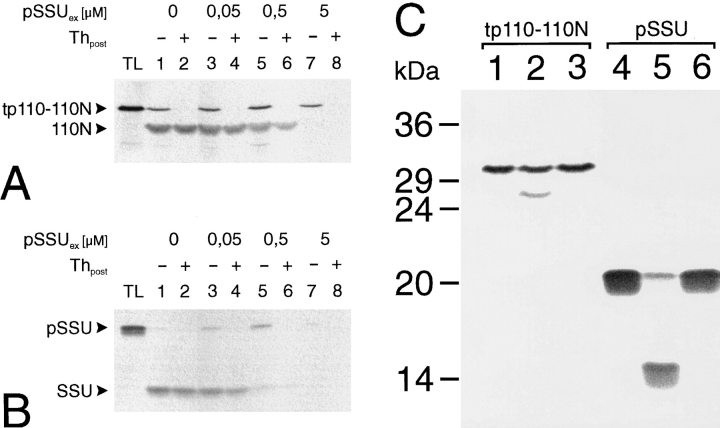
Further evidence for this conclusion results from studies that demonstrate that tp110-110N import is competed for by the addition of an excess pSSU (Fig. 4 A). A similar competition for the import yield can be observed between the 35S-labeled pSSU translation product and an excess of E. coli–expressed pSSU (Fig. 4 B). E. coli–expressed pSSU was introduced into the import reaction as a ureadenatured protein. While a competition at the level of translocation was always observed, the binding of reticulocyte lysate–synthesized translation products to the outer envelope membrane is not competed for to the same extent (Fig. 4, A vs. B). This might be caused by a certain amount of nonspecific binding to the chloroplast surface that cannot be competed for by denatured (i.e., unfolded) pSSU, which might bypass the primary binding site and thus compete only at the level of translocation. The results (Fig. 4, A and B), however, clearly demonstrate that pSSU and tp110-110N use common components of the chloroplast import machinery.
Figure 4.
IEP110 uses the general import and processing components of chloroplasts. (A) tp110-110N import into pea chloroplasts was assayed in the absence or presence of increasing pSSU concentration. pSSUex was synthesized in E. coli and added as a urea-denatured protein to the import assay (final urea concentration 80 mM). Import experiments were initiated by the addition of organelles (equivalent to 30 μg chl). (B) E. coli–synthesized pSSUex competes the import of reticulocyte lysate made 35S-labeled pSSU with similar efficiency as tp110-110N. Experimental conditions are as desribed in A. (C) Tp110-110N and pSSU are both processed in vitro by a stromal extract. A stromal processing extract was incubated for either 0 min (lanes 1 and 4) or 90 min (lanes 2, 3 and 5, 6) with the respective translation product in the absence (lane 2 and 5) or presence (lane 3 and 6) of 10 mM 1,10 o-phenanthroline.
Translocation across the envelope membranes is followed by processing in the stroma. The stromal processing protease is a metalloenzyme and can be specifically inhibited by o-phenanthroline (Abad et al., 1989). In vitro processing of pSSU and tp110-110N by a stromal extract demonstrated that both precursor proteins could be converted to their respective mature forms (Fig. 4 C, lanes 1, 2, and 4, 5). Furthermore, the processing of both preproteins was inhibited by o-phenanthroline (Fig. 4 C, lanes 3 and 6). Together, the data presented in Fig. 4, A–C, strongly indicate that pIEP110 uses the general import machinery and processing system of the majority of chloroplast-targeted preproteins.
Inner Envelope Targeting of IEP110.
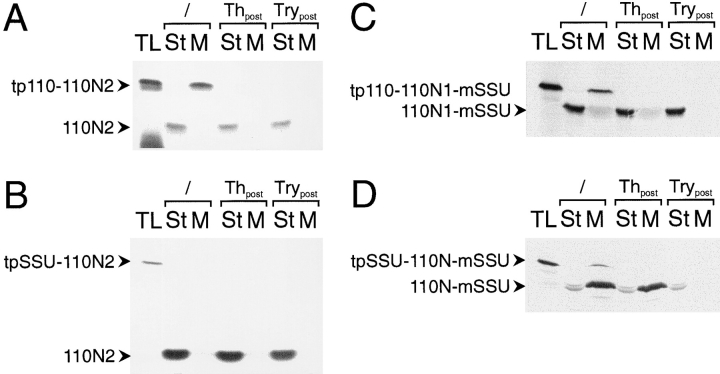
As shown above, the 110N region contained all the sequence determinants necessary for targeting and insertion into the inner envelope membrane. To distinguish domains within this region that could be involved in either targeting or insertion, 110N was subdivided into N1, which contained the most NH2-proximal amino acids of 110N, and N2, which contained the COOH-proximal portion of the 110N region (details are outlined in Fig. 1 A). The sequence of the N2 subdomain was expressed in frame with tp110 or tpSSU, resulting in the hybrid proteins tp110110N2 and tpSSU-110N2. When import reactions were performed with either tp110-110N2 or tpSSU-110N2, processed 110N2 was recovered exclusively in the soluble stromal phase (Fig. 5, A and B, lanes 2 and 3), as was also indicated by its resistance to either thermolysin or trypsin treatment (Fig. 5, A and B, lanes 4–7). The results suggested that the 110N2 domain did not contain sufficient targeting information for the inner envelope membrane.
Figure 5.
The N1 and N2 subdomains of IEP110 are both necessary for efficient envelope targeting and insertion. (A and B) amino acids 150–269 of IEP110 were fused to the presequence of either IEP110 or SSU, resulting in tp110-110N2 and tpSSU-110N2, respectively. 35S-labeled translation product was incubated with intact pea chloroplasts (equivalent to 30 μg chl), and the localization of processed, mature 110N2 was tested by chloroplast subfractionation before (lanes 2 and 3) or after treatment with the proteases thermolysin (lanes 4 and 5) or trypsin (lanes 6 and 7). All other experimental conditions are as decribed in Fig. 2. (C) The tp110-110N1-mSSU translation product is imported into pea chloroplasts. Chloroplasts were subfractionated into a soluble stroma phase (lanes 2, 4, and 6) and a membrane fraction (lanes 3, 5, and 7) either before (lanes 2 and 3) or after treatment of the organelles with the proteases thermolysin (lanes 4 and 5) or trypsin (lanes 6 and 7). (D) The tpSSU-110N-mSSU translation product is imported into the chloroplasts under standard conditions. Chloroplasts are fractionated either before or after protease treatment, as outlined above. All experimental conditions and abbreviations are as outlined in Fig. 2.
The 110N1 domain, which also comprises the putative membrane-anchoring α helix, could therefore be solely responsible for membrane targeting. A hybrid protein was synthesized which consisted of tp110 fused to the N1 domain, which contained SSU as a COOH-terminal extension and putative reporter for intermembrane space location. When the tp110-110N1-mSSU translation product was imported into chloroplasts, 110N1-mSSU was again largely recovered in the soluble stromal phase. About 10% of 110N1-mSSU was found in the membrane fraction (Fig. 5 C, lanes 2 and 3). Soluble mature 110N1-mSSU was resistant to the proteases thermolysin and trypsin. However, membrane-bound 110N1-mSSU was resistant to thermolysin but accessible to trypsin, i.e., 110N1 functions as the correct topogenic signal, albeit with low efficiency. The efficiency of membrane targeting and insertion might be strongly enhanced by the 110N2 domain.
To test this notion, tpSSU-110N-mSSU, which contained both the N1 and N2 subdomains, was synthesized in reticulocyte lysate and used in a standard import experiment. Interestingly, processed, mature 110N-mSSU was largely recovered in the membrane fraction (Fig. 5 D). 110N-mSSU was resistant to thermolysin but accessible to trypsin, demonstrating that the hybrid protein attained an in situ–like membrane orientation; i.e., the COOH-proximal portion of 110N-mSSU is exposed to the intermembrane space. Between 10 and 20% of processed, mature 110N-mSSU was recovered as a soluble protein in a trypsin-inaccessible location, i.e., in the stromal phase. Up to this point, the data indicate that 110N1 contains the information that anchors the protein into the membrane but functions with low efficiency, while 110N2 in combination with 110N1 strongly increases the yield of membraneinserted 110N, maybe because 110N2 contains essential targeting or sorting information that only works in a cooperative way with 110N1.
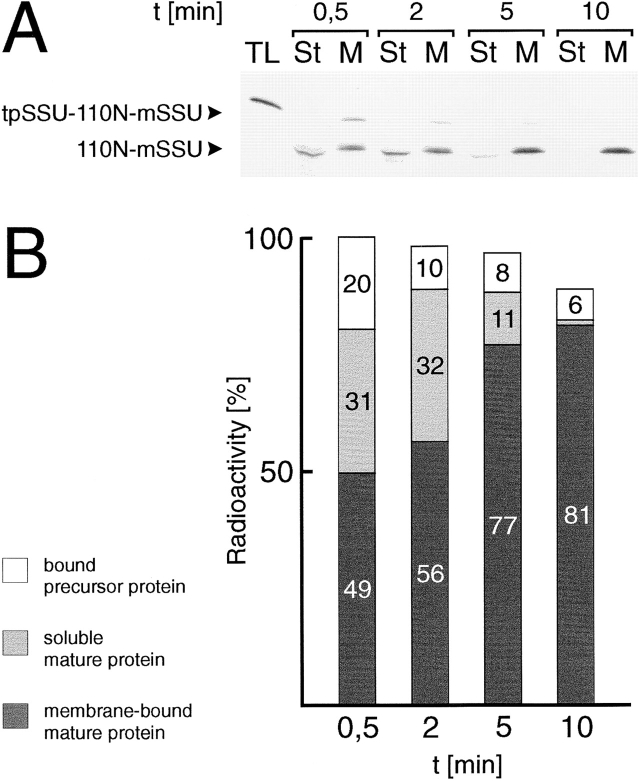
Since processed, mature 110N-mSSU was detectable in the membrane as well as in the soluble phase, the following question arose: does 110N-mSSU integration into the inner envelope occur via a soluble intermediate, or is a significant portion of 110N-mSSU missorted to the stroma? Therefore, 35S-labeled tpSSU-110N-mSSU was incubated with intact chloroplasts in the presence of 3 mM ATP for 5 min at 0°C to allow binding but no translocation. Chloroplasts were separated from unbound precursors by centrifugation. Organelles were resuspended in 25°C tempered fresh import buffer in the presence of 3 mM ATP, and were incubated further at 25°C for various times (Fig. 6, A and B). At the earliest time point during the chase period (30 s), a significant portion (20%) of the labeled protein was recovered as membrane-bound precursor, while about one third was found as soluble processed mature form in the stroma. About 50% of the protein had already reached the inner envelope membrane. Most of the chloroplastbound precursor was imported during the chase period, and only ∼5% of the total labeled protein remained on the chloroplast surface. Furthermore, the proportion of soluble 110N-mSSU intermediate declined steadily and was at the brink of detectability after 10 min. Simultaneously, the membrane-recovered portion of 110N-mSSU increased to more than 80% of the initially introduced labeled proteins. Because of the removal of unbound precursor before the beginning of the chase period, we analyzed only the import and insertion of prebound molecules; i.e., the degradation of soluble 110N-mSSU can be excluded because, otherwise, we should not be able to detect an almost constant amount of radioactivity during the entire chase period (Fig. 6 B). In the attempts to observe more of the soluble 110N-mSSU protein in the stroma, the chase period was initiated by resuspending the chloroplasts in ice-cold import buffer, followed by a slow warming period. In general, the results were similar to those described in Fig. 6, except that at the early time points, 30 s and 2 min, bound tpSSU-110N-mSSU made up more than 50% of the total labeled protein, while processed 110NmSSU showed a 1:1 distribution between the stroma and the inner envelope (data not shown). The final result of the chase period was identical to that described in Fig. 6. These data clearly demonstrate that a soluble stroma localized intermediate can be chased to a membrane-bound location with an orientation that most of the protein is exposed to the intermembrane space (see also Fig. 4).
Figure 6.
110N-mSSU can be translocated into the inner envelope membrane via a soluble stromal intermediate. (A) 35S-labeled tpSSU-110N-mSSU translation was incubated with intact chloroplasts (equivalent to 200 μg chl in 500 μl reaction volume) at 0°C to allow binding but not import. After 5 min, the chloroplasts were reisolated through a 40% Percoll cushion, washed, and resuspended in fresh 25°C tempered import buffer. Aliquots (equivalent to 30 μg chl) were taken at the time intervals indicated, and the translocation reaction was halted by the addition of ice-cold import buffer. Chloroplasts were separated into soluble and total membrane fractions. Products were analysed by SDS-PAGE and fluorography. (B) Quantification of the fluorogram shown in A. Different exposures of the x-ray film were quantified using a laser densitometer. The sum of radioactivity found in either the soluble or membrane fraction at 0.5 min was set to 100%.
Discussion
Our data demonstrate that chloroplasts have the capability to reexport proteins into the inner envelope after their complete import into and processing in the stroma. This is not only a membrane insertion process, but the translocation machinery involved can export a large polypeptide chain across the membrane into the intermembrane space. The SSU reporter peptide in 110N-mSSU has a length of 140 amino acids, which is completely translocated across the inner envelope into the intermembrane space, as indicated by its complete sensitivity to trypsin. The components of an export machinery are unknown at the present time, and it remains to be seen if it shares common constituents with the inner envelope import machinery. The identification of a chloroplast-coded, inner envelope– localized protein (Sasaki et al., 1993) had indicated previously that an independent transport pathway should exist for this membrane. No data exist as to whether this protein translocates co- or posttranslationally into the inner envelope.
tpSSU-110N-mSSU is translocated into the inner envelope after processing in the stroma as the mature form; i.e., the export signal is not present in the transit sequence, but internal targeting information is necessary. This topogenic signal is localized in amino acids 38–269, most likely as a bipartite signal. The NH2-proximal part (N1, amino acids 38–149), which also contains the putative membrane anchor region of IEP110, is sufficient for targeting to the inner membrane and proper insertion, albeit with low efficiency. The N2 region of IEP110 (amino acids 150–269) cannot direct proteins to the inner envelope, but remains in the stroma upon import. Compartimentation of N1 or N2 upon import is most likely to be independent of the stroma-targeting signal used, since both tpSSU and tp110 contain only envelope transfer stroma-targeting information. In combination with the N1 region, N2 directs the protein quantitatively to the inner envelope. We propose that N1 contacts the membrane surface very early and that N2 subsequently initiates the translocation process, e.g., as a start transfer signal.
pIEP110 takes the general chloroplast import pathway, as shown by the mutual exchange of transit sequences between pIEP110 and pSSU. tp110 seems to be less efficient than tpSSU, which supports two- to threefold higher import yields. Simultaneously, more binding to the chloroplast surface is generally detected in constructs that contain tp110. The reason for this is unclear at the moment. Furthermore, the stromal processing peptidase seems to also process pIEP110. Import into chloroplasts and integration into the inner envelope of pIEP110 and tpSSU110N are rapid processes, and a soluble intermediate was beyond detectability. Only when we used tpSSU-110NmSSU was a soluble intermediate in the translocation pathway clearly visible. While ∼50% of mature 110NmSSU has already reached the inner envelope after a 30-s chase period, the results in Fig. 5 clearly establish that soluble mature 110N-mSSU can be exported into and across the membrane. Two reasons could explain these differences: (a) the passenger protein mSSU retards the integration and translocation process because its primary sequence or secondary structure are not well suited for the export machinery; and (b) the mSSU part in 110N-mSSU interacts transiently with its “natural” partners of the pSSU import and assembly pathway, e.g., chaperonin 60 or the large subunit of ribulose-1,5-bisphosphate carboxylase. Independently of the reason for the kinetic differences in 110N-mSSU export, our results show very clearly that the signal responsible for reexport to the inner envelope protein is present in 110N and is “stronger” than the potential interaction of mSSU with stromal proteins. At the moment, it cannot be excluded that a fraction of 110NmSSU is never released from the membrane before insertion, indicating that parallel mechanisms that are used alternatively might exist, or that the currently available experimental approaches are not elaborate enough to resolve this problem. Lowering the temperature during the critical chase period did not result in a block of membrane insertion; only in a general slowdown of the entire process. Therefore, specific inhibitors that block single steps in the process are needed to describe the sequence of translocation and insertion in detail. However, our results demonstrate that a reexport machinery exists in the chloroplasts' stroma that can cooperate faithfully with an inner envelope machinery. In the future, this soluble intermediate will be used as a tool to search for the components of the reexport pathway. The initial protein import experiments of tpSSU-110N-mSSU in the presence of NaN3 or guanosine nucleotides did not show significant alterations in the yield of inner envelope–localized 110N-mSSU (Lübeck, J., and J. Soll, unpublished data).
Studies of sorting of proteins to the inner mitochondrial membrane have suggested that the yeast cytochrome oxidase subunit Va uses a “stop-transfer” mechanism, i.e., one that never leaves the inner membrane, even during processing by the matrix processing protease (Miller and Cumsky, 1993; see also Glick et al., 1992). Sorting of subunit 9 of Fo-ATPase to the inner membrane of Neurospora mitochondria, however, was proposed to occur after complete translocation into the organelle via a conservative sorting mechanism (Rojo et al., 1993). The soluble matrix localization of processed mature subunit 9 of Fo-ATPase was not analyzed (Rojo et al., 1993; see also Stuart and Neupert, 1996). The soluble intermediate we describe in this study for the translocation of 110N-mSSU into the inner envelope might involve a sorting system that is not present in mitochondria, e.g., a Sec, NSF, or SRP homologue–dependent pathway. More work is required to answer these questions related to chloroplastic biogenesis.
Acknowledgments
This work was supported by the Deutsche Forschungsgemeinschaft.
Abbreviations used in this paper
- chl
chlorophyll
- IEP
inner envelope protein
- IEP110
chloroplastic IEP of 110 kD
- m
mature form of
- p
precursor of
- SSU
small subunit of ribulose-1,5-bisphosphate carboxylase
- tp
transit peptide of
- 110N
NH2-proximal part of mature IEP110
Footnotes
Address all correspondence to Professor Jürgen Soll, Botanisches Institut, Christian-Albrechts-Universität, D-24118 Kiel, Germany. Tel.: (49) 431880-4210. Fax: (49) 431-880-1527. E-mail: jsoll@bot.uni-kiel.de
References
- Abad MS, Clark SE, Lamppa GK. Properties of a chloroplast enzyme that cleaves the chlorophyll a/b-binding precursor. Optimization of an organelle-free reaction. Plant Physiol. 1989;90:117–124. doi: 10.1104/pp.90.1.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon DJ. Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. . Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berghöfer J, Karnauchov I, Herrmann RG, Klösgen RB. Isolation and characterization of a cDNA encoding the secA protein from spinach chloroplasts. Evidence for azide resistance of sec-dependent protein translocation across thylakoid membranes in spinach. J Biol Chem. 1995;270:18341–18346. doi: 10.1074/jbc.270.31.18341. [DOI] [PubMed] [Google Scholar]
- Bonner WM, Laskey RA. A film detection method for tritiumlabelled proteins and nucleic acids in polyacrylamide gels. Eur J Biochem. 1974;46:84–88. doi: 10.1111/j.1432-1033.1974.tb03599.x. [DOI] [PubMed] [Google Scholar]
- Brink S, Fischer K, Klösgen RB, Flügge UI. Sorting of nuclearencoded chloroplast membrane proteins to the envelope and the thylakoid membrane. J Biol Chem. 1995;270:20808–20815. doi: 10.1074/jbc.270.35.20808. [DOI] [PubMed] [Google Scholar]
- Cline K, Henry R. Import and routing of nucleus-encoded chloroplast proteins. Annu Rev Cell Dev Biol. 1996;12:1–26. doi: 10.1146/annurev.cellbio.12.1.1. [DOI] [PubMed] [Google Scholar]
- Cline K, Henry H, Li C, Yuan J. Multiple pathways for protein transport into or across the thylakoid membrane. EMBO (Eur Mol Biol Organ) J. 1993;12:4108–4114. doi: 10.1002/j.1460-2075.1993.tb06094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline K, Werner-Washburne M, Andrews J, Keegstra K. Thermolysin is a suitable protease for probing the surface of intact pea chloroplasts. Plant Physiol. 1984;75:675–678. doi: 10.1104/pp.75.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flügge U-I, Fischer K, Gross A, Sebald W, Lottspeich F, Eckerskorn C. The triose phosphate-3-phosphoglycerate-phosphate translocator of spinach chloroplasts: nucleotide sequence of a full-length cDNA clone and import of the in vitro synthesized precursor protein into chloroplasts. EMBO (Eur Mol Biol Organ) J. 1989;8:39–46. doi: 10.1002/j.1460-2075.1989.tb03346.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin AE, Hoffmann NE. Chloroplasts contain a homolog of the 54 kD subunit of the signal recognition particle. J Biol Chem. 1993;268:22175–22180. [PubMed] [Google Scholar]
- Glick B, Beasley EM, Schatz G. Protein sorting in mitochondria. Trends Biol Sci. 1992;17:453–459. doi: 10.1016/0968-0004(92)90487-t. [DOI] [PubMed] [Google Scholar]
- Grossmann A, Bartlett S, Chua N-H. Energy-dependent uptake of cytoplasmically synthesized polypeptides by chloroplasts. Nature (Lond) 1980;285:625–628. [Google Scholar]
- Hageman J, Baecke C, Ebskamp M, Pilon R, Smeekens S, Weisbeek P. Protein import into and sorting inside the chloroplast are independent processes. Plant Cell. 1990;2:479–494. doi: 10.1105/tpc.2.5.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi, R. 1990. Recombinant PCR. In PCR Protocols: A Guide to Methods and Applications. M.A. Innis., D.H. Gelfand, J.J. Sninsky, and T.J. White, editors. Academic Press, Inc., San Diego, CA. 177–183.
- Hoffmann NE, Franklin AE. Evidence for a stromal GTP requirement for the integration of a chlorophyll a/b-binding polypeptide into thylakoid membranes. Plant Physiol. 1994;105:295–304. doi: 10.1104/pp.105.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugueney P, Bouvier F, Badillo A, d'Harlingue A, Kuntz M, Camara B. Identification of a plastid protein involved in vesicle fusion and/ or membrane protein translocation. Proc Natl Acad Sci USA. 1995;92:5630–5634. doi: 10.1073/pnas.92.12.5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyard J, Block MA, Douce R. Molecular aspects of plastid envelope biochemistry. Eur J Biochem. 1991;199:489–509. doi: 10.1111/j.1432-1033.1991.tb16148.x. [DOI] [PubMed] [Google Scholar]
- Joyard J, Billecocq A, Bartlett SG, Block MA, Chua N-H, Douce R. Localization of polypeptides to the cytosolic side of the outer envelope membrane of spinach chloroplasts. J Biol Chem. 1983;258:10000–10006. [PubMed] [Google Scholar]
- Keegstra K, Youssif AE. Isolation and characterization of chloroplast envelope membranes. Methods Enzymol. 1986;118:316–325. [Google Scholar]
- Kessler F, Blobel G. Interaction of the protein import and folding machineries in the chloroplasts. Proc Natl Acad Sci USA. 1996;93:7684–7689. doi: 10.1073/pnas.93.15.7684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein PR, Salvucci ME. Photoaffinity labelling of mature and precursor forms of the small subunit of ribulose-1,5-biphosphate carboxylase/ oxygenase after expression in Escherichia coli. . Plant Physiol. 1992;98:546–553. doi: 10.1104/pp.98.2.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight JS, Gray JC. The N-terminal hydrophobic region of the mature phosphate translocator is sufficient for targeting to the chloroplast inner envelope membrane. Plant Cell. 1995;7:1421–1432. doi: 10.1105/tpc.7.9.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott TG, Robinson C. The secA inhibitor, azide, reversibly blocks the translocation of a subset of proteins across the chloroplast thylakoid membrane. J Biol Chem. 1994;269:7843–7846. [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the heads of bacteriophage T4. Nature (Lond) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laidler V, Chaddock AM, Knott TG, Walker D, Robinson C. A secY homolog in Arabidopsis thaliana.Sequence of a full-length cDNA clone and import of the precursor protein into chloroplasts. J Biol Chem. 1995;270:17664–17667. doi: 10.1074/jbc.270.30.17664. [DOI] [PubMed] [Google Scholar]
- Lübeck, J., L. Heins, and J. Soll. 1997. Protein import into chloroplasts. Physiol. Plant. In press.
- Lübeck J, Soll J, Akita M, Nielsen E, Keegstra K. Topology of IEP110, a component of the chloroplastic protein import machinery present in the inner envelope membrane. EMBO (Eur Mol Biol Organ) J. 1996;15:4230–4238. [PMC free article] [PubMed] [Google Scholar]
- Malhotra V, Orci L, Glick BS, Block MR, Rothmann JE. Role of an N-ethylmaleimide-sensitive transport component in promoting fusion of transport vesicles with cisternea of the Golgi stack. Cell. 1988;58:221–227. doi: 10.1016/0092-8674(88)90554-5. [DOI] [PubMed] [Google Scholar]
- Marshall JS, DeRocher AE, Keegstra K, Vierling E. Identification of heat shock protein hsp70 homologues in chloroplasts. Proc Natl Acad Sci USA. 1990;87:374–378. doi: 10.1073/pnas.87.1.374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michl D, Robinson C, Shackleton JB, Herrmann RG, Klösgen RB. Targeting of proteins to the thylakoids by bipartite presequences: Cfo II is imported by a novel, third pathway. EMBO (Eur Mol Biol Organ) J. 1994;13:1310–1317. doi: 10.1002/j.1460-2075.1994.tb06383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BR, Cumsky MG. Intramitochondrial sorting of the precursor to yeast cytochrome c oxidase subunit Va. J Cell Biol. 1993;121:1021–1029. doi: 10.1083/jcb.121.5.1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohara T, Nakai M, Goto A, Endo T. Isolation and characterization of the cDNA for pea chloroplast secA. Evolutionary conservation of the bacterial-type secA-dependent protein transport within chloroplasts. FEBS Lett. 1995;364:305–308. doi: 10.1016/0014-5793(95)00415-6. [DOI] [PubMed] [Google Scholar]
- Robinson C, Cai D, Hulford A, Brock IW, Michl D, Hazell L, Schmidt I, Herrmann R, Klösgen RB. The presequence of a chimeric construct dictates which of two mechanisms are utilized for translocation across the thylakoid membrane: evidence for the existence of two distinct translocation systems. EMBO (Eur Mol Biol Organ) J. 1994;13:279–285. doi: 10.1002/j.1460-2075.1994.tb06260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo EE, Stuart RA, Neupert W. Conservative sorting of F0-ATPase subunit 9: export from matrix requires ΔpH across inner membrane and matrix ATP. EMBO (Eur Mol Biol Organ) J. 1995;14:3445–3451. doi: 10.1002/j.1460-2075.1995.tb07350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon M, Fischer K, Flügge U-I, Soll J. Sequence analysis and protein import studies of an outer chloroplast envelope polypeptide. Proc Natl Acad Sci USA. 1990;87:5778–5782. doi: 10.1073/pnas.87.15.5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Soulsen AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, Y., K. Sekiguchi, Y. Nagano, and R. Matsuno. 1993. Chloroplast envelope protein encoded by chloroplast genome. FEBS Lett. 316:93–98. [DOI] [PubMed]
- Stuart RA, Neupert W. Topogenesis of inner membrane proteins of mitochondria. Trends Biol Sci. 1996;21:261–267. [PubMed] [Google Scholar]
- Tranel PJ, Froehlich J, Goyal A, Keegstra K. A component of the chloroplastic protein import apparatus is targeted to the outer envelope membrane via a novel pathway. EMBO (Eur Mol Biol Organ) J. 1995;14:2436–2446. doi: 10.1002/j.1460-2075.1995.tb07241.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waegemann K, Soll J. Characterization of the protein import apparatus in isolated outer envelopes of chloroplasts. Plant J. 1991;1:149–158. [Google Scholar]
- Waegemann K, Soll J. Characterization and isolation of the chloroplast protein import machinery. Methods Cell Biol. 1995;50:235–267. doi: 10.1016/s0091-679x(08)61035-3. [DOI] [PubMed] [Google Scholar]
- Yuan J, Henry R, Cline K. Stromal factors play an essential role in protein integration into thylakoids that cannot be replaced by unfolding or by heat shock protein hsp70. Proc Natl Acad Sci USA. 1993;90:8552–8556. doi: 10.1073/pnas.90.18.8552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Henry R, McCafferty M, Cline K. SecA homolog in protein transport within chloroplasts: evidence for endosymbiont-derived sorting. Science (Wash DC) 1994;266:796–798. doi: 10.1126/science.7973633. [DOI] [PubMed] [Google Scholar]