Abstract
Mutations in the essential Drosophila melanogaster gene zw10 disrupt chromosome segregation, producing chromosomes that lag at the metaphase plate during anaphase of mitosis and both meiotic divisions. Recent evidence suggests that the product of this gene, DmZW10, acts at the kinetochore as part of a tension-sensing checkpoint at anaphase onset. DmZW10 displays an intriguing cell cycle–dependent intracellular distribution, apparently moving from the centromere/kinetochore at prometaphase to kinetochore microtubules at metaphase, and back to the centromere/kinetochore at anaphase (Williams, B.C., M. Gatti, and M.L. Goldberg. 1996. J. Cell Biol. 134:1127-1140).
We have identified ZW10-related proteins from widely diverse species with divergent centromere structures, including several Drosophilids, Caenorhabditis elegans, Arabidopsis thaliana, Mus musculus, and humans. Antibodies against the human ZW10 protein display a cell cycle–dependent staining pattern in HeLa cells strikingly similar to that previously observed for DmZW10 in dividing Drosophila cells. Injections of C. elegans ZW10 antisense RNA phenocopies important aspects of the mutant phenotype in Drosophila: these include a strong decrease in brood size, suggesting defects in meiosis or germline mitosis, a high percentage of lethality among the embryos that are produced, and the appearance of chromatin bridges at anaphase. These results indicate that at least some aspects of the functional role of the ZW10 protein in ensuring proper chromosome segregation are conserved across large evolutionary distances.
Eukaryotic organisms show wide diversity in the manner by which they organize the centromere/kinetochore of their chromosomes. The chromosomes of the budding yeast Saccharomyces cerevisiae have a well-defined point centromere: all centromeric functions are restricted to a short stretch of DNA ∼125 bp in length (for review see Clarke, 1990). Most other eukaryotes have more diffuse regional centromeres that differ considerably both in terms of their component DNA and in the ultrastructure of their associated kinetochores (for review see Pluta et al., 1995). In the fission yeast Schizosaccharomyces pombe, regional centromeres span 40–100 kb of partially repetitive DNA (Marschall and Clarke, 1995), but no well-defined kinetochore structure is visible in the electron microscope (Ding et al., 1993). Mammalian centromeres, which contain hundreds of kilobases of repetitive, heterochromatic, α-satellite DNA, are found at the major constriction of mitotic chromosomes where they specify a trilaminar kinetochore. While some insects like Drosophila also have a regional centromere/kinetochore (Goldstein, 1981), other insects and some nematodes, including Caenorhabditis elegans, organize holokinetochores with continuous trilaminar plates along the entire length of the chromosome that are visible during mitosis (Albertson and Thomson, 1982). Interestingly, in C. elegans and most other organisms with holocentric chromosomes, no kinetochore is visible during meiosis, and the microtubules appear to project directly into chromatin (Pimpinelli and Goday, 1989; Albertson and Thomson, 1993). Finally, higher plants have yet different regional centromeres that organize kinetochores in a ball and cup–type fashion (Bajer and Mole-Bajer, 1969).
Given that all centromeres must connect chromosomes to the spindle apparatus, help power chromosome movements relative to the spindle apparatus, maintain sister chromatid cohesion before anaphase, and prevent anaphase onset in the absence of proper bipolar spindle attachments (for review see Gorbsky, 1995; Pluta et al., 1995), why have so many structurally dissimilar kinds of centromeres evolved? Although the answer to this question is currently not known, a comparison of the molecular components characterizing various centromere types will clearly be an essential part of the solution. In this paper we describe the molecular evolution of one centromere/kinetochore component, originally identified as the protein product of the Drosophila melanogaster gene l(1)zw10, hereafter called DmZW10.
Mutations in the l(1)zw10 gene (zw10) disrupt chromosome segregation during mitosis and both meiotic divisions. Mitotic missegregation in mutants produces high levels of aneuploid cells and consequent lethality during late larval/pupal stages after maternal stores of the DmZW10 protein are depleted (Smith et al., 1985; Williams et al., 1992, 1996). Chromosomal segregation defects in mutants are first detected during anaphase: the separation of sister chromatids (during mitosis and meiosis II) or homologous chromosomes (during meiosis I) occurs asynchronously. As a result, some lagging chromatids or chromosomes remain in the vicinity of the metaphase plate during anaphase. The DmZW10 protein displays an intriguing cell cycle–dependent intracellular distribution, appearing to move from the centromere/kinetochore at prometaphase to kinetochore microtubules at metaphase, and back to the centromere/kinetochore at anaphase (Williams et al., 1992, 1996; Williams and Goldberg, 1994).
DmZW10 localization to the centromere/kinetochore at prometaphase and anaphase is correlated with proper functioning of the centromere. For example, the DmZW10 protein is found only at a single site on a transmissible dicentric chromosome in which one of the two centromeric regions is inactivated (Ault and Lyttle, 1988; Williams et al., 1996). This correlation is also observed in the analysis of animals carrying mutations in other genes that disrupt chromosome segregation. In Dub mutant meiotic divisions (Moore et al., 1995) and in aar mutant mitotic divisions (Gomes et al., 1993), DmZW10 does not associate with anaphase chromosomes that lag at the position of the metaphase plate. However, the protein localizes correctly to the centromere/kinetochore of chromosomes in the same cell that properly migrate to the spindle poles (Williams et al., 1994; Williams and Goldberg, 1996). It is of further interest that DmZW10 localizes, at least during meiotic divisions in the male, to derivatives of minichromosomes that are devoid of centromeric DNA sequences, but that are nonetheless transmitted with considerable efficiency through male meiosis. The behavior of these acentric minichromosomes presumably results from the acquisition of “neocentromere” activity by the subtelomeric heterochromatin they contain (Murphy and Karpen, 1995; Williams, B.C., T.D. Murphy, M.L. Goldberg, and G.H. Karpen, manuscript submitted for publication). This latter result indicates that association of DmZW10 with a chromosome reflects centromeric function rather than any particular DNA sequence at the centromere.
Recent evidence suggests that DmZW10 may act as part of, or immediately downstream of, the tension-sensing checkpoint that has recently been proposed to render anaphase onset dependent upon bipolar tension exerted across all centromeres (Li and Nicklas, 1995; Nicklas et al., 1995; Rieder et al., 1995). In zw10 mutants, sister chromatids separate prematurely in the presence of microtubule-depolymerizing drugs, representing malfunction of an anaphase onset checkpoint (Smith et al., 1985; Williams et al., 1992). Furthermore, the localization pattern of DmZW10 with respect to each chromosome is influenced by the presence or absence of tension across the centromere. While DmZW10 normally appears at metaphase of the first meiotic division to move from the centromere/kinetochores of bivalent chromosomes under bipolar tension to the attached kinetochore microtubules, the protein remains at the centromere/kinetochore of univalents in the same cell that are attached only to a single spindle pole (Williams et al., 1996). It is thus conceivable that the movements of DmZW10 between chromosomes and the spindle during the cell cycle might provide a physical basis for the measurement of tension.
We report here that DmZW10 homologues are present in widely diverse species, including C. elegans (CZW-1), Arabidopsis thaliana (AtZW10), mice (MmZW10), and humans (HZW10). We also show evidence of at least partial functional homology between ZW10 proteins in two ways. First, we demonstrate that antibodies against the HZW10 protein reveal a cell cycle–dependent staining pattern in human tissue culture cells strikingly similar to that of DmZW10 in dividing Drosophila cells. Second, some defects caused by zw10 mutations in D. melanogaster can be phenocopied in C. elegans by antisense czw-1 RNA injections. These results suggest that the functional role of ZW10 proteins in ensuring proper chromosome segregation is conserved in higher eukaryotes.
Materials and Methods
Cloning of zw10 Homologues
Drosophila pseudoobscura genomic DNA (from C. Aquadro, Cornell University, Ithaca, NY) was digested with SacI and EcoRI. After a low stringency Southern blot analysis with a D. melanogaster zw10 cDNA probe to identify the size of homologous restriction fragments, fragments of ∼5 kb were extracted from preparative agarose gels and cloned into pBluescript (Stratagene, La Jolla, CA) digested with the same enzymes. From this minilibrary, clones encoding D. pseudoobscura ZW10 were isolated by low stringency hybridization with the D. melanogaster zw10 cDNA probe; the same technique was used to isolate clones containing the gene for Drosophila grimshawi ZW10 from a genomic D. grimshawi library provided by M.P. Kambysellis (New York University). Sequences for these Drosophila genes have been deposited in GenBank/EMBL/ DDBJ under the following accession numbers: D. melanogaster (X64390), D. pseudoobscura (U54997), and D. grimshawi (U54998).
The existence of homologues of zw10 in other organisms was first inferred from the results of tblastn searches of GenBank (Gish and States, 1993), which detected a human expressed sequence tag (EST)1 (I.M.A.G.E. Consortium cDNA clone yd42e12.r1; Lennon et al., 1996), a mouse EST (I.M.A.G.E. Consortium cDNA clone me35a05.r1; Lennon et al., 1996), a C. elegans EST (yk32e2 from Yuji Kohara, National Institute of Genetics, Mishima, Japan), and an A. thaliana EST (88F18T7 from the Arabidopsis Biological Resource Center, Ohio State University, Columbus; Newman et al., 1994), all encoding peptides with a high level of similarity to the carboxyl terminus of DmZW10. Partial cDNAs containing these ESTs were obtained from the appropriate genome centers. In addition, the human EST yd42e12.r1, the C. elegans EST yk32e2, and the Arabidopsis EST 88F18T7 were used to isolate a near full-length HZW10 cDNA from a HeLa cell S3 Unizap cDNA library (Stratagene), additional CZW-1 cDNAs from a λZAP C. elegans library (Barstead and Waterson, 1989), and a near full-length AtZW10 cDNA from a λ-gt11 root library (Peterman and Goodman, 1991), respectively. cDNAs were sequenced on both strands by the ddNTP chain termination method with the Sequenase kit (United States Biochemical Corp., Cleveland, OH). The protein sequences were aligned by ClustalW using the Megalign program (Lasergene, Madison, WI). A human multiple tissue Northern blot (Clonetech, Palo Alto, CA) was probed with cDNAs for HZW10 or actin according to the manufacturer's protocol.
Antibody Production
A chimeric gene encoding a glutathione-S-transferase/HZW10 (amino acids 92–779) fusion protein was constructed in the vector pGEX-2T (Pharmacia, Uppsala, Sweden) and expressed in Escherichia coli XL1-Blue (Stratagene) after induction with 1 mM isopropylthio-β-d-galactoside (IPTG). Extracts enriched for inclusion bodies (Harlow and Lane, 1988) were further purified by SDS-PAGE. The gel was stained in 50% methanol with Coomassie blue and destained in distilled water. The 100-kD fusion protein was cut out of the gel and collected by electroelution into dialysis bags. Rabbits were injected three times with 200 μg of purified fusion protein to raise polyclonal antibodies. Serum was affinity purified against the fusion protein on a CNBr-activated Sepharose 4B column (Pharmacia).
To assess the efficacy of the antibody, a Western blot was performed on whole HeLa cell extracts that were separated by SDS-PAGE using 7.5% acrylamide and electroblotted onto a nylon membrane (Amersham Corp., Arlington Heights, IL). Alternatively, HeLa cells were fractionated into soluble and insoluble fractions by centrifugation at 15,000 g for 15 min after lysis in a Dounce homogenizer in buffer (150 mM NaCl, 50 mM Tris, pH 8.0, 1 mM DTT, and 40 μg/ml PMSF) with or without 1% Triton X-100 (Sigma Chemical Co., St. Louis, MO). Membranes were probed with affinity-purified anti-HZW10 at a dilution of 1:100 followed by donkey anti–rabbit IgG secondary antibody coupled to HRP (Amersham Corp.) at a dilution of 1:2,000, which was detected using the enhanced chemiluminescence kit (Amersham Corp.).
Tissue Culture and Immunofluorescence
HeLa cells (the gift of E. Keller, Cornell University) were grown in DME supplemented with 10% FBS, 1,000 U/ml penicillin G sodium, and 1 mg/ ml streptomycin sulfate at 37°C in 5% CO2 (all tissue culture media from Life Technologies, Grand Island, NY). In some experiments, HeLa cells were transiently transfected by the CaPO4 precipitation method at 35°C in 3% CO2 (Chen and Okayama, 1987) with a construct containing the S65T variant of the green fluorescent protein (GFP; Heim et al., 1995) fused to the amino terminus of HZW10 under the control of the cytomegalovirus promoter in the vector pWS4 (Sheay et al., 1993). Western blots of transfected cell extracts were performed as above. In other experiments, HeLa cells were synchronized at the start of S phase by two 16-h blocks with 2.5 mM thymidine separated by 9 h of normal growth (Yen et al., 1992). Cyclin B1 antibody (GNS1; Santa Cruz Biotechnology, Santa Cruz, CA) was used on Western blots at a 1:200 dilution as a control to show that the cells were synchronized.
Metaphase-arrested chromosome spreads were made by treating HeLa cells overnight in 10 μg/ml colcemid (Sigma Chemical Co.) or 1 μg/ml nocodazole (Sigma Chemical Co.), swelling in 0.8% sodium citrate for 20 min, and spreading by centrifugation briefly at 500 g (Earnshaw and Rattner, 1991). The spreads were fixed in 4% paraformaldehyde in PBS for 10 min. To visualize GFP, cells were preextracted in 0.5% Triton X-100 as described (Echeverri et al., 1996) before fixation as above. Unsynchronized HeLa cells were grown on coverslips, further attached by centrifugation at 500 g for 1 min, fixed in 4% paraformaldehyde in PBS for 10 min, and permeabilized in PBS + 0.2% Triton X-100 for 5 min. Primary antibodies (affinity-purified anti-HZW10 at a dilution of 1:250 [see Figs. 4, 6, and 7]; human CREST serum NR [from W. Earnshaw, University of Edinburgh, UK] at 1:1,000 [Figs. 4–6]; mAb M154 against CENP-E [the gift of T. Yen, Fox Chase Cancer Center, Philadelphia, PA] at 1:500 [see Fig. 4]; or mAb 4A-1 against tubulin [from M. Fuller, Stanford University, Palo Alto, CA] at 1:20 [see Fig. 7]) were added in PBS to fixed cells for 1 h. After washing 15 min in PBS, the secondary antibodies (TRITC-conjugated goat anti–rabbit IgG [see Figs. 4, 6, and 7], FITC-conjugated goat anti–human IgG [see Figs. 4, 6, and 7], FITC-conjugated goat anti–mouse IgG [see Figs. 4 and 7], or TRITC-conjugated goat anti–human IgG [see Fig. 5]) were added at a dilution of 1:100 in PBS for 1 h and washed for 30 min in PBS (all secondary antibodies were from Jackson ImmunoResearch Laboratories, West Grove, PA). DNA was stained with 0.05 μg/ml Hoechst 33258 (Sigma Chemical Co.) for 5 min. Coverslips were mounted in 2% N-propyl gallate and 80% glycerol, and then examined either on a Zeiss Axioskop (Zeiss, Oberkochen, Germany) or with a Bio-Rad MRC600 confocal system (Bio-Rad Laboratories, Hercules, CA). GFP and anti-HZW10 signals were pseudocolored red, while NR and CENP-E signals were pseudocolored green using Adobe Photoshop (Adobe Systems, Inc., Mountain View, CA).
Figure 4.
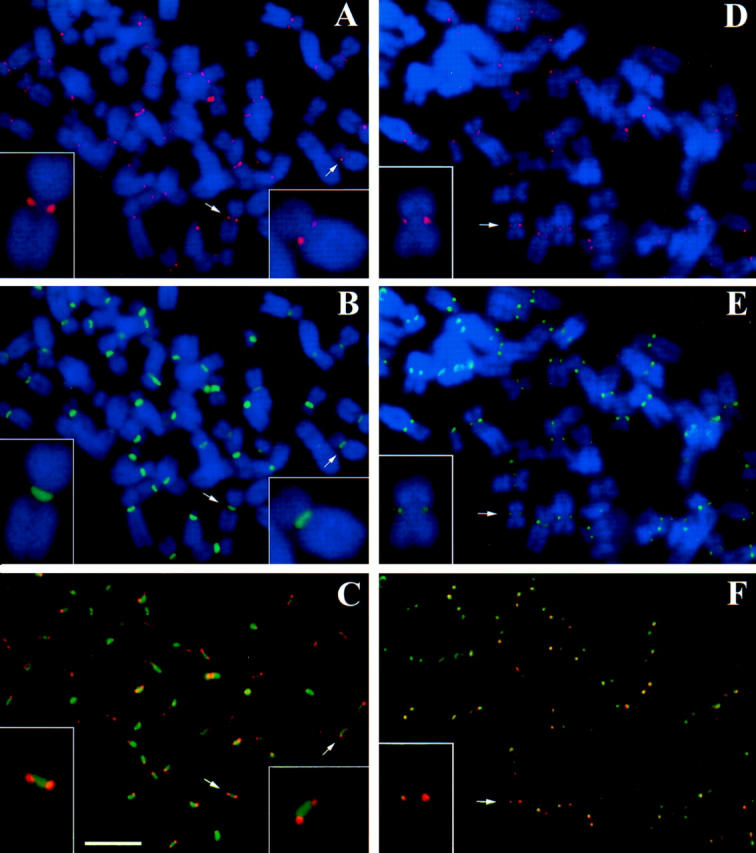
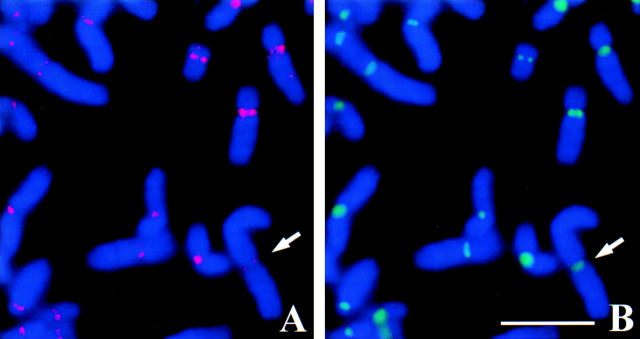
HZW10 immunolocalization in metaphase-arrested HeLa cell chromosome spreads. (A–C) Chromosome spread of a HeLa cell stained with HZW10 (red) and centromeres stained by NR autoimmune CREST serum (green). (D–F) Chromosome spread stained with HZW10 (red), and kinetochores stained by CENP-E (green). DNA is stained in blue in A, B, D, and E. (Arrows) Chromosomes enlarged in the insets. Bar, 5 μm.
Figure 6.
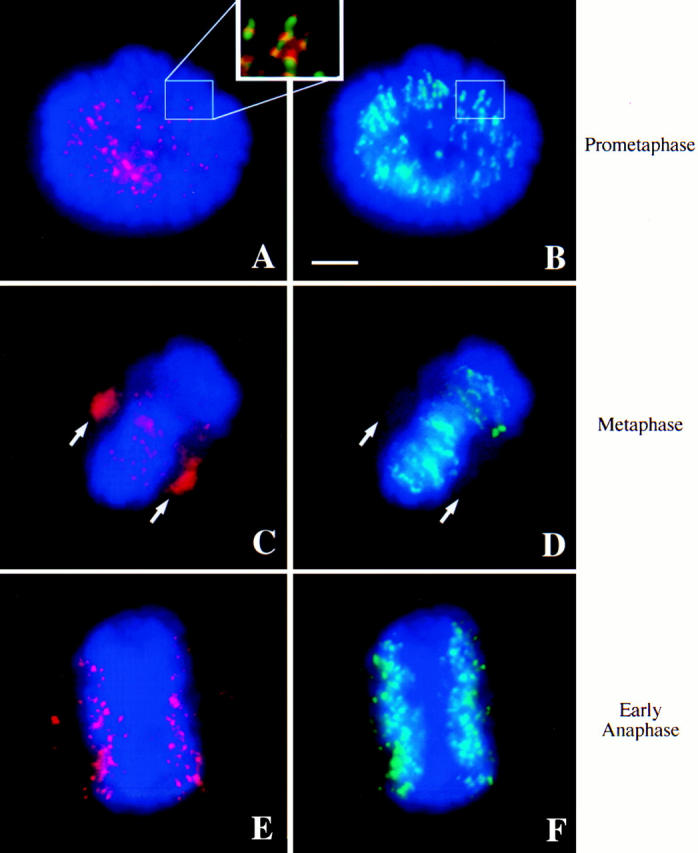
HZW10 immunolocalization in HeLa cells. DNA is colored blue in all panels; in A, C, and E, HZW10 is colored red; in B, D, and F, centromeres as recognized by NR autoimmune serum are green. (A and B) A prometaphase cell. (Inset) The colocalization of HZW10 and NR. (C and D) A metaphase cell. Arrows point to the position of the spindle poles. (E and F) An early anaphase cell. Bar, 5 μm.
Figure 7.
HZW10 colocalization with the spindle in metaphase HeLa cells. Anti-HZW10 staining (A) mostly overlaps with the spindle as revealed with antibodies against tubulin (B). Punctate anti-HZW10 staining near the center of the spindle either represents residual kinetochore localization at metaphase or discrete sites of staining along kinetochore microtubules. Bar, 5 μm.
Figure 5.
Chromosome spread of a cell transiently transfected with a GFP/HZW10 fusion protein construct. GFP fluorescence (A, pseudocolored red) and centromeres stained by NR (B, green). DNA is colored blue. Arrows point to the same chromosome for orientation; note the partial overlap of GFP and NR signals. Bar, 5 μm.
C. elegans Antisense mRNA Injections
The C. elegans ZW10 EST yk32e2 was cloned into the EcoRI and KpnI sites of pBluescript SK (Stratagene), which placed the antisense strand under control of the T7 promoter. The plasmid was linearized at the 5′ end of the cDNA by EcoRI digestion, and antisense RNA was made using the mCAP RNA capping kit (Stratagene) with T7 RNA polymerase. The RNA was injected into N2 (wild-type) hermaphrodite gonads at 1 mg/ml in diethyl pyrocarbonate–treated water as previously described (Guo and Kemphues, 1995). Water was injected as a negative control, and par-2 sense RNA (Boyd et al., 1996) was injected at 1 mg/ml as a positive control (gift of L. Boyd, Cornell University). Individual injected worms were plated at 20°C and transferred to new plates each day. Embryonic lethality (failure to hatch) and brood size (numbers of embryos produced) were monitored for 24–48 h after eggs were laid. Embryos from some injected worms were fixed and stained with 4′6-diamidino-2-phenylindole as previously described (Etemad-Moghadam et al., 1995) to observe mitotic chromatin.
Results
Cloning ZW10 Homologues
DmZW10 is a “pioneer” protein whose sequence does not suggest any obvious functional domains. In an effort to better understand the function of DmZW10, we searched for evolutionarily conserved domains in related genes from other species. We were able to clone by low stringency hybridization zw10 homologues from two diverse Drosophila species: D. pseudoobscura, which diverged from D. melanogaster ∼25 million years ago, and D. grimshawi , a Hawaiian species that diverged from D. melanogaster ∼32 million years ago (Russo et al., 1995). The sequences of the zw10 genes in these three Drosophila species (data not shown; see Materials and Methods for GenBank accession numbers) are quite similar, sharing >65% amino acid identity. Since conservation is fairly uniform throughout the length of these proteins, with no regions showing markedly higher or lower degrees of similarity, we next wanted to expand the search for important domains by examining homologous genes in more distantly related species.
BLAST searches of GenBank found ESTs predicting polypeptides with a high level of similarity to the carboxyl terminus of DmZW10 in C. elegans (CZW-1, following C. elegans convention), humans (HZW10), mice (MmZW10), and A. thaliana (AtZW10). We then isolated corresponding clones from C. elegans, human, and Arabidopsis cDNA libraries and determined their sequence, conceptual translations of which are shown in Fig. 1. Although our longest CZW-1 cDNA was not full length, the C. elegans genome project (Wilson et al., 1994) subsequently sequenced the entire czw-1 gene, located in the vicinity of unc-24 on linkage group IV. Their data verified our cDNA sequence, yielding the open reading frame of CZW-1 presented in Fig. 1. The 2,884-bp HZW10 cDNA we isolated from a HeLa cell cDNA library is near full length because it hybridizes to a single poly A+ RNA species of ∼2.9 kb in all of eight different human cancer cell lines on a Northern blot (three of which are shown in Fig. 2 A). Human ESTs related to zw10 have been found in cDNA libraries from a variety of human cell types (infant brain, fetal liver and spleen, pancreatic carcinoma, and uteral), suggesting that the human gene is expressed widely during development. We sequenced a 1,240-bp cDNA containing the mouse EST me35a05.r1 in its entirety; this represents the carboxyl 256 residues of the mouse protein that are 92% identical to the carboxyl HZW10 sequence. The AtZW10 cDNA we have obtained is probably full length, based on its similarity to other zw10 sequences and the presence of an in-frame stop codon 45 bp upstream of the predicted initiation codon.
Figure 1.

Multiple sequence alignment of ZW10 homologues. Homologues to DmZW10 were found in humans (HZW10), C. elegans (CZW-1), and A. thaliana (AtZW10). These four proteins are shown aligned throughout their entire coding sequences. Identical amino acids between any two of the four sequences are shaded. Dashed lines represent gaps required for maximal sequence alignment. Homologues from D. pseudoobscura, D. grimshawi, and mice are not shown. Sequences for these zw10 genes have been deposited in GenBank/ EMBL/DDBJ under the following accession numbers: D. melanogaster (X64390), D. pseudoobscura (U54997), D. grimshawi (U54998), C. elegans (CELF20D12), A. thaliana (U80984), M. musculus (AF003951), and H. sapiens (U54996).
Figure 2.
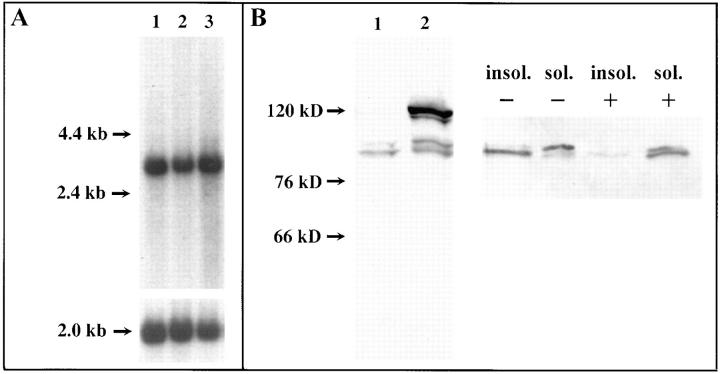
The human zw10 transcript and HZW10 gene product. (A) Human zw10 cDNA hybridizes to a single 2.9-kb transcript on a Northern blot of RNAs from three cancer cell lines. Lanes 1–3 contain 2 μg of poly A+ RNA from promyelocytic leukemia HL-60, HeLa S3, and chronic myelogenous leukemia K-562 cell lines, respectively. RNA size marker bands are shown at the left. The lower part of the figure shows the same blot probed with actin as a loading control. (B) Western blots of HeLa cell protein extracts were analyzed with HZW10 affinity-purified antibody. Left blot: (lane 1) whole extracts from untransfected cells; (lane 2) whole extracts from cells transfected with a GFP/HZW10 construct. Right blot: HZW10 protein in soluble and insoluble fractions of HeLa cell extracts made in the presence (+) or absence (−) of 1% Triton X-100. The positions of protein standards (E. coli β-galactosidase, 120 kD; rabbit fructose-6-phosphate kinase, 76 kD; and chicken pyruvate kinase, 66 kD) are shown at the left.
Pairwise comparisons of the conceptual translations of these cDNAs reveal that the amino acid sequences of the ZW10 proteins from D. melanogaster, C. elegans, humans, and Arabidopsis are on average between 17–26% identical, and 27–35% similar, over the entire protein. The predicted sequences of all the ZW10 homologues can be aligned throughout their length (Fig. 1), but they are particularly well conserved in the carboxyl third of the protein. For example, 41 of the 98 amino acids at the carboxyl termini of the D. melanogaster and human proteins are identical. It is of interest that no other proteins or domains with significant homology to any of these ZW10 proteins were identified in database searches.
Preliminary Characterization of HZW10
To allow detection of HZW10 protein, we raised polyclonal antibodies against a glutathione-S-transferase/HZW10 fusion protein. The affinity-purified serum recognizes a doublet of approximately the expected size (M r ∼90 kD) on a Western blot of HeLa cell extracts (Fig. 2 B, lane 1), which is not recognized by the preimmune serum (data not shown). To determine whether this antibody in fact recognizes HZW10 epitopes, cells were transiently transfected with a construct encoding the 30-kD GFP fused in frame to the amino terminus of HZW10. The anti-HZW10 antibody recognizes a second doublet at the predicted size of M r ∼120 kD in these transfected cells (Fig. 2 B, lane 2). We therefore conclude that the purified antiserum exclusively recognizes HZW10 and suggest that this protein is subject to modification in HeLa cells, causing it to behave as a doublet upon electrophoresis in SDS-PAGE gels. Interestingly, the two bands in the doublet can be separated from each other on the basis of their solubility. The upper band of the doublet is found in soluble fractions of HeLa cell extracts made in the absence of nonionic detergent, whereas the lower band only becomes soluble when extracts are made in the presence of detergent (Fig. 2 B, right). Even though we do not know the biochemical nature of the modification responsible for the two bands, it is unlikely to be phosphorylation, as treatment with λ protein phosphatase did not affect the behavior of the doublet on Western blots while it did dephosphorylate a control phosphoprotein (data not shown).
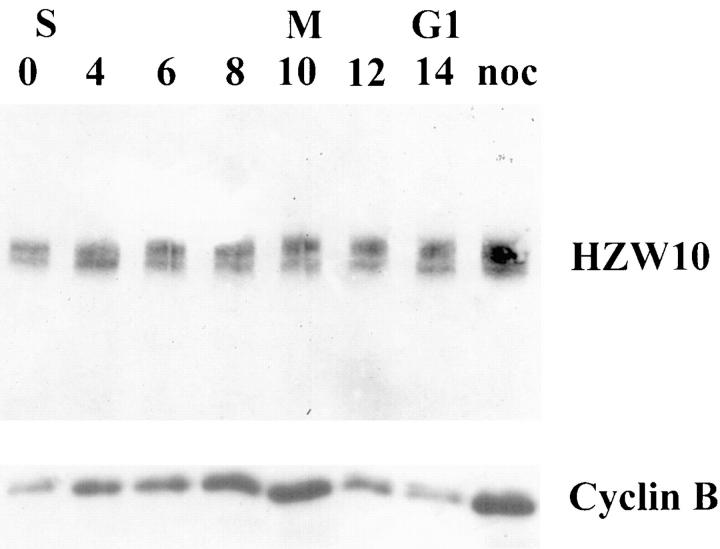
To determine if levels or modifications of HZW10 are controlled in a cell cycle–dependent manner, we synchronized HeLa cells at the G1/S transition by a double thymidine block and harvested samples at several time points after release. Fig. 3 shows that HZW10 protein bands do not change significantly in relative intensity during the cell cycle, whereas cyclin B levels in the same samples cycle as expected (Murray and Kirschner, 1989).
Figure 3.
Lack of variation in HZW10 protein levels during the cell cycle. Total HeLa cell protein extracts were prepared from the indicated cell cultures; equal aliquots of each sample were transferred to two Western blots that were analyzed separately with HZW10 antibody (top) or with cyclin B antibody (bottom). HeLa cells synchronized at G1/S phase were released from a thymidine block at time 0, and fractions were collected 4, 6, 8, 10, 12, and 14 h later. The lane at the right is from cells treated with nocodazole from time 0–14 h. Approximate stage of the cell cycle (S, M, and G1) as judged by the mitotic index is shown above the blot. Subtle changes in the relative intensities of the HZW10 bands in certain samples are not reproducible.
Immunolocalization of HZW10
If HZW10 is a functional homologue of DmZW10, the two proteins should have analogous subcellular distributions. We thus used the affinity-purified anti-HZW10 antibodies used in Fig. 2 B as a probe for the immunolocalization of the protein; this antibody preparation recognized structures in HeLa cells (see Figs. 4, 6 and 7) that were not recognized by the preimmune serum (data not shown). Anti-HZW10 antibodies localize to the vicinity of the centromere/kinetochore in chromosome spreads from HeLa cells arrested in a metaphase-like state by the microtubule-depolymerizing drug colcemid. To visualize the centromere in the same chromosomes, we employed serum from an autoimmune patient (NR) with scleroderma spectrum disease (CREST; Moroi et al., 1980). The NR serum (gift from W. Earnshaw) recognizes three centromere proteins: CENP-A, CENP-B, and CENP-C. HZW10 recognizes two punctate dots that partially overlap with, and extend ∼0.2 μm to the outside of, NR-reactive sites (Fig. 4, A–C). This is the predicted pattern of localization for a component of the kinetochore or the fibrous corona immediately distal to the kinetochore. To further refine the localization pattern of HZW10, other chromosome spreads were simultaneously stained for HZW10 and for CENP-E, a component of the kinetochore (Liao et al., 1994). HZW10 and CENP-E completely colocalize as two punctate dots, suggesting that HZW10 is a component of the kinetochore or the fibrous corona in arrested HeLa cell chromosomes (Fig. 4, D–F).
To confirm this localization of HZW10, we examined chromosomes from HeLa cells that were transiently transfected with a GFP/HZW10 fusion protein construct and subsequently subjected to metaphase arrest with colcemid. GFP fluorescence is concentrated at the kinetochore in the same pattern revealed by anti-HZW10 immunofluorescence (Fig. 5). It should be noted that GFP/HZW10 signals in the vicinity of the centromere can be observed only if the cells are preextracted with nonionic detergent before fixation (see Echeverri et al., 1996 for a description of this treatment), to diminish very high levels of signal in the cytoplasm, which we assume are caused by protein overexpression from the transfected construct.
Since other proteins have been found to associate artifactually with the centromere in metaphase-arrested cells (Compton et al., 1991), we examined the distribution of HZW10 in untreated, asynchronous HeLa cells. Cells were fixed without prior detergent extraction and were stained with anti-HZW10 (Fig. 6). The protein displays a dynamic pattern of localization during the cell cycle similar to that previously noted for DmZW10 in Drosophila neuroblasts, embryos, and spermatocytes (Williams et al., 1992, 1996; Williams and Goldberg, 1994). HZW10 is first seen on the centromeres as punctate dots in what we presume to be G2 cells, as only a small proportion of interphase cells reveals this staining (not shown). The other interphase cells show only diffuse cytoplasmic staining with anti-HZW10. In prometaphase HeLa cells, anti-HZW10 antibodies reveal a strong, punctate staining pattern expected for a kinetochore binding protein (Fig. 6, A and B). These signals are immediately adjacent to (and partially overlap with) sites of NR staining, again suggesting that HZW10 is a component of the kinetochore (Fig. 6, A and B, inset). At metaphase, some HZW10 remains near the centromeres, but most relocalizes to the spindle (Figs. 6, C and D, and 7) in a pattern of variably sized speckles that appears to follow the spindle fibers. Although it is clear that much of the HZW10 in metaphase cells is associated with the spindle, because of the poor cytology in HeLa cells we cannot determine if HZW10 localizes specifically to the kinetochore microtubules, as appears to be the case for DmZW10 in Drosophila. In fact, some proportion of HZW10 seems to be concentrated closer to the spindle poles than we would have expected from the Drosophila precedent. In very early anaphase, discrete sites of HZW10 localization are found in the vicinity of the centromeres (Fig. 6, E and F), although at weaker levels than those seen in prometaphase cells. As anaphase progresses, HZW10 is no longer localized near chromosomes or the mitotic apparatus, but instead is present diffusely in the cytoplasm (data not shown). This analysis was verified by the observation that asynchronous HeLa cells transfected with GFP/HZW10 showed a similar cell cycle–dependent distribution of GFP fluorescence (data not shown), although signal quality was poor because of the necessity for detergent preextraction (see above).
Effects of Antisense mRNA Injections in C. elegans
To test whether CZW-1 and DmZW10 have similar functions, we wanted to determine whether aspects of the Drosophila zw10 mutant phenotype could be phenocopied by CZW-1 antisense RNA injections in nematodes. Injections of either sense or antisense RNA into gonads of wild-type worms have been shown to give gene-specific loss-of-function phenotypes in embryos, presumably by depleting the maternally provided gene product (Guo and Kemphues, 1995). In these experiments, water was injected as a negative control and sense par-2 RNA, which is known to cause a strong embryonic lethality (Boyd et al., 1996), was injected as a positive control. Three major phenotypes result from injection of hermaphrodites with CZW-1 antisense RNA. First, compared with worms injected with water or par-2 sense RNA, these animals show a significant decrease in brood size: i.e., they produce far fewer embryos (Table I). Second, the injection of CZW-1 antisense RNA causes high levels of embryonic lethality among those embryos that are produced. Finally, in one-third (27/81) of the anaphase figures observed in embryos derived from worms injected with CZW-1 antisense RNA, chromatin bridges were observed between the two complements of separating sister chromatids at anaphase (Fig. 8), as compared with 3% (3/96) of anaphases in water-injected controls. It should be noted that chromatin bridges at anaphase are also observed in Drosophila embryos lacking maternal ZW10 protein (Williams and Goldberg, 1994).
Table I.
Effects of Injection of Antisense RNA in Caenorhabditis elegans
| RNA injected | Number of worms injected | Average percentage of lethality ± SE | Average brood size ± SE | Abnormal anaphase figures/number observed | ||||
|---|---|---|---|---|---|---|---|---|
| Antisense CZW-1 | 25 | 19 ± 2.2 | 55.4 ± 4.2 | 27/81 (33%) | ||||
| Sense par-2 | 8 | 47 ± 91 | 146.1 ± 16.3 | ND | ||||
| Water | 11 | 0.4 ± 0.1 | 173.5 ± 6.8 | 3/96 (3%) |
Figure 8.
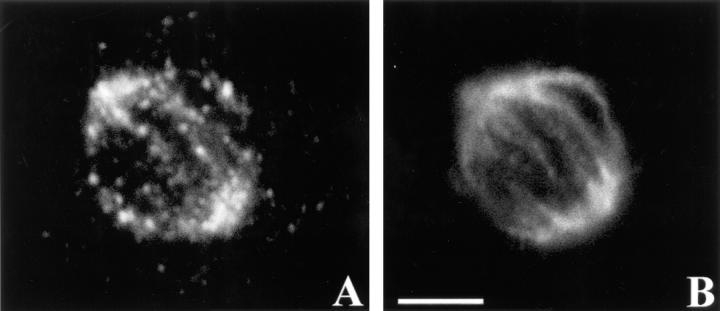
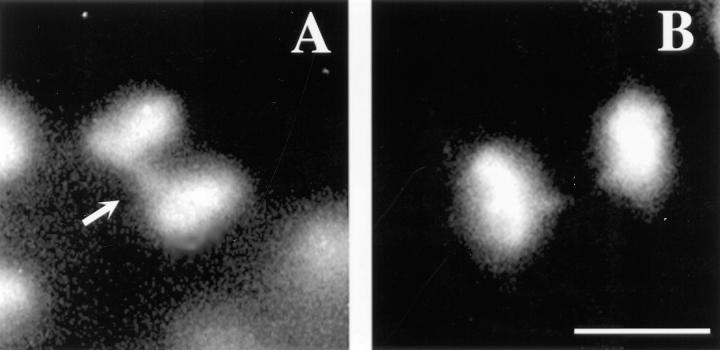
Chromatin bridges in anaphase figures of embryos from worms injected with CZW-1 antisense RNA. (A) Defective embryonic anaphase figure from a worm injected with CZW-1 antisense RNA. The arrow points to an abnormal chromatin bridge. (B) Normal embryonic anaphase figure from a worm injected with water. Chromatin is stained with 4′,6-diamidino-2-phenylindole. Bar, 5 μm.
Discussion
Structural and Functional Conservation of zw10-related Genes
We have identified homologues of zw10 by sequence similarity in a diverse range of species that last shared a common ancestor perhaps one billion years ago (Doolittle et al., 1996). It is therefore likely that zw10-related genes exist in all plants and animals. It is noteworthy that zw10-related genes are found in organisms with diverse kinetochore structures. Human and Drosophila chromosomes have regional centromeres with a trilaminar plate kinetochore at the pairing point of sister chromatids (for review see Pluta et al., 1995), higher plants have a ball and cup– structured kinetochore at a regional centromere (Bajer and Mole-Bajer, 1969), and C. elegans chromosomes have holokinetochores that extend along their entire length (Albertson and Thomson, 1982). Of course, sequence similarity only suggests the possibility that genes have homologous functions in different organisms. In this paper we describe two lines of evidence that ZW10 proteins in various species indeed play similar intracellular roles.
First, functional homologues should be found in analogous positions in the cell throughout the cell cycle, which we have shown to be true for ZW10 in Drosophila and humans. In cells arrested with microtubule poisons, both DmZW10 (Williams and Goldberg, 1994) and HZW10 (Fig. 4) are found in the vicinity of the centromere/kinetochore. This position is likely to correspond to the kinetochore or the fibrous corona, as its position slightly overlaps but is mostly immediately outside of CREST staining of the centromere (Figs. 4, 5, and 6, A and B). Moreover, HZW10 colocalizes with the kinetochore protein CENP-E, which is found just outside sites of CENP-B staining (Shelby et al., 1996; Fig. 4). In cycling HeLa cells, HZW10 first associates with the kinetochore in late G2, the time at which other proteins are recruited to the centromere/kinetochore, including CENP-F (Liao et al., 1995), CENP-E (Yen et al., 1991), and cytoplasmic dynein (Pfarr et al., 1990; Steuer et al., 1990). Just like DmZW10, HZW10 remains at the kinetochore throughout prometaphase until metaphase when, again like DmZW10, HZW10 is primarily found on the spindle, although some may remain at the kinetochores (Figs. 6, C and D, and 7). Because of the relatively small size of the spindle in HeLa cells, we are unable to resolve whether HZW10 is associated with the spindle as a whole, with the spindle poles, or with a spe-cific subset of spindle microtubules (presumably the kineto-chore microtubules) as is the case for DmZW10 (Williams et al., 1992, 1996). Both DmZW10 and HZW10 primarily stain kinetochores in early anaphase (Fig. 6, E and F), but HZW10 is lost from the kinetochores and becomes cytoplasmic as anaphase progresses (not shown). This differs from the situation in Drosophila, where DmZW10 remains at the kinetochore throughout anaphase. However, if as we have proposed, ZW10 plays its essential role at anaphase onset (Williams and Goldberg, 1994; Williams et al., 1996), this difference in localization in late anaphase may not be significant. In any event, the overall similarities between the patterns of ZW10 localization in Drosophila and human cells are striking.
A second argument for the functional conservation of ZW10 proteins is our demonstration that injection of CZW-1 antisense RNA into C. elegans hermaphrodites disrupts cell division in a manner reminiscent of the effects of zw10 mutations in Drosophila. The decreased yield of embryos (brood size) generated by injected animals suggests that depletion of CZW-1 interferes with mitotic or meiotic germline divisions. Furthermore, the anaphase chromatin bridges in those embryos that are produced are analogous to the lagging chromatids seen in Drosophila cells lacking DmZW10 protein. C. elegans chromosomes are holokinetic, with microtubule attachments along the whole length of the chromosome (Albertson and Thomson, 1982). A holokinetic chromatid improperly attached to both spindle poles would be effectively dicentric, and thus would produce a chromatin bridge in anaphase. By causing a failure in the anaphase checkpoint, the CZW-1 antisense RNA may trigger anaphase before proper attachments of holokinetochores to single spindle poles can be established. Given the invariant cell lineage pattern characterizing C. elegans development, failure to resolve the resultant anaphase bridges in even a few cells would cause the embryonic lethality we have observed after injection of CZW-1 antisense RNA (Sulston et al., 1983).
HZW10 Protein Modifications
Figs. 2 B and 3 show that HZW10 runs as two electrophoretic bands. These bands do not represent alternative genes or alternative splicing patterns of a single primary transcript because transfection of cells with an epitope-tagged cDNA construct produces two novel bands, each of a molecular weight predicted from the addition of the epitope tag to the two original bands. In addition, HZW10 appears to be encoded by only a single resolvable species of poly A+ RNA (Fig. 2 A). It is thus likely that the HZW10 polypeptide is subject to posttranslational modification. We do not currently know the nature of this presumed modification, although results with phosphatase treatment suggest that it is not phosphorylation (data not shown). Fig. 3 indicates that the modifications are not cell cycle regulated; indeed, HZW10 protein itself is not obviously degraded at the conclusion of mitosis as is cyclin B and other centromere/kinetochore components such as CENP-E (Yen et al., 1992) and CENP-F (Liao et al., 1995). These findings suggest that the redistributions of HZW10 during the cell cycle are not due to changes in the protein's modification, but are instead mediated by other unknown components with which HZW10 interacts. The modifications we have observed may nonetheless have important functional significance as they appear at a minimum to affect the protein's solubility properties.
Evolution of Centromere/Kinetochore Proteins
All centromeres must perform the similar functions of connecting chromosomes to the spindle apparatus, helping to power chromosome movements relative to the spindle apparatus, maintaining sister chromatid cohesion before anaphase, and preventing anaphase onset in the absence of proper bipolar spindle attachments (for review see Gorbsky, 1995; Pluta et al., 1995). Thus, it is expected that many molecules present at the centromere/kinetochore have been conserved throughout the evolution of eukaryotic organisms. The evidence for such conservation is still somewhat fragmentary and anecdotal, in large part because of a lack of information on the molecular makeup of centromeres in organisms other than yeast and humans. S. cerevisiae chromosomes have a well-defined point centromere to which a number of proteins, including the CBF3 complex, bind in vitro (Clarke, 1990; Pluta et al., 1995). Thanks mainly to serum from patients with the autoimmune disease CREST, many molecular components have been localized to the human centromere/kinetochore (for review see Pluta et al., 1995). However, far less is known of the molecular constitution of other regional centromeres/kinetochores. For example, in higher plants, only three antibodies, CREST sera EK and S174 (Mole-Bajer et al., 1990; Houben et al., 1995) and mAb 6C6 (Schmit et al., 1994), have been shown to recognize centromere antigens; to our knowledge, no antigens have been shown yet to recognize a holokinetochore.
To date, only a few centromere/kinetochore components have been shown to be well conserved between yeast and higher eukaryotes, although many more are likely to be identified as genome projects advance. The SKP1 gene product of S. cerevisiae, which is required for ubiquitin-mediated proteolysis at G1/S and G2/M and is a component of the centromere binding CBF3 complex, has homologues in C. elegans, A. thaliana, and humans (Bai et al., 1996; Connelly and Hieter, 1996). It has not yet been demonstrated, however, whether these plant and animal homologues are also centromere components, or are instead members of another ubiquinating complex. The MAD2 checkpoint gene has also been shown to be conserved between S. cerevisiae, C. elegans, Xenopus, and humans (Chen et al., 1996; Li and Benezra, 1996); the human and Xenopus MAD2 proteins associate with the centromere/kinetochore during prometaphase, although the localization of the S. cerevisiae protein has not been determined. Two other yeast proteins, which are likely on the basis of genetic criteria to function at the centromere, have domains with limited homology to known human centromere components. These are CSE4, which shares a histone H3-like domain with CENP-A (Stoler et al., 1995), and MIF2, which contains motifs also found in CENP-C (Meluh and Koshland, 1995).
Even if many components and aspects of centromere function will be found to be widely conserved, evolutionary differences in centromere organization suggest that significant organism-specific differences in centromere/kinetochore proteins must also exist. In this light, it is important to note that although ZW10 proteins are highly similar in a variety of animals and plants, particularly in their COOH-terminal regions, we failed to detect any zw10- homologous regions in the complete S. cerevisiae genomic sequence (Dujon, 1996), even when searching with short regions of all available zw10 genes. It of course remains possible that a yeast protein may contain small, undetectable motifs that provide ZW10-like function; however, we must nonetheless entertain the possibility that ZW10 is not a constituent of the point centromeres of S. cerevisiae. If so, S. cerevisiae centromeres differ from those of plants and animals not only in the amount of DNA they contain, the number of microtubules they bind, and in ultrastructure (Pluta et al., 1995), but also in the identity of at least one of their associated proteins that we have shown to be critical for proper chromosome segregation in Drosophila (Williams et al., 1992, 1996) and in C. elegans (this paper). It will be of interest to determine whether ZW10-like proteins are found in other unicellular eukaryotes that have more extensive centromeric regions than S. cerevisiae, such as S. pombe.
Despite the importance of ZW10 proteins in Drosophila and nematodes, and presumably in all plants and animals, this protein may be dispensable in yeast. Although we do not yet fully understand the precise molecular function of ZW10, it seems likely that other proteins in higher organisms are likely to be involved in the same process. For example, mutations in the Drosophila gene rough deal (rod) cause phenotypes indistinguishable from those associated with mutant alleles of zw10 (Karess and Glover, 1989); we have established that ZW10 fails to localize to the centromere or the spindle in rod mutants (Williams and Goldberg, 1994). We find it interesting that, like ZW10, the ROD protein is conserved between Drosophila, C. elegans, and humans, and no yeast protein that is homologous to ROD has been detected in computer database searches (Karess, R., personal communication). In addition, several other human antigens have been previously described whose intracellular distributions shift between the centromere and spindle during the cell cycle in a fashion similar to that of ZW10 depicted in Fig. 6. These include antigens recognized by mAbs raised by Compton et al. (1991), CENP-E (Yen et al., 1991), and components of dynein (Pfarr et al., 1990; Steuer et al., 1990) and dynactin (Echeverri et al., 1996). That ZW10 may be only part of a multicomponent system needed for accurate chromosome segregation during cell division is further suggested by our recent finding that at least some Drosophila ZW10 protein is found as part of a large (19S) soluble complex in embryonic extracts (Starr, D.A., T. Hays, and M.L. Goldberg, unpublished data). Our future efforts will be devoted to a further characterization of this complex.
Acknowledgments
We thank E. Keller and D. Dean for use of tissue culture facilities; A. Mackay, W. Earnshaw, G. Chan, and T. Yen for advice on tissue culture techniques, reagents, and helpful comments; L. Boyd for par-2 RNA; C. Aquadro, F. Piano, and M. Kambysellis for Drosophila DNA and advice on sequence comparisons; and R. Karess for communicating unpublished results. We are indebted to K. Kemphues, in whose laboratory some of this work was performed.
Abbreviations used in this paper
- EST
expressed sequence tag
- GFP
green fluorescent protein
Footnotes
Please address all correspondence to Michael L. Goldberg, Section of Genetics and Development, Cornell University, 425 Biotechnology Building, Ithaca, NY 14853-2703. Tel.: (607) 254-4802. Fax: (607) 255-6249. E-mail: MLG11@cornell.edu
This research was supported by grant GM48430 from the National Institutes of Health to M.L. Goldberg and NIH training grant GM07617 to the Field of Genetics and Development at Cornell University.
Received for publication 29 January 1997 and in revised form 3 July 1997.
References
- Albertson DG, Thomson JN. The kinetochores of Caenorhabditis elegans. . Chromosoma (Berl) 1982;86:409–428. doi: 10.1007/BF00292267. [DOI] [PubMed] [Google Scholar]
- Albertson DG, Thomson JN. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans. . Chromosome Res. 1993;1:15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- Ault JG, Lyttle TW. A transmissible dicentric chromosome in Drosophila melanogaster. . Chromosoma (Berl) 1988;97:71–79. [Google Scholar]
- Bai C, Sen P, Hofmann K, Ma L, Gobel M, Harper JW, Elledge SJ. SKP1connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- Bajer A, Mole-Bajer J. Formation of spindle fibers, kinetochore orientation, and behavior of the nuclear envelope during mitosis in endosperm. Chromosoma (Berl) 1969;27:448–484. [Google Scholar]
- Barstead RJ, Waterson RH. The basal component of the nematode dense-body is vinculin. J Biol Chem. 1989;264:10177–10185. [PubMed] [Google Scholar]
- Boyd L, Guo S, Levitan D, Stinchcomb DT, Kemphues KJ. PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C. elegansembryos. Development (Camb) 1996;122:3075–3084. doi: 10.1242/dev.122.10.3075. [DOI] [PubMed] [Google Scholar]
- Chen C, Okayama H. High efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science (Wash DC) 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Clarke L. Centromeres of budding and fission yeasts. Trends Genet. 1990;6:150–154. doi: 10.1016/0168-9525(90)90149-z. [DOI] [PubMed] [Google Scholar]
- Compton DA, Yen TJ, Cleveland DW. Identification of novel centromere/kinetochore-associated proteins using monoclonal antibodies generated against human mitotic chromosome scaffolds. J Cell Biol. 1991;112:1083–1097. doi: 10.1083/jcb.112.6.1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly C, Hieter P. Budding yeast SKP1encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell. 1996;86:275–285. doi: 10.1016/S0092-8674(00)80099-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, McDonald KL, McIntosh JR. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast Schizosaccharomyces pombe. . J Cell Biol. 1993;120:141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doolittle RF, Feng D, Tsang S, Cho G, Little E. Determining divergence times of the major kingdoms of living organisms with a protein clock. Science (Wash DC) 1996;271:470–477. doi: 10.1126/science.271.5248.470. [DOI] [PubMed] [Google Scholar]
- Dujon R. The yeast genome project: what did we learn? . Trends Genet. 1996;12:263–270. doi: 10.1016/0168-9525(96)10027-5. [DOI] [PubMed] [Google Scholar]
- Earnshaw, W.C., and J.B. Rattner. 1991. The use of autoantibodies in the study of nuclear and chromosomal organization. In Functional Organization of the Nucleus. B.A. Hamkalo and S.C.R. Elgin, editors. Academic Press, San Diego, CA. 136–176.
- Echeverri CJ, Paschal BM, Vaughan KT, Vallee RB. Molecular characterization of the 50-kD subunit of dynactin reveals function for the complex in chromosome alignment and spindle organization during mitosis. J Cell Biol. 1996;132:617–633. doi: 10.1083/jcb.132.4.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemad-Moghadam B, Guo S, Kemphues KJ. Asymmetrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C. elegansembryos. Cell. 1995;83:743–752. doi: 10.1016/0092-8674(95)90187-6. [DOI] [PubMed] [Google Scholar]
- Gish W, States DJ. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- Goldstein LSB. Kinetochore structure and its role in chromosome orientation during the first meiotic division in male Drosophila melanogaster. . Cell. 1981;25:591–602. doi: 10.1016/0092-8674(81)90167-7. [DOI] [PubMed] [Google Scholar]
- Gomes R, Karess RE, Ohkura H, Glover DM, Sunkel CE. Abnormal anaphase resolution (aar): a locus required for progression through in Drosophila. . J Cell Sci. 1993;104:583–593. doi: 10.1242/jcs.104.2.583. [DOI] [PubMed] [Google Scholar]
- Gorbsky GJ. Kinetochores, microtubules and the metaphase checkpoint. Trends Cell Biol. 1995;5:143–148. doi: 10.1016/s0962-8924(00)88968-0. [DOI] [PubMed] [Google Scholar]
- Guo S, Kemphues KJ. par-1, a gene required for establishing polarity in C. elegansembryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell. 1995;81:611–620. doi: 10.1016/0092-8674(95)90082-9. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and D. Lane. 1988. Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY. 726 pp.
- Heim R, Cubitt AB, Tsien RY. Improved green fluorescence. Nature (Lond) 1995;373:663–664. doi: 10.1038/373663b0. [DOI] [PubMed] [Google Scholar]
- Houben A, Guttenbach M, Kreb W, Pich U, Schubert I, Schmid M. Immunostaining and interphase arrangement of field bean kinetochores. Chromosome Res. 1995;3:27–31. doi: 10.1007/BF00711158. [DOI] [PubMed] [Google Scholar]
- Karess RE, Glover DM. rough deal: a gene required for proper mitotic segregation in Drosophila. . J Cell Biol. 1989;109:2951–2961. doi: 10.1083/jcb.109.6.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lennon, G., C. Aufrray, M. Polymeropoulos, and M.B. Soares. 1996. The I.M.A.G.E. consortium: an integrated molecular analysis of genomes and their expression. Genomics. 151–152. [DOI] [PubMed]
- Li X, Nicklas RB. Mitotic forces control a cell cycle checkpoint. Nature (Lond) 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Li Y, Benezra R. Identification of a human mitotic checkpoint gene: hsMAD2. . Science (Wash DC) 1996;274:246–248. doi: 10.1126/science.274.5285.246. [DOI] [PubMed] [Google Scholar]
- Liao H, Li G, Yen TJ. Mitotic regulation of microtubule cross-linking activity of CENP-E kinetochore protein. Science (Wash DC) 1994;265:394–398. doi: 10.1126/science.8023161. [DOI] [PubMed] [Google Scholar]
- Liao H, Winkfein RJ, Mack G, Rattner JB, Yen TJ. CENP-F is a protein of the nuclear matrix that assembles onto kinetochores at late G2 and is rapidly degraded after mitosis. J Cell Biol. 1995;130:507–518. doi: 10.1083/jcb.130.3.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall LG, Clarke L. A novel cis-acting centromeric DNA element affects S. pombecentromeric chromatin structure at a distance. J Cell Biol. 1995;128:445–454. doi: 10.1083/jcb.128.4.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiaeencodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mole-Bajer J, Bajer AS, Zinkowski RP, Balczon RD, Brinkley BR. Autoantibodies from a patient with scleroderma CREST recognized kinetochores of the higher plant Haemanthus. . Proc Natl Acad Sci USA. 1990;87:3599–3603. doi: 10.1073/pnas.87.9.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore DP, Miyazaki WY, Tomkiel JE, Orr-Weaver TL. Double or nothing: a Drosophilamutation affecting meiotic chromosome segregation in both males and females. Genetics. 1995;136:953–964. doi: 10.1093/genetics/136.3.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moroi Y, Peebles MJ, Fritzler J, Steigerwald J, Tan EM. Autoantibody to centromere (kinetochore) in scleroderma sera. Proc Natl Acad Sci USA. 1980;77:1627–1631. doi: 10.1073/pnas.77.3.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy TD, Karpen GH. Location of centromere function in a Drosophilaminichromosome. Cell. 1995;82:599–609. doi: 10.1016/0092-8674(95)90032-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray AW, Kirschner MW. Dominoes and clocks: the union of two views of the cell cycle. Science (Wash DC) 1989;246:614–621. doi: 10.1126/science.2683077. [DOI] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green P, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Somerville S, Thomashow M, et al. Genes galore: a summary of methods for accessing results from large-scale partial sequencing of anonymous cDNA clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas RB, Ward SC, Gorbsky GJ. Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint. J Cell Biol. 1995;130:929–939. doi: 10.1083/jcb.130.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterman TK, Goodman HM. The glutamine synthetase gene family of Arabidopsis thalianalight-regulation and differential expression in leaves, roots, and seeds. J Mol Gen Genet. 1991;230:145–154. doi: 10.1007/BF00290662. [DOI] [PubMed] [Google Scholar]
- Pfarr CM, Coue M, Grissom PM, Hays TS, Porter ME, McIntosh JR. Cytoplasmic dynein is localized to kinetochores during mitosis. Nature (Lond) 1990;345:263–265. doi: 10.1038/345263a0. [DOI] [PubMed] [Google Scholar]
- Pimpinelli S, Goday C. Unusual kinetochores and chromatin diminution in Parascaris. . Trends Genet. 1989;5:310–315. doi: 10.1016/0168-9525(89)90114-5. [DOI] [PubMed] [Google Scholar]
- Pluta AF, Mackay AM, Ainsztein AM, Goldberg IG, Earnshaw WC. The centromere: hub of chromosomal activities. Science (Wash DC) 1995;270:1591–1594. doi: 10.1126/science.270.5242.1591. [DOI] [PubMed] [Google Scholar]
- Rieder CL, Cole RW, Khodjakov A, Sluder G. The checkpoint delaying anaphase in response to chromosome monoorientation is mediated by inhibitory signal produced by unattached kinetochores. J Cell Biol. 1995;130:941–948. doi: 10.1083/jcb.130.4.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo CAM, Takezaki N, Nei M. Molecular phylogeny and divergence times of Drosophilid species. Mol Biol Evol. 1995;12:391–404. doi: 10.1093/oxfordjournals.molbev.a040214. [DOI] [PubMed] [Google Scholar]
- Schmit A, Stoppin V, Chevrier V, Job D, Lambert A. Cell cycle dependent distribution of a centrosomal antigen at the perinuclear MTOC or at the kinetochores of higher plant cells. Chromosoma (Berl) 1994;103:343–351. doi: 10.1007/BF00417882. [DOI] [PubMed] [Google Scholar]
- Sheay W, Nelson S, Martinez I, Chu T-HT, Bhatia S, Dornberg R. Downstream insertion of the adenovirus tripartite leader sequence enhances expression in universal eukaryotic vectors. Biotechniques. 1993;15:856–862. [PubMed] [Google Scholar]
- Shelby RD, Hahn KM, Sullivan KF. Dynamic elastic behavior of α-satellite DNA domains visualized in situ in living human cells. J Cell Biol. 1996;135:545–557. doi: 10.1083/jcb.135.3.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DA, Baker BS, Gatti M. Mutations in genes controlling essential mitotic functions in Drosophila melanogaster. . Genetics. 1985;110:647–670. doi: 10.1093/genetics/110.4.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuer ER, Wordeman L, Schroer TA, Sheetz MP. Localization of cytoplasmic dynein to mitotic spindles and kinetochores. Nature (Lond) 1990;345:266–268. doi: 10.1038/345266a0. [DOI] [PubMed] [Google Scholar]
- Stoler S, Keith KC, Curnick KE, Fitzgerald-Hayes M. A mutation in CSE4, an essential gene encoding a novel chromatin-associated protein in yeast, causes chromosome nondisjunction and cell cycle arrest at mitosis. Genes Dev. 1995;9:573–586. doi: 10.1101/gad.9.5.573. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. . Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Williams BC, Gatti M, Goldberg ML. Bipolar spindle attachments affect redistributions of ZW10, a Drosophilacentromere/kinetochore component required for accurate chromosome segregation. J Cell Biol. 1996;134:1127–1140. doi: 10.1083/jcb.134.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BC, Goldberg ML. Determinants of Drosophilazw10 protein localization and function. J Cell Sci. 1994;107:785–798. doi: 10.1242/jcs.107.4.785. [DOI] [PubMed] [Google Scholar]
- Williams BC, Karr TL, Montgomery JM, Goldberg ML. The Drosophila l(1)zw10gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J Cell Biol. 1992;118:759–773. doi: 10.1083/jcb.118.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, et al. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. . Nature (Lond) 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Wise D, Zinkowski RP, Brinkley BR, Earnshaw WC, Cleveland DW. CENP-E, a novel human centromere-associated protein required for progression from metaphase to anaphase. EMBO (Eur Mol Biol Organ) J. 1991;10:1245–1254. doi: 10.1002/j.1460-2075.1991.tb08066.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Gang L, Scharr BT, Szilak I, Cleveland DW. CENP-E is a putative kinetochore motor that accumulates just before mitosis. Nature (Lond) 1992;359:536–539. doi: 10.1038/359536a0. [DOI] [PubMed] [Google Scholar]